Significance
Calpains are a family of intracellular proteases that have in common their dependence on Ca2+ and their ability to make a small number of specific cuts in their natural protein substrates. Although many calpain-generated protein fragments have been identified, understanding their functions is just beginning. This study shows that natural C-terminal fragments of a variety of proteins, including transmembrane ion channels, transcriptional regulators, apoptosis controllers, kinases, and phosphatases, are short-lived substrates of the N-end rule pathway, a ubiquitin-dependent processive proteolytic system that recognizes destabilizing N-terminal residues of protein substrates. This advance illuminates functional aspects of specific calpain fragments and suggests a specific molecular basis for many of the previously identified, mechanistically unclear roles of the N-end rule pathway.
Keywords: proteolysis, calpain substrates, ubiquitin
Abstract
Calpains are Ca2+-dependent intracellular proteases. We show here that calpain-generated natural C-terminal fragments of proteins that include G protein–coupled receptors, transmembrane ion channels, transcriptional regulators, apoptosis controllers, kinases, and phosphatases (Phe-GluN2a, Lys-Ica512, Arg-Ankrd2, Tyr-Grm1, Arg-Atp2b2, Glu-Bak, Arg-Igfbp2, Glu-IκBα, and Arg-c-Fos), are short-lived substrates of the Arg/N-end rule pathway, which targets destabilizing N-terminal residues. We also found that the identity of a fragment’s N-terminal residue can change during evolution, but the residue’s destabilizing activity is virtually always retained, suggesting selection pressures that favor a short half-life of the calpain-generated fragment. It is also shown that a self-cleavage of a calpain can result in an N-end rule substrate. Thus, the autoprocessing of calpains can control them by making active calpains short-lived. These and related results indicate that the Arg/N-end rule pathway mediates the remodeling of oligomeric complexes by eliminating protein fragments that are produced in these complexes through cleavages by calpains or other nonprocessive proteases. We suggest that this capability of the Arg/N-end rule pathway underlies a multitude of its previously known but mechanistically unclear functions.
Calpains are a family of intracellular proteases that have in common their dependence on Ca2+, their belonging to the superfamily of cysteine proteases, and their ability to make a small number of specific cuts in their natural protein substrates (1–3). Of the 14 distinct human calpains, the ubiquitously expressed calpain-1 (μ-calpain) and calpain-2 (m-calpain) are the major species. Natural calpain substrates were identified thus far largely by serendipity, as distinguished from proteome-scale surveys. The number of known calpain substrates in mammals is roughly 200. Calpain-mediated cuts have been accurately mapped in fewer than 100 of these proteins. The actual diversity of calpain substrates encoded by a mammalian genome is unknown. It may significantly exceed 1,000 different proteins (2).
The functions of calpains are many and stem from calpain-mediated cleavages of specific proteins at defined positions, usually within unstructured loops (1, 2). These cleavages are controlled by the levels of Ca2+, by phosphorylation of calpains and/or their substrates, by natural calpain inhibitors such as calpastatin (4), and by a regulated localization (e.g., membrane localization) of specific calpains. For example, cell movements involve calpain-mediated, spatiotemporally controlled cleavages of cytoskeletal proteins (5). Proteolysis by calpains plays many other roles as well, including their major functions in the brain and muscle (6–8). Despite the obvious biological importance of calpains, the functional understanding of calpain-generated specific protein fragments and their effects on oligomeric protein complexes (in which these fragments are formed and initially reside) is just beginning.
In vivo cleavages of cytosolic and nuclear proteins by nonprocessive proteases such as separases, caspases, and (some) deubiquitylases can generate C-terminal fragments that bear destabilizing N-terminal residues, i.e., the ones that can be recognized by the N-end rule pathway, a processive proteolytic system. These fragments have been shown to be short-lived N-end rule substrates, with either demonstrated or predicted functional ramifications (9–14). In the present work, we extended these insights to a number of natural calpain substrates, thereby linking the N-end rule pathway to biological consequences of calpain activity.
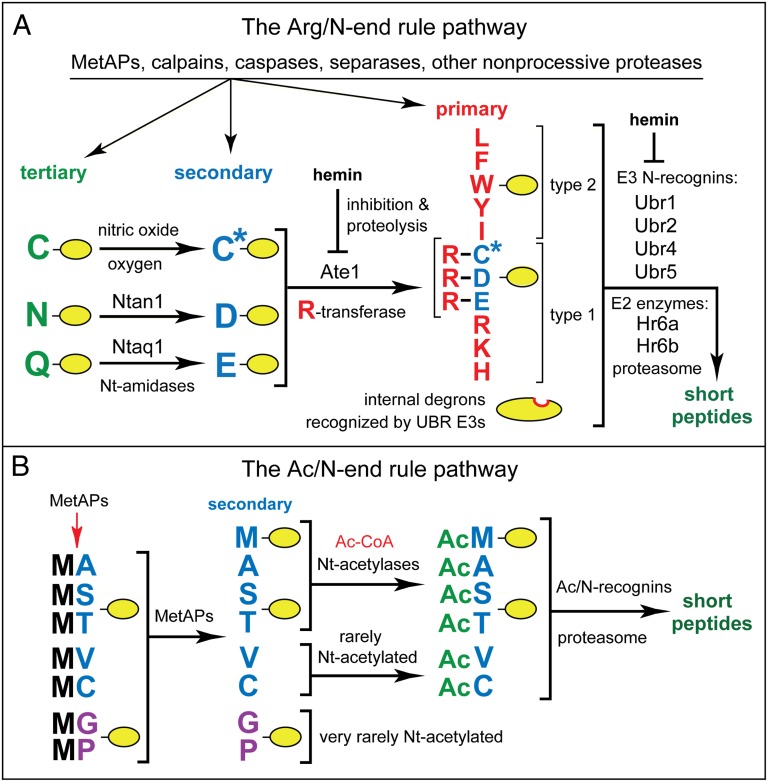
The N-end rule pathway recognizes proteins containing N-terminal degradation signals called N-degrons, polyubiquitylates these proteins, and thereby causes their degradation by the proteasome (Fig. 1) (9, 11, 12, 14–23). The main determinant of an N-degron is a destabilizing N-terminal residue of a protein. Recognition components of the N-end rule pathway are called N-recognins. In eukaryotes, N-recognins are E3 ubiquitin (Ub) ligases that can target N-degrons (Fig. 1).
Fig. 1.
The mammalian N-end rule pathway. See Introduction for descriptions of the pathway’s mechanistic aspects and biological functions. N-terminal residues are denoted by single-letter abbreviations. A yellow oval denotes the rest of a protein substrate. (A) The Arg/N-end rule pathway. It recognizes proteins through their specific unacetylated N-terminal residues (9, 19). (B) The Ac/N-end rule pathway. It recognizes proteins through their Nα-terminally acetylated (Nt-acetylated) residues (17, 23, 24). Red arrow on the left indicates the removal of the N-terminal Met residue by Met-aminopeptidases (MetAPs). N-terminal Met is retained if a residue at position 2 is larger than Val (56). See SI Text for a detailed supplementary legend and supplementary references to this figure.
