Abstract
Forty-two different sense codons, coding for all 20 amino acids, were placed at the ribosomal E site location, two codons upstream of a UGA or UAG codon. The influence of these variable codons on readthrough of the stop codons was measured in Escherichia coli. A 30-fold difference in readthrough of the UGA codon was observed. Readthrough is not related to any property of the upstream codon, its cognate tRNA or the nature of its codon-anticodon interaction. Instead, it is the amino acid corresponding to the second upstream codon, in particular the acidic/basic property of this amino acid, which seems to be a major determinant. This amino acid effect is influenced by the identity of the A site stop codon and the efficiency of its decoding tRNA, which suggests a correlation with ribosomal pausing. The magnitude of the amino acid effect is in some cases different when UGA is decoded by a wildtype form of tRNA(Trp) as compared with a suppressor form of the same tRNA. This indicates that the structure of the A site decoding tRNA is also a determinant for the amino acid effect.
Full text
PDF



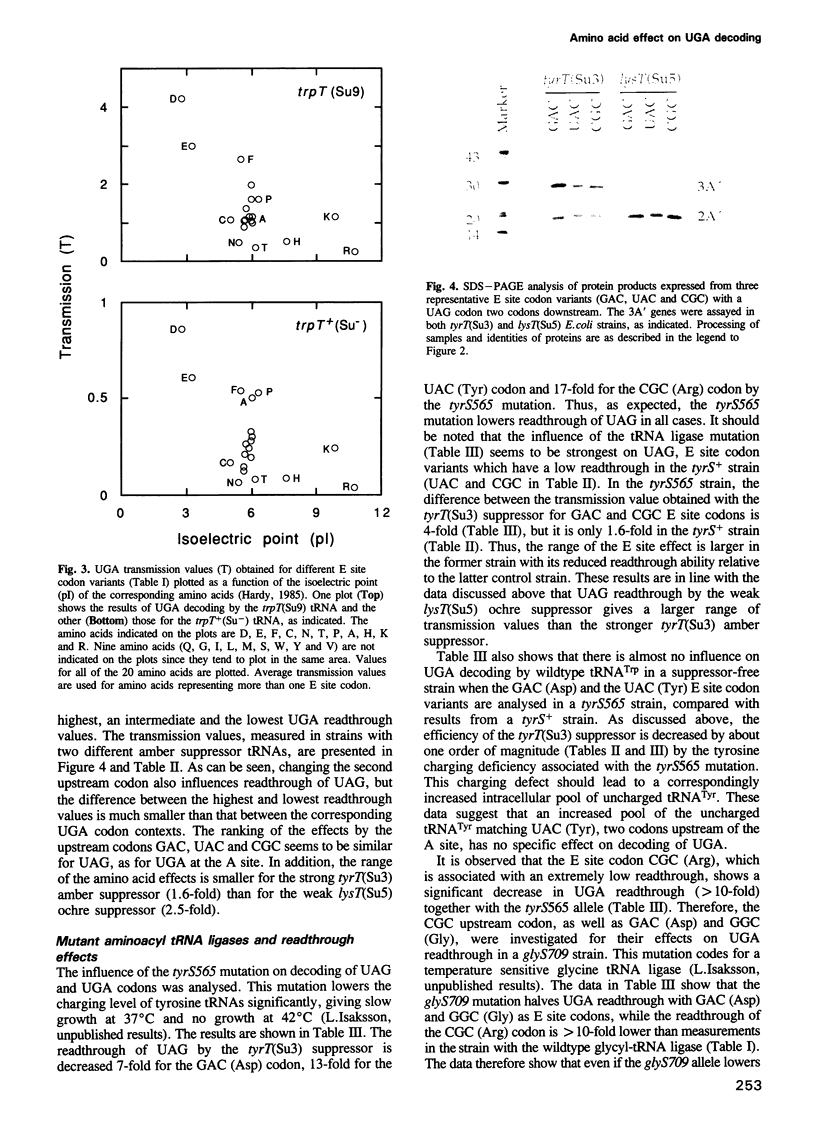
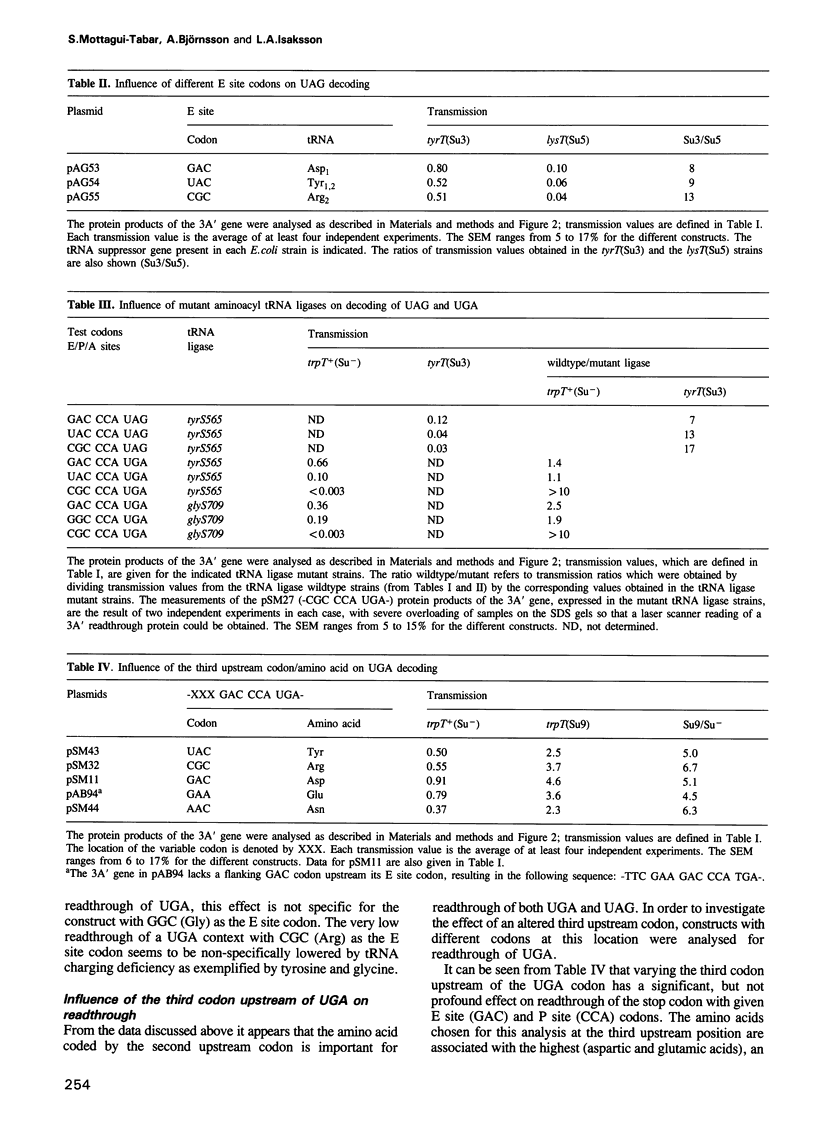



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahmsén L., Moks T., Nilsson B., Uhlén M. Secretion of heterologous gene products to the culture medium of Escherichia coli. Nucleic Acids Res. 1986 Sep 25;14(18):7487–7500. doi: 10.1093/nar/14.18.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S. G., Buckingham R. H., Kurland C. G. Does codon composition influence ribosome function? EMBO J. 1984 Jan;3(1):91–94. doi: 10.1002/j.1460-2075.1984.tb01766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Thompson S., Gesteland R. F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu Rev Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- Baron C., Heider J., Böck A. Interaction of translation factor SELB with the formate dehydrogenase H selenopolypeptide mRNA. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4181–4185. doi: 10.1073/pnas.90.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990 Apr 25;18(8):2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham R. H., Sörensen P., Pagel F. T., Hijazi K. A., Mims B. H., Brechemier-Baey D., Murgola E. J. Third position base changes in codons 5' and 3' adjacent UGA codons affect UGA suppression in vivo. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):259–262. doi: 10.1016/0167-4781(90)90177-4. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988 Sep 5;203(1):75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Donly B. C., Edgar C. D., Adamski F. M., Tate W. P. Frameshift autoregulation in the gene for Escherichia coli release factor 2: partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 1990 Nov 25;18(22):6517–6522. doi: 10.1093/nar/18.22.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V., Näslund A. K., Kurland C. G. Growth-rate-dependent accumulation of twelve tRNA species in Escherichia coli. J Mol Biol. 1993 Mar 20;230(2):483–491. doi: 10.1006/jmbi.1993.1165. [DOI] [PubMed] [Google Scholar]
- Faxén M., Plumbridge J., Isaksson L. A. Codon choice and potential complementarity between mRNA downstream of the initiation codon and bases 1471-1480 in 16S ribosomal RNA affects expression of glnS. Nucleic Acids Res. 1991 Oct 11;19(19):5247–5251. doi: 10.1093/nar/19.19.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A., Geigenmüller U., Rheinberger H. J., Nierhaus L. H. The allosteric three-site model for the ribosomal elongation cycle. Analysis with a heteropolymeric mRNA. J Biol Chem. 1989 May 5;264(13):7291–7301. [PubMed] [Google Scholar]
- Grajevskaja R. A., Ivanov Y. V., Saminsky E. M. 70-S ribosomes of Escherichia coli have an additional site for deacylated tRNA binding. Eur J Biochem. 1982 Nov;128(1):47–52. doi: 10.1111/j.1432-1033.1982.tb06929.x. [DOI] [PubMed] [Google Scholar]
- Grosjean H. J., de Henau S., Crothers D. M. On the physical basis for ambiguity in genetic coding interactions. Proc Natl Acad Sci U S A. 1978 Feb;75(2):610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider J., Baron C., Böck A. Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J. 1992 Oct;11(10):3759–3766. doi: 10.1002/j.1460-2075.1992.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Gold L. Translation of the UGA triplet in vitro by tryptophan transfer RNA's. J Mol Biol. 1971 Jun 14;58(2):459–468. doi: 10.1016/0022-2836(71)90363-9. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Kato M., Nishikawa K., Uritani M., Miyazaki M., Takemura S. The difference in the type of codon-anticodon base pairing at the ribosomal P-site is one of the determinants of the translational rate. J Biochem. 1990 Feb;107(2):242–247. doi: 10.1093/oxfordjournals.jbchem.a123033. [DOI] [PubMed] [Google Scholar]
- Kirillov S. V., Makarov E. M., Semenkov YuP Quantitative study of interaction of deacylated tRNA with Escherichia coli ribosomes. Role of 50 S subunits in formation of the E site. FEBS Lett. 1983 Jun 27;157(1):91–94. doi: 10.1016/0014-5793(83)81122-3. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Kopelowitz J., Hampe C., Goldman R., Reches M., Engelberg-Kulka H. Influence of codon context on UGA suppression and readthrough. J Mol Biol. 1992 May 20;225(2):261–269. doi: 10.1016/0022-2836(92)90920-f. [DOI] [PubMed] [Google Scholar]