Regulated degradation of proteins or their fragments by the N-end rule pathway mediates a strikingly broad range of biological functions, including the sensing of heme, NO, oxygen, and short peptides; the control, through subunit-selective degradation, of the input stoichiometries of subunits in oligomeric protein complexes; the elimination of misfolded or otherwise abnormal proteins; the degradation of specific proteins after their retrotranslocation to the cytosol from mitochondria or other membrane-enclosed compartments; the repression of apoptosis and neurodegeneration; the regulation of chromosome transcription, repair, replication, and cohesion/segregation; the regulation of G proteins, autophagy, peptide import, meiosis, immunity, fat metabolism, cell migration, actin filaments, cardiovascular development, spermatogenesis, neurogenesis, and memory; and the regulation of many processes in plants (Fig. 1 and SI Text).
In eukaryotes, the N-end rule pathway consists of two branches. One of them, called the Arg/N-end rule pathway, targets specific unacetylated N-terminal residues (Fig. 1A) (11, 15, 23). The primary destabilizing N-terminal residues Arg, Lys, His, Leu, Phe, Tyr, Trp, and Ile are directly recognized by N-recognins. In contrast, N-terminal Asn, Gln, Asp, and Glu (as well as Cys under some metabolic conditions) are destabilizing owing to their preliminary enzymatic modifications, which include Nt-deamidation and Nt-arginylation (Fig. 1A). In the yeast Saccharomyces cerevisiae, the Arg/N-end rule pathway is mediated by the Ubr1 N-recognin, a 225-kDa RING-type E3 Ub ligase and a part of the targeting complex comprising the Ubr1-Rad6 and Ufd4-Ubc4/5 holoenzymes (9, 16, 19). In multicellular eukaryotes, several E3 Ub ligases (including Ubr1) function as N-recognins of the Arg/N-end rule pathway (Fig. 1A). It was recently shown that both yeast and mammalian Ubr1 can recognize not only the unacetylated primary destabilizing N-terminal residues cited above, but also the unacetylated N-terminal Met residue, if it is followed by a hydrophobic residue. This capability of the Arg/N-end rule pathway greatly expands the range of its substrates, as virtually all nascent proteins bear N-terminal Met (23).
The other branch, called the Ac/N-end rule pathway, recognizes proteins through their Nα-terminally acetylated (Nt-acetylated) residues (Fig. 1B) (17, 23, 24). The degradation signals and E3 Ub ligases of the Ac/N-end rule pathway are called Ac/N-degrons and Ac/N-recognins, respectively. Nt-acetylation of cellular proteins is apparently irreversible, in contrast to acetylation-deacetylation of internal Lys residues. The bulk of Nt-acetylation is cotranslational, being mediated by ribosome-associated Nt-acetylases. Approximately 90% of human proteins are Nt-acetylated (25, 26). Many, possibly most, Nt-acetylated proteins contain Ac/N-degrons (Fig. 1B) (17, 23, 24).
Our surveys of the previously characterized natural calpain substrates in mammals identified ∼50 proteins (about 40% of the total number of actually mapped substrates) whose calpain-generated C-terminal fragments bear destabilizing N-terminal residues recognized by the Arg/N-end rule pathway. These proteins include transcriptional and translational regulators, transmembrane ion channels, cytoskeletal proteins, kinases, phosphatases, and other enzymes (Fig. 2). We selected from this set, essentially at random, 11 natural fragments (Phe1279-GluN2a, Lys609-Ica512, Arg103-Ankrd2, Tyr937-Grm1, Arg1091-Atp2b2, Glu16-Bak, Arg181-Igfbp2, Glu51-IκBα, Arg91-c-Fos, Asp142-Capns1, and Leu28-Capn1) (Fig. 2, #1 and #4–13) and examined them for actually being N-end rule substrates. It should be noted that none of the earlier studies that initially described these or other calpain-generated protein fragments considered the possibility of their regulation through degradation by the N-end rule pathway.
Fig. 2.
Calpain-generated C-terminal fragments of mammalian proteins that are identified or predicted substrates of the Arg/N-end rule pathway. The entries whose numbers are in green are the experimentally characterized and validated (most of them in the present study) substrates of the Arg/N-end rule pathway. The entries whose numbers are in black are predicted Arg/N-end rule substrates. Each entry cites a calpain-generated C-terminal fragment of a protein and the fragment’s N-terminal residue (using three-letter abbreviations for amino acids), followed by a description of uncleaved (full-length) precursor protein. A calpain cleavage site, denoted by an arrowhead, is denoted using single-letter abbreviations for amino acids. An enlarged P1′ residue (in red) becomes N-terminal on the cleavage. The indicated residue numbers are the number of the first shown residue of a full-length protein and the number of its last residue, respectively. All entries are mouse proteins, with the exception of #14 and #27, which are human proteins. See SI Text for a detailed supplementary legend to this data-rich figure.
We show here that all of these fragments are, in fact, short-lived substrates of the Arg/N-end rule pathway. As discussed below, calpains join other proteases, including Met-aminopeptidases, separases and caspases, as substrate-generating upstream components of the Arg/N-end rule pathway (Fig. 1A). Specifically, cleavages by these and other nonprocessive proteases can produce protein-size fragments that bear active N-degrons. We discuss the biological ramifications of this advance and the resulting connection between the Arg/N-end rule pathway and the functions of calpains.
Results
Destabilizing N-Terminal Residues of Calpain-Generated Fragments.
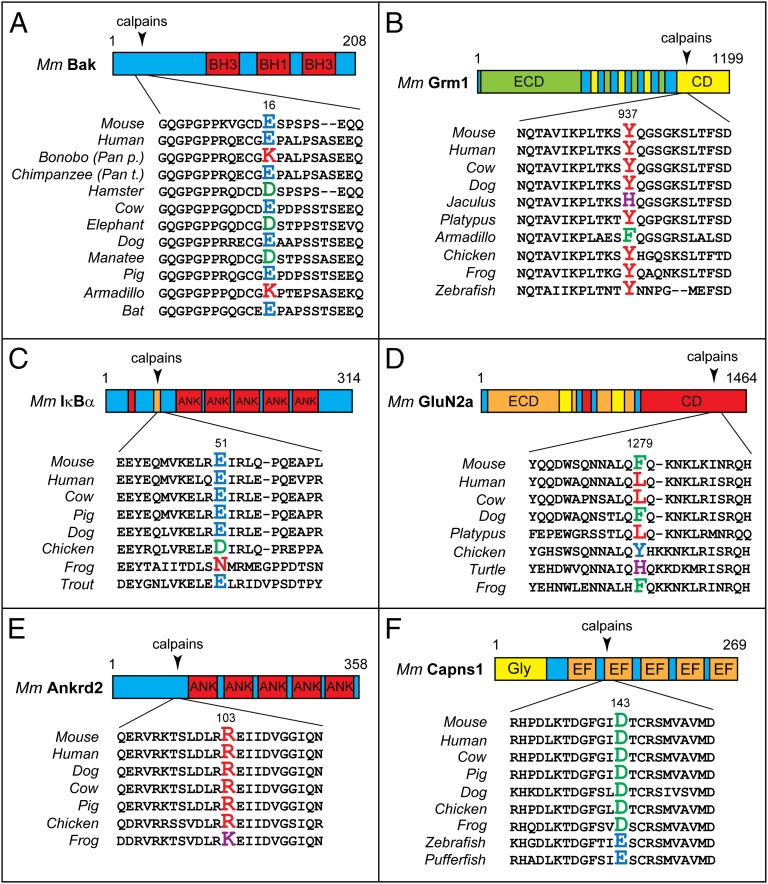
A P1′ residue of a cleavage site is the residue that becomes N-terminal upon cleavage. The identities of P1′ residues in full-length proteins that give rise to the Arg91-c-Fos, Lys609-Ica512, Arg1091-Atp2b2, and Leu28-Capn1 fragments were unchanged during evolution of examined vertebrates (Fig. S1 A, B, D, and E). In contrast, N-terminal Arg of the Arg103-Ankrd2 fragment is conserved among mammals and birds but changes to Lys in amphibians (Fig. 3E). N-terminal Glu of Glu16-Bak is conserved in humans, dogs, cows, and bats, but changes to Asp in hamsters and manatees and to Lys in armadillos and bonobos (Pan paniscus), a species of chimpanzee (Fig. 3A). Remarkably, this residue of Bak is Glu (i.e., the same as in human Glu16-Bak) in the other chimpanzee species, Pan troglodytes, strongly suggesting that the Glu16→Lys16 mutation in Bak of P. paniscus occurred after divergence of these chimpanzee lineages, i.e., less than 2 Mya (Fig. 3A). N-terminal Asp of Asp143-Capns1 is conserved in vertebrates other than fishes but changes to Glu in fishes (Fig. 3F). N-terminal Arg of Arg181-Igfbp2 is conserved from mammals to fishes but changes to Lys in lizards (Fig. S1C). N-terminal Glu of Glu51-IκBα is conserved among mammals and fishes but changes to Asp in chickens and to Asn in amphibians (Fig. 3C). N-terminal Tyr of Tyr937-Grm1 is conserved in most vertebrates but changes to Phe in armadillos and to His in Jaculus, a family of small rodents (Fig. 3B). N-terminal Phe of mouse Phe1279-GluN2a changes to Leu in humans, cows, and platypuses, to Tyr in chickens, and to His in turtles (Fig. 3D).
Fig. 3.
Evolutionary conservation of destabilizing activity of P1′ residues in calpain cleavage sites. Arrowheads indicate calpain cleavage sites. P1′ residues, which become N-terminal upon the cleavage, are larger and colored. The diagrams and indicated residue numbers are of mouse [Mus musculus (Mm)] proteins. (A) Bak. (B) Grm1. (C) IκBα. (D) GluN2a. (E) Ankrd2. (F) Capsn1. See the main text for descriptions of these proteins and the drift-with-constraint evolutionary patterns of P1′ residues in calpain cleavage sites.
Remarkably, this seeming jumble of evolutionary changes of P1′ residues in calpain substrates (Fig. 3 and Fig. S1) exhibits a telling constraint: all of the above P1′ residues (future N-terminal residues), including the ones that became altered during evolution, are destabilizing in the Arg/N-end rule pathway (Fig. 1A). In other words, the observed changes of P1′ residues in these calpain substrates always retained the ability of a fragment’s N-terminal residue to be targeted by the Arg/N-end rule pathway (Figs. 1A and 3 and Fig. S1). A constraint of this kind would be expected if a short in vivo half-life of a fragment (rather than the exact identity of its N-terminal residue) is a fitness-increasing property of this fragment, favored by natural selection.
Calpain-Generated Protein Fragments as N-End Rule Substrates.
Previous analyses of cleavage sites in calpain substrates were performed in vitro, using purified calpains and (usually) higher concentrations of Ca2+ than the physiologically relevant (low micromolar) calpain-activating levels of Ca2+. These analyses indicated that some proteins can be cleaved in vitro by calpains at multiple sites (typically two, occasionally three or more). In such cases only one, usually the predominant site, was examined in the present study (Figs. 4 and 5). In vivo, the same proteins would encounter lower levels of calpains and lower durations of calpain-activating concentrations of Ca2+. In addition, natural calpain substrates are usually subunits of oligomeric complexes. For all these reasons, calpain-mediated cleavages of proteins in vivo are likely to be more confined in extent and scope than the observed in vitro cleavages. Having mentioned limitations of in vitro analyses, we should emphasize that all calpain-generated fragments cited in Fig. 2 have been detected in vivo as well (Introduction).
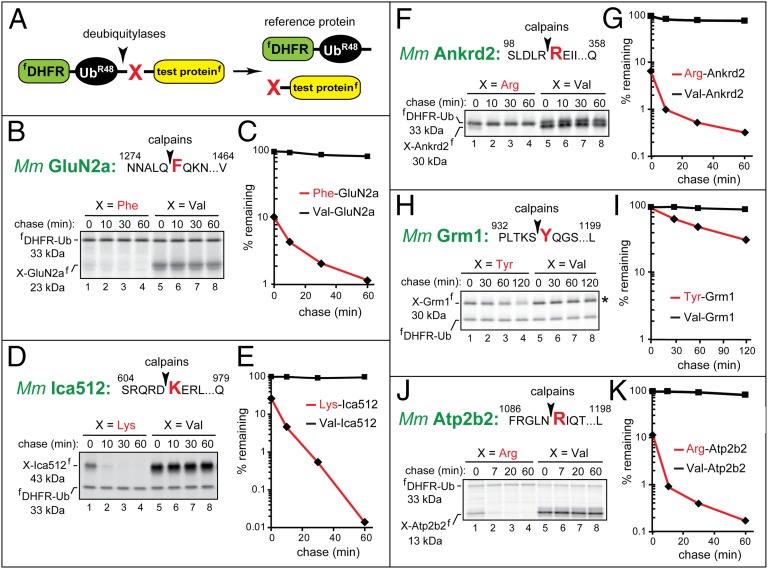
Fig. 4.
The Phe-GluN2a, Lys-Ica512, Arg-Ankrd2, Tyr-Grm1, and Arg-Atp2b2 fragments as short-lived substrates of the Arg/N-end rule pathway. The molecular mass values for untagged test proteins were ∼1 kDa smaller than their indicated sizes, as these fragments contained the ∼1-kDa flag epitope. Designations are as in Fig. 1. (A) The ubiquitin reference technique (URT). (B) Phe1279-GluN2a. (C) Quantification of data in B. (D) Lys609-Ica512. (E) Quantification of data in D. (F) Arg103-Ankrd2. (G) Quantification of data in F. (H) Tyr937-Grm1. (I) Quantification of data in H. (J) Arg1091-Atp2b2. (K) Quantification of data in J.
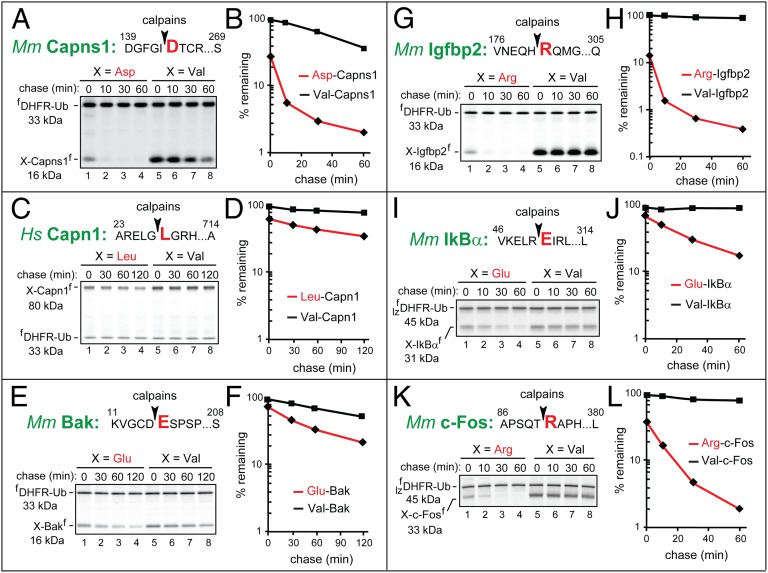
Fig. 5.