- Lill R., Robertson J. M., Wintermeyer W. tRNA binding sites of ribosomes from Escherichia coli. Biochemistry. 1984 Dec 18;23(26):6710–6717. doi: 10.1021/bi00321a066. [DOI] [PubMed] [Google Scholar]
- Martin R., Weiner M., Gallant J. Effects of release factor context at UAA codons in Escherichia coli. J Bacteriol. 1988 Oct;170(10):4714–4717. doi: 10.1128/jb.170.10.4714-4717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B., Moks T., Jansson B., Abrahmsén L., Elmblad A., Holmgren E., Henrichson C., Jones T. A., Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987 Feb-Mar;1(2):107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- Pedersen W. T., Curran J. F. Effects of the nucleotide 3' to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J Mol Biol. 1991 May 20;219(2):231–241. doi: 10.1016/0022-2836(91)90564-m. [DOI] [PubMed] [Google Scholar]
- Picking W. D., Picking W. L., Odom O. W., Hardesty B. Fluorescence characterization of the environment encountered by nascent polyalanine and polyserine as they exit Escherichia coli ribosomes during translation. Biochemistry. 1992 Mar 3;31(8):2368–2375. doi: 10.1021/bi00123a023. [DOI] [PubMed] [Google Scholar]
- Potapov A. P. A stereospecific mechanism for the aminoacyl-tRNA selection at the ribosome. FEBS Lett. 1982 Sep 6;146(1):5–8. doi: 10.1016/0014-5793(82)80693-5. [DOI] [PubMed] [Google Scholar]
- Remme J., Margus T., Villems R., Nierhaus K. H. The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur J Biochem. 1989 Aug 1;183(2):281–284. doi: 10.1111/j.1432-1033.1989.tb14925.x. [DOI] [PubMed] [Google Scholar]
- Rheinberger H. J., Sternbach H., Nierhaus K. H. Three tRNA binding sites on Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5310–5314. doi: 10.1073/pnas.78.9.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydén S. M., Isaksson L. A. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol Gen Genet. 1984;193(1):38–45. doi: 10.1007/BF00327411. [DOI] [PubMed] [Google Scholar]
- Smith D., Yarus M. tRNA-tRNA interactions within cellular ribosomes. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4397–4401. doi: 10.1073/pnas.86.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986 Aug 26;14(16):6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Weber K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat New Biol. 1971 Sep 15;234(50):206–209. doi: 10.1038/newbio234206a0. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Huang W. M., Dunn D. M. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990 Jul 13;62(1):117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoni F., Heider J., Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]