The Asp-Capns1, Leu-Capn1, Glu-Bak, Arg-Igfbp2, Glu-IκBα, and Arg-c-Fos fragments as short-lived substrates of the Arg/N-end rule pathway. Designations are as in Figs. 1 and 3. (A) Asp142-Capns1. (B) Quantification of data in A. (C) Leu28-Capn1. (D) Quantification of data in C. (E) Glu16-Bak. (F) Quantification of data in E. (G) Arg181-Igfbp2. (H) Quantification of data in G. (I) Glu51-IκBα. (J) Quantification of data in I. (K) Arg91-c-Fos. (L) Quantification of data in K.
The N-end rule degradation assays used the Ub reference technique (URT) (Fig. 4A) (27). The cotranslational cleavage of a URT-based fusion by deubiquitylases produces, at the initially equimolar ratio, a test protein with a desired N-terminal residue and a reference protein such as fDHFR-UbR48, a flag-tagged derivative of dihydrofolate reductase (Fig. 4A). In URT-based assays, a labeled test protein is quantified by measuring its level relative to the level of a stable reference at the same time point (12, 14).
URT-based 35S-pulse-chases were performed in a transcription-enabled rabbit reticulocyte extract, which has been extensively used to analyze the Arg/N-end rule pathway (9). Specific URT fusions were labeled with 35S-Met/Cys for 10 min at 30 °C, followed by a chase, immunoprecipitation with anti-flag antibody, SDS/PAGE, autoradiography, and quantification (Figs. 4 and 5). The logic of these assays involves a comparison between the degradation rates of a protein bearing a destabilizing N-terminal residue and an otherwise identical protein with an N-terminal residue such as Val, which is not recognized by the Arg/N-end rule pathway (Fig. 1A). In addition to being more accurate than pulse-chases without a stable reference, URT makes it possible to measure the degradation of a test protein during the pulse, i.e., before the chase (12, 14).
Phe1279-GluN2a.
GluN2a (NMDA-R2a) is a subunit of the NMDA receptor (NMDAR), a glutamate receptor that can function as a ligand-gated Ca2+ channel (28). The 23-kDa calpain-generated mouse Phe1279-GluN2a fragment (Figs. 2, #13, and 3D) (29) was short-lived in reticulocyte extract (initial postpulse t1/2 of ∼8 min), in striking contrast to the otherwise identical Val1279-GluN2a fragment, which was nearly completely stable (Fig. 4 B and C). Moreover, ∼90% of 35S-labeled Phe1279-GluN2a was degraded by the Arg/N-end rule pathway before the chase (during the 10-min pulse), in contrast to Val1279-GluN2a (Fig. 4C).
Lys609-Ica512.
Ica512 (Ptprn) is a member of the receptor protein phosphatase (PTP) family of integral membrane proteins, except that the cytosolic domain of Ica512 lacks phosphatase activity (30). Ica512 is expressed largely in neurons and neuroendocrine cells and is a major target of autoreactive antibodies in type I diabetes. The 43-kDa calpain-generated mouse Lys609-Ica512 fragment (Fig. 2, #12, and Fig. S1D) enters the nucleus and acts as a transcriptional regulator (30, 31). Lys609-Ica512 was short-lived in reticulocyte extract (postpulse t1/2 of ∼6 min), in contrast to Val609-Ica512, which was completely stable (Fig. 4 D and E). Moreover, ∼75% of 35S-labeled Lys609-Ica512 was degraded before the chase, in contrast to Val609-Ica512 (Fig. 4E).
Arg103-Ankrd2.
Ankrd2 (Marp2, Arpp) is a member of the MARP (muscle ankyrin repeat protein) family. Ankrd2 is preferentially expressed in the skeletal and cardiac muscle and functions as a negative regulator of muscle differentiation (32). The 30-kDa calpain-generated mouse Arg103-Ankrd2 fragment (Figs. 2, #10, and 3E) (33) was short-lived in reticulocyte extract (initial postpulse t1/2 of ∼5 min), in contrast to Val103-Ankrd2, which was nearly completely stable (Fig. 4 F and G). In addition, ∼94% of 35S-labeled Arg103-Ankrd2 was degraded by the Arg/N-end rule pathway before the chase, in contrast to Val103-Ankrd2 (Fig. 4G).
Tyr937-Grm1.
Grm1 (mGluR1α, mGlu1α) is an isoform of a member of the group I subfamily of metabotropic glutamate receptors (mGluRs). Grm1 is activated by glutamate, couples to Gq-type G proteins, and mediates a variety of functions, including (indirectly) Ca2+ transients (34). The 30-kDa calpain-generated mouse Tyr937-Grm1 fragment (Figs. 2, #11, and 3B) (35) was short-lived in reticulocyte extract (postpulse t1/2 of ∼45 min), in contrast to Val937-Grm1, which was stable (Fig. 4 H and I). One illustration of differences among the proteins characterized in this study was the slower (but still entirely N-end rule specific) degradation of Tyr937-Grm1, whose t1/2 of ∼45 min (Fig. 4 H and I) was at least fivefold longer than the half-lives of other calpain-generated Arg/N-end rule substrates (Fig. 4 B–G). In addition, little degradation of this fragment was observed before the chase (Fig. 4I), in contrast to the massive prechase degradation (75–94%) of more unstable N-end rule substrates (Fig. 4 C, E, and G).
Arg1091-Atp2b2.
The plasma membrane Ca2+ ATPase (PMCA) is a transmembrane pump that ejects Ca2+ from the cytosol to extracellular space and is encoded by the functionally overlapping Atp2b1-Atp2b4 genes (36). The 13-kDa calpain-generated mouse Arg1091-Atp2b2 fragment (Fig. 2, #8, and Fig. S1E) (37) was short-lived in reticulocyte extract (initial postpulse t1/2 of ∼5 min), in contrast to Val1091-Atp2b2, which was nearly completely stable (Fig. 4 J and K). In addition, ∼89% of 35S-labeled Arg1091-Atp2b2 was degraded before the chase, in contrast to Val1091-Atp2b2 (Fig. 4K).
Glu16-Bak.
Bak is a conditionally proapoptotic member of the Bcl-2 superfamily of apoptosis regulators. Owing to its C-terminal transmembrane domain, Bak resides in the outer mitochondrial and ER membranes. Under proapoptotic conditions, Bak contributes to the mitochondrial outer membrane permeabilization (MOMP), which can result in apoptosis (38). The proapoptotic cleavage of Bak by calpains generates the 16-kDa Glu16-Bak fragment lacking the 15-residue N-terminal region of Bak that appears to interact with membranes in full-length Bak (39). Mouse Glu16-Bak (Figs. 2, #1, and 3A) was short-lived in reticulocyte extract (initial postpulse t1/2 of ∼20 min), in contrast to Val16-Bak, which was still unstable but to a much lower extent than Glu16-Bak (postpulse t1/2 > 2 h), indicating the presence of a relatively weak internal degron (Fig. 5 E and F). In addition, ∼45% of 35S-labeled Glu16-Bak was degraded before the chase, in contrast to Val16-Bak (Fig. 5F).
Arg181-Igfbp2.
Insulin-like growth factor-binding proteins (IGFBPs) regulate insulin-like growth factors (IGFs) through interactions with IGFs that compete with their binding to IGF receptor. Although a large fraction of IGFBPs is extracellular, these proteins and their fragments have also been detected in the cytosol and the nucleus (40). The 16-kDa mouse Arg181-Igfbp2 fragment (Fig. 2, #6 and Fig. S1C) (41) was short-lived in reticulocyte extract (postpulse t1/2 of ∼5 min), in contrast to Val181-Igfbp2, which was completely stable (Fig. 5 G and H). In addition, ∼85% of 35S-labeled Arg181-Igfbp2 was degraded before the chase, in contrast to Val181-Igfbp2 (Fig. 5).
Glu51-IκBα.
IκBα binds to the multifunctional transcriptional regulator NFκB and retains it in the cytosol. The subunit-selective and conditional degradation of IκBα in the IκBα-NFκB complex activates NFκB (42). The degradation of IκBα is mediated by two pathways, regulated by different inputs. Specifically, phosphorylation of IκBα activates its phosphodegron and results in its Ub-dependent degradation by a pathway distinct from the N-end rule pathway. IκBα can also be cleaved by calpains, resulting in the 31-kDa Glu51-IκBα fragment (Figs. 2, #5, and 3C) (43). Mouse Glu51-IκBα was short-lived in reticulocyte extract (postpulse t1/2 of ∼14 min), in contrast to Val51-IκBα, which was nearly completely stable (Fig. 5 I and J). In addition, ∼35% of 35S-labeled Glu51-IκBα was degraded before the chase, in contrast to Val51-IκBα (Fig. 5J).
Arg91-c-Fos.
c-Fos is a part of major transcriptional regulators, through its interactions with other transcription factors, including c-Jun. c-Fos can be conditionally destroyed by pathways of the Ub system that are distinct from the Arg/N-end rule pathway (44). c-Fos can also be cleaved by calpains, resulting in the 33-kDa Arg91-c-Fos fragment (Fig. 2, #4, and Fig. S1A) (45). Mouse Arg91-c-Fos was short-lived in reticulocyte extract, with an initial postpulse t1/2 of ∼9 min, in contrast to Val91-c-Fos, which was nearly completely stable (Fig. 5 K and L). In addition, ∼60% of 35S-labeled Arg91-c-Fos was degraded before the chase, in contrast to Val91-c-Fos (Fig. 5L).
Autoprocessed Calpain Subunits as N-End Rule Substrates.
Both Ca2+-activated calpain-1 (the catalytic subunit Capn1 plus the small subunit Capns1) and calpain-2 (Capn2 plus Capns1) can undergo a limited autoproteolysis (46, 47). These N-terminal truncations of mouse Capn1, Capn2, and Capns1 generate, respectively, the Leu28-Capn1, Lys10-Capn2, and Asp142-Capns1 fragments, all of which bear destabilizing N-terminal residues (Figs. 1A, 2, #7, #9, and 3F and Fig. S1B). Calpain autoproteolysis is illustrated in Fig. S2D. Purified human calpain-1 was incubated in the presence of 40 μM CaCl2, followed by SDS/PAGE, which revealed the characteristic conversion of full-length Capn1 to a faster-migrating species (Fig. S2D) (47). The D. melanogaster calpain-B can also be autoproteolyzed, resulting in the active Asn224-calpain-B fragment (48) bearing N-terminal Asn, a destabilizing residue (Fig. 1A).
N-terminal autotruncations decrease the levels of Ca2+ required for half-maximal calpain activity, a finding consistent with the possibility that autoproteolysis is functionally relevant (49). In addition, autotruncations of calpains can also occur in living cells (50). At the same time, other studies presented strong in vitro arguments that Ca2+-mediated activation of calpains does not require their autoproteolysis (1, 51). If specific autotruncated calpain fragments were short-lived substrates of the Arg/N-end rule pathway, such a finding, in conjunction with the observed evolutionary conservation of the calpain auto-cleavage sites and P1′ residues (Fig. 3F and Fig. S1B), would suggest that the limited autoproteolysis of calpains is functionally relevant.
Leu28-Capn1.
Capn1 is the catalytic subunit of calpain-1. The 80-kDa autogenerated human Leu28-Capn1 fragment (Fig. 2, #9, and Fig. S1B) (46) was degraded particularly rapidly either during or shortly after its synthesis (before the chase) in reticulocyte extract, and was moderately short-lived afterward (postpulse t1/2 of ∼140 min), in contrast to Val28-Capn1, which was nearly completely stable (Fig. 5 C and D). Approximately 40% of 35S-labeled Leu28-Capn1 was degraded before the chase, in contrast to Val28-Capn1 (Fig. 5D).
Asp142-Capns1.
Capns1 is the noncatalytic subunit of calpain-1 and calpain-2. The 16-kDa calpain-generated mouse Asp142-Capns1 fragment (Figs. 2, #7, and 3F) (52) was short-lived in reticulocyte extract (initial postpulse t1/2 of ∼7 min), in contrast to Val142-Capns1, which was still unstable but much longer-lived (t1/2 of ∼45 min; Fig. 5 A and B). In addition, ∼70% of 35S-labeled Asp142-Capns1 was degraded before the chase, in contrast to Val142-Capns1 (Fig. 5B).
N-End Rule Substrate Produced by Calpain from a Full-Length Protein.
To probe degradation of a calpain-generated C-terminal fragment in living cells, we analyzed Phe1279-GluN2a. As described above, this fragment of a subunit of the NMDAR receptor was rapidly destroyed by the Arg/N-end rule pathway in reticulocyte extract (Figs. 2, #13, 3D, and 4 B and C). A URT-based pulse-chase (Fig. 4A) in transfected HEK293T human cells produced similar results. Specifically, Phe1279-GluN2a was short-lived in HEK293T cells, in contrast to Val1279-GluN2a (Fig. S2 A and B). The latter fragment, although not as stable as it was in reticulocyte extract (Fig. 4 B and C), was degraded in vivo much more slowly (t1/2 > 2 h) than the otherwise identical Phe1279-GluN2a fragment (initial postpulse t1/2 of ∼15 min; Fig. S2 A and B). Moreover, and also similarly to the results with reticulocyte extract, ∼70% of 35S-labeled Phe1279-GluN2a was degraded before the chase (during the pulse), in contrast to Val1279-GluN2a (Fig. S2 A and B).
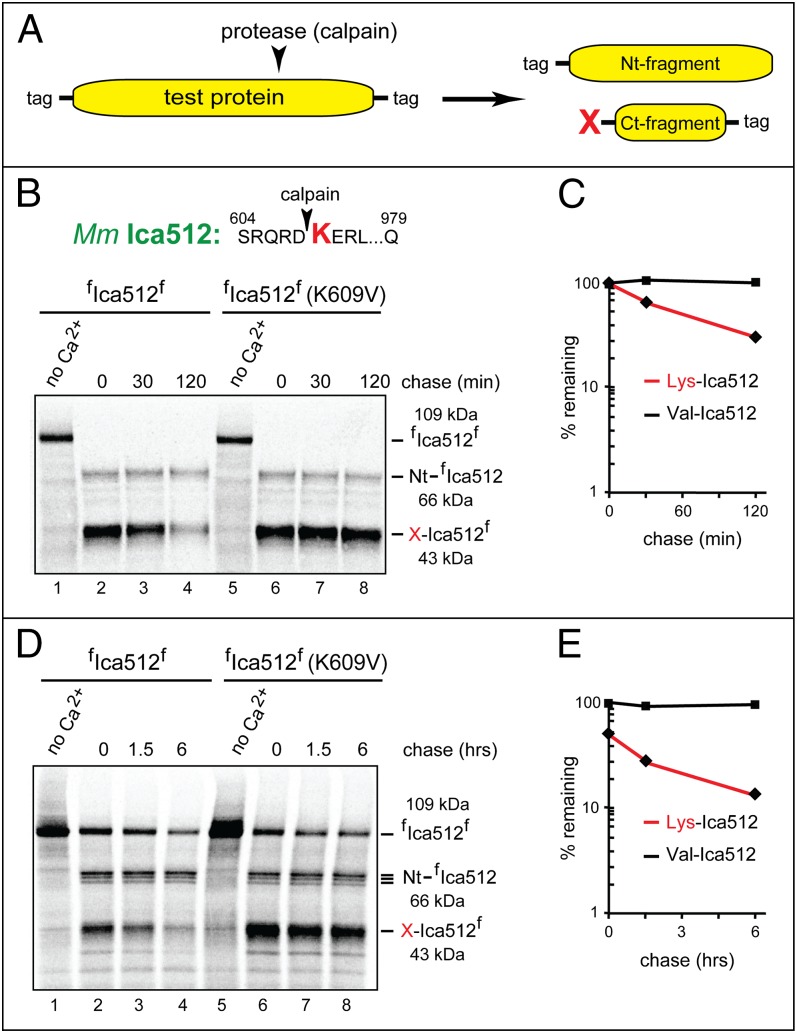
The Arg/N-end rule substrates examined thus far (Figs. 2–5 and Fig. S2A) were produced through the Ub fusion technique, which makes it possible to form, cotranslationally, specific calpain-generated fragments without using calpains (Fig. 4A). Can similar results be obtained through the actual calpain-mediated cleavage of a full-length protein? To address this question, we used the N-terminally and C-terminally flag-tagged full-length Ica512 (Ptprn) (Figs. 2, #12, and 4 D and E, and Fig. S1D), termed fIca512f. A mutant (control) derivative of fIca512f, termed fIca512f-K609V, contained Val, instead of WT Lys, at the P1′ position 609. In the plasmids that expressed fIca512f and fIca512f-K609V, the encoded N-terminal flag was at the position immediately downstream of the signal sequence of fIca512f. This arrangement allowed the retention of N terminus-proximal flag on the natural processing and transmembrane localization of fIca512f and fIca512f-K609V in HEK293T cells. Specifically, the flag tag became N-terminal after the ER import of fIca512f and fIca512f-K609V in HEK293 cells and the removal of their signal sequence by signal peptidase.
Calpain-based 35S-pulse-chase assays with full-length fIca512f and fIca512f-K690V were initially performed in reticulocyte extract. Proteins were synthesized and labeled for 60 min with [35S]methionine (Fig. 6 B and C). The cleavage of 35S-labeled fIca512f and fIca512f-K609V was initiated by the addition of CaCl2 and purified human calpain-1. After 30 min, the calpain inhibitor z-VF was added, followed by a chase for 30 and 120 min, immunoprecipitation with anti-flag antibody, SDS/PAGE, autoradiography, and quantification. The calpain-mediated cleavage of 35S-fIca512f and fIca512f-K609V reached completion during the 30-min incubation with calpain-1 (Fig. 6B, lanes 2 and 6). The relative stabilities of the resulting C-terminal fragments were as would be predicted from assays with Lys609-Ica512 vs. Val609-Ica512 that had been produced through the Ub fusion technique. Specifically, the calpain-generated Val609-Ica512f fragment (produced from full-length fIca512f-K609V) was completely stable in reticulocyte extract, whereas the otherwise identical WT Lys609-Ica512f was short-lived (t1/2 of ∼40 min; Fig. 6 B and C). As could be expected, the posttranslationally generated (from full-length fIca512f) Lys609-Ica512f fragment was degraded less rapidly than the otherwise identical fragment that had been cotranslationally produced through the Ub fusion technique (Fig. 6 B and C vs. Fig. 4 D and E).
Fig. 6.
Calpain-mediated generation of Arg/N-end rule substrates. (A) The N-terminal/C-terminal double tagging of a test protein to enable the detection of both fragments that result from an in vitro or in vivo protein cleavage. (B) Calpain-mediated generation of the Lys609-Ica512f fragment from full-length fIca512f and specific degradation of Lys609-Ica512f in reticulocyte extract. Lane 1, the doubly flag-tagged full-length fIca512f protein was labeled for 60 min with [35S]methionine in reticulocyte extract, followed by immunoprecipitation with anti-flag antibody, SDS/PAGE, and autoradiography. Lane 2, same as lane 1, but after the addition of CaCl2 and purified human calpain-1, followed by incubation for 30 min and the addition of the z-VF calpain inhibitor. Lanes 3 and 4, same as lane 2 but a chase for 30 and 120 min, respectively, before immunoprecipitation with anti-flag antibody SDS/PAGE, and autoradiography. Lanes 5–8, same as lanes 1–4, respectively, but with the mutant fIca512f-K609V protein. (C) Quantification of data in B. (D) Lane 1, HEK293T cells transiently expressing fIca512f were labeled for 60 min with [35S]methionine, followed by protein extraction, immunoprecipitation with anti-flag antibody, SDS/PAGE, and autoradiography. Lane 2, same as lane 1, but after incubation of 35S-labeled cells for 60 min with 2.5 mM unlabeled l-methionine, 3 mM CaCl2, and 50 µM A23184, a Ca2+ ionophore. Lanes 3 and 4, same as lane 2 but a chase for 1.5 and 6 h, respectively, before immunoprecipitation with anti-flag antibody, SDS/PAGE, and autoradiography. Lanes 5–8, same as lanes 1–4, respectively, but with the mutant fIca512f-K609V protein (main text). (E) Quantification of data in D.
To verify membrane localization of fIca512f expressed in HEK293T cells, their detergent-free extract was fractionated into soluble cytosol (SC) and membrane (M) fractions. The bulk of fIca512f was recovered in the insoluble (membrane) fraction (Fig. S2C). For calpain-based 35S-pulse-chases with fIca512f and fIca512f-K690V, HEK293T cells expressing these proteins were labeled for 60 min at 37 °C with [35S]methionine, followed by a 60-min incubation in the presence of 2.5 mM unlabeled l-methionine, 3 mM CaCl2, and 50 µM A23184, a Ca2+ ionophore. The cell-penetrating z-VF calpain inhibitor was then added, followed by a chase for 1.5 and 6 h, extraction of proteins, immunoprecipitation, SDS/PAGE, autoradiography, and quantification. The results were similar to those with reticulocyte extract, except for two differences (Fig. 6 D and E. vs. Fig. 6 B and C). First, the calpain-mediated cleavage of full-length fIca512f and fIca512f-K690V was complete in reticulocyte extract but partial in HEK293T cells. Second, there was a considerable degradation of the calpain-generated Lys609-Ica512f fragment (in comparison with Val609-Ica512f) by the Arg/N-end rule pathway before the chase in HEK293T cells, similarly to the early-degradation pattern of Lys609-Ica512 that had been produced through the Ub fusion technique in reticulocyte extract (Fig. 6 D and E. vs. Figs. 4 D and E and 6 B and C).
These assays confirmed that the degradation of a calpain-generated C-terminal protein fragment that had been derived posttranslationally from a full-length precursor in reticulocyte extract or HEK293T cells was qualitatively similar to the degradation of the otherwise identical fragment that had been produced through the Ub fusion technique (Figs. 4–6 and Fig. S2 A and B). The cotranslationally generated Arg/N-end rule substrate such as Lys609-Ica512 was degraded faster than the otherwise identical substrate that had been produced posttranslationally (Fig. 4 D and E vs. Fig. 6 A–C). These results were consistent with documented patterns of more efficacious degradation, by the Arg/N-end rule pathway, of conformationally immature proteins (9).
Discussion
Previous studies have shown that C-terminal fragments of intracellular proteins that can be generated by separases and caspases often bear destabilizing N-terminal residues. As a result, these fragments can be recognized and degraded by the Arg/N-end rule pathway (Fig. 1A) (9–13).
For example, the high fidelity of chromosome cohesion/segregation in S. cerevisiae requires the destruction, by the Arg/N-end rule pathway, of a C-terminal fragment of the Scc1/Rad21 subunit of cohesin, a protein whose ring-shaped molecules hold together sister chromatids (10). The cleavage of Scc1, mediated by a protease called separase, opens the cohesin ring and allows separation of the chromatids. The normally short-lived C-terminal fragment of Scc1 becomes long-lived in ubr1Δ cells, which lack the Arg/N-end rule pathway. Ubr1-lacking cells exhibit a strikingly elevated frequency of chromosome loss. This genomic instability was traced to a long in vivo half-life of the normally short-lived C-terminal Scc1 fragment, whose increased levels in ubr1Δ cells impair chromosome mechanics (10).
More recently, we discovered that the Arg/N-end rule pathway is also a functionally important repressor of apoptosis, through its ability to destroy and thereby to down-regulate a dozen or more of specific proapoptotic protein fragments that bear destabilizing N-terminal residues and are generated by activated caspases from full-length precursor proteins (11). We also found that the previously identified, neurodegeneration-associated C-terminal fragments of Tau, Tdp43, and α-synuclein (these fragments are produced through cleavages by a variety of nonprocessive proteases) can be destroyed through their N-degrons, suggesting that one function of the Arg/N-end rule pathway is to counteract neurodegeneration (12).
In the present study, the range of physiological Arg/N-end rule substrates was expanded to include a number of calpain-generated protein fragments that bear destabilizing N-terminal residues. We selected, nearly at random, 11 C-terminal fragments of natural calpain substrates from a larger set of such substrates whose fragments were predicted to bear N-degrons (Fig. 2 and the Introduction). All of the examined fragments were found to be efficacious targets of the Arg/N-end rule pathway. Moreover, in most of them, their N-degrons were, to a good approximation, either the sole or major degradation signals (Figs. 2 and 4–6 and Fig. S2 A and B). Given these results, it is likely that many (possibly most) of the currently unverified calpain-generated protein fragments that are classed as predicted N-end rule substrates (Fig. 2, #14–34) will eventually prove to be such on direct examination.
The set of natural calpain-generated C-terminal protein fragments that have been identified in the present study as short-lived Arg/N-end rule substrates comprises Phe-GluN2a, the fragment of the subunit of the NMDAR; Lys-Ica512, the fragment of a receptor protein phosphatase-like protein; Arg-Ankrd2, the fragment of a negative regulator of muscle differentiation; Tyr-Grm1, the fragment of a metabotropic glutamate receptor; Arg-Atp2b2, the fragment of a transmembrane pump that ejects Ca2+ from the cytosol; Glu-Bak, the proapoptotic fragment of an apoptosis regulator; Arg-Igfbp2, the fragment of an insulin-like growth factor-binding protein; Glu-IκBα, the fragment of the inhibitory subunit of the IκBα-NFκB complex; Arg-c-Fos, the fragment of a major transcriptional regulator; Leu28-Capn1, the autogenerated, catalytically active fragment of calpain-1; and Asp142-Capns1, the fragment of the noncatalytic subunit of calpain-1 (Figs. 2, #1 and #4–13, and 3–6 and Figs. S1 and S2).
Now that a number of functionally diverse proteins have been identified as precursors of calpain-generated fragments that are targeted for degradation by the Arg/N-end rule pathway (Figs. 2–6), the next step is to analyze the fragments themselves, their formation within cognate oligomeric complexes in vivo, the kinetics of fragments’ degradation under physiologically relevant conditions, the remodeling of oligomeric complexes in which the fragments initially reside, and the functional significance of fragments vis-á-vis specific biological circuits that involve these (and analogous) fragments, as well as corresponding precursor proteins.
One verifiable possibility, stated here in the context of the IκBα precursor protein (Figs. 2, #5, 3C, and 5 I and J) and likely to be relevant to other calpain substrates as well, is as follows. The calpain-mediated cleavage of IκBα, which yields the Glu51-IκBα fragment and the corresponding N-terminal fragment, may not suffice, by itself, for a rapid-enough in vivo dissociation of (at least) the Glu51-IκBα C-terminal fragment from the cleaved IκBα-NFκB complex. If so, the subunit selectivity of the Arg/N-end rule pathway (9) that presumably involves a mechanochemical dislodging of the Glu51-IκBα fragment from the cleaved IκBα-NFκB complex before the fragment’s processive degradation may be required for an optimal rate of liberation of the NFκB transcriptional activator. This rate would determine, in turn, the activity of NFκB circuits that involve the calpain-mediated cleavage of IκBα.
Both the proven examples above (the fidelity of chromosome segregation; the repression of apoptosis) and analogous but unexamined scenarios, such as the one about the Glu51-IκBα fragment, imply a mode of action by the Arg/N-end rule pathway that can recur in a broad variety of contexts. Confining this understanding to calpains, we suggest that the Arg/N-end rule pathway can use its subunit selectivity (9, 53) to reset the state of a biological circuit by making possible an efficacious, degradation-mediated replacement of a cleaved subunit by the intact one in a protein complex that had sustained a calpain-mediated cleavage. If the efficacy of this subunit’s replacement is a part of a circuit’s output signal, the rate of degradation of a subunit’s fragment by the Arg/N-end rule pathway may act as a timer, by analogy to other relatively slow and irreversible transitions, such as, for example, the hydrolysis of GTP by G-type proteins that mediates a variety of functions, from kinetic proofreading in protein synthesis to receptor desensitization.
Described in Results and illustrated in Fig. 3 and Fig. S1C are several examples of the “drift-with-constraint” evolutionary pattern of P1′ residues in calpain substrates (a P1′ residue becomes N-terminal upon cleavage of a polypeptide). For instance, the evolutionary changes of the P1′ residue in the cleavage site of Bak, a calpain substrate and regulator of apoptosis, are confined solely to Glu, Asp, and Lys, all of which are destabilizing residues (Figs. 1A and 3A). These evolutionary patterns and the metabolic instability of the mouse Glu16-Bak fragment (Fig. 5 E and F) strongly suggest that the degradation of this calpain-generated fragment serves to down-regulate its proapoptotic activity. The instability of proapoptotic Glu16-Bak that has been demonstrated in the present study is yet another illustration of the previously discovered function of the Arg/N-end rule pathway in which it acts as a repressor of apoptosis, by selectively destroying protease-generated proapoptotic protein fragments, the ones analogous to but distinct from Glu16-Bak (11).
We also found that the self-cleavages of calpain-1, either of its catalytic (Capn1) or noncatalytic (Capns1) subunits, yield C-terminal fragments (catalytically active in the case of Capn1) that are targeted for degradation by the Arg/N-end rule pathway (Fig. 5 A–D). Thus, the autoprocessing of calpains can control them by making active calpains short-lived. The activation of calpains under conditions of traumatic or hypoxic brain injury is accompanied by the in vivo autoproteolysis of calpain-1 and has been shown to result in a preferential degradation of the autotruncated calpain-1 (54), the latter finding being in agreement with our data (Figs. 3F and 5 A–D and Fig. S1B). Given the current uncertainty about biological roles of catalytically active calpain fragments (Results), key questions for future studies in this domain include the extent of calpain autoproteolysis in vivo under normal or stressful conditions; the contribution of resulting calpain fragments to the overall proteolytic activity of a specific calpain; and the role of the Arg/N-end rule pathway in the degradation of natural calpain fragments.
Both our findings and the earlier understanding of the dynamics of protein fragments vis-à-vis the Arg/N-end rule pathway (7, 9, 12, 14, 23) indicate that the topologically unique nature of N-degrons (their main determinant is a single N-terminal residue) has significantly contributed to coevolution of nonprocessive proteases and their cleavage sites in cellular proteins. As described above, these cleavage sites tend to be conserved in evolution, and their P1′ residues often yield C-terminal fragments whose N-terminal residues are recognized by the Arg/N-end rule pathway (Figs. 1A, 2, 4, and 5). Moreover, although the exact identity of a P1′ residue in a cleavage site can change during evolution, the residue’s destabilizing nature is virtually always retained (Fig. 3 and Fig. S1), suggesting that a short in vivo half-life of a fragment (rather than the exact identity of its N-terminal residue) is a fitness-increasing property of this fragment, favored by natural selection.
In sum, the degradation of protease-generated protein fragments by the Arg/N-end rule pathway (Fig. 1A) has been identified as a significant part of cellular regulation. Examples, in addition to the present results (Figs. 1–6), include the aforementioned maintenance of the fidelity of chromosome cohesion/segregation and the function of the Arg/N-end rule pathway in repressing apoptosis and neurodegeneration (10, 12, 14). Another example is the control of error-prone DNA synthesis, a part of DNA damage response. The Usp1 deubiquitylase (its functions include the control of DNA replication through unrepaired DNA lesions) can cleave itself but remains, transiently, an active deubiquitylase, as the two fragments of self-cleaved Usp1 stay together for some time (14, 55). The C-terminal fragment of the self-cleaved Usp1 has been shown to be a short-lived physiological substrate of the Arg/N-end rule pathway (14). The rate of processive degradation of this fragment may act as a timer that determines the rate of cessation of deubiquitylation by self-cleaved Usp1.
An Nt-acetylated protein fragment is a potential target of the Ac/N-end rule pathway (Fig. 1B) (23, 24). An Nt-acetylated N-terminal fragment can be produced posttranslationally, by a calpain or another nonprocessive protease upon cleavage of a cotranslationally Nt-acetylated full-length protein. This cleavage would also yield a C-terminal fragment, the one that can be recognized and degraded by the Arg/N-end rule pathway, as described in the present study (Figs. 2–6). In addition, a C-terminal fragment bearing an N-terminal residue that is permissive for Nt-acetylation (25) may be posttranslationally Nt-acetylated and thereby become a substrate of the Ac/N-end rule pathway. The latter possibility remains to be addressed experimentally.
The ability of the Arg/N-end rule and Ac/N-end rule pathways (Fig. 1) to target proteins for degradation by recognizing their destabilizing N-terminal residues is a unique attribute of these pathways vis-à-vis the rest of the Ub system. This capability, in conjunction with the subunit selectivity of proteolysis by the N-end rule pathway (9, 53), plays a major role in protein remodeling through the selective elimination of protein fragments from oligomeric complexes in which these fragments are generated and transiently reside. Nonprocessive intracellular proteases that produce such fragments include calpains, caspases, separases, and secretases. The regulated destruction of protein fragments is likely to underlie a number of diverse biological functions of the N-end rule pathway that are cited in the Introduction. Many of these functions have been discovered through genetic (nonmechanistic) approaches. Fragments produced through cleavages by calpains are a major part of natural protein fragments under both normal and stressful conditions. The present results will facilitate the functional understanding of the striking diversity of calpain-generated fragments in the context of the N-end rule pathway (Fig. 2 and Introduction). Most of these fragments are still understood in descriptive (nonfunctional) terms.
Materials and Methods
Plasmids, cDNAs, and Primers.
URT-based plasmids and other plasmids, constructed by standard methods, are described in Table S1. PCR primers are described in Table S2. Specific cDNAs encoding test proteins of the present study were either amplified from mouse cDNA libraries or purchased from Open Biosystems.
In Vitro and in Vivo Degradation Assays.
The in vitro transcription-translation-degradation assays used the TNT T7 Coupled Transcription/Translation System (Promega) (11). Human HEK293T cells were transfected with specific plasmids using Lipofectamine-2000 (Invitrogen).
These and other technical details of the present study are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank E. Udartseva for excellent technical assistance; members of the A.V. laboratory for advice and help; and Brandon Wadas and Tri Vu for comments on the manuscript. This study was supported by National Institutes of Health Grants DK039520 and GM031530 (to A.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401639111/-/DCSupplemental.
References
- 1.Campbell RL, Davies PL. Structure-function relationships in calpains. Biochem J. 2012;447(3):335–351. doi: 10.1042/BJ20120921. [DOI] [PubMed] [Google Scholar]
- 2.Ono Y, Sorimachi H. Calpains: An elaborate proteolytic system. Biochim Biophys Acta. 2012;1824(1):224–236. doi: 10.1016/j.bbapap.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83(3):731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron. 2013;80(5):1175–1189. doi: 10.1016/j.neuron.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 5.Franco SJ, Huttenlocher A. Regulating cell migration: Calpains make the cut. J Cell Sci. 2005;118(Pt 17):3829–3838. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 6.Baudry M, Bi X, Gall C, Lynch G. The biochemistry of memory: The 26year journey of a ‘new and specific hypothesis’. Neurobiol Learn Mem. 2011;95(2):125–133. doi: 10.1016/j.nlm.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varshavsky A. Augmented generation of protein fragments during wakefulness as the molecular cause of sleep: A hypothesis. Protein Sci. 2012;21(11):1634–1661. doi: 10.1002/pro.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorimachi H, Ono Y. Regulation and physiological roles of the calpain system in muscular disorders. Cardiovasc Res. 2012;96(1):11–22. doi: 10.1093/cvr/cvs157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410(6831):955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 11.Piatkov KI, Brower CS, Varshavsky A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc Natl Acad Sci USA. 2012;109(27):E1839–E1847. doi: 10.1073/pnas.1207786109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brower CS, Piatkov KI, Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell. 2013;50(2):161–171. doi: 10.1016/j.molcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Payoe R, Fahlman RP. The C-terminal proteolytic fragment of the breast cancer susceptibility type 1 protein (BRCA1) is degraded by the N-end rule pathway. J Biol Chem. 2012;287(10):7495–7502. doi: 10.1074/jbc.M111.301002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piatkov KI, Colnaghi L, Békés M, Varshavsky A, Huang TT. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol Cell. 2012;48(6):926–933. doi: 10.1016/j.molcel.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 16.Hwang C-S, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat Cell Biol. 2010;12(12):1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shemorry A, Hwang C-S, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50(4):540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varshavsky A. Discovery of cellular regulation by protein degradation. J Biol Chem. 2008;283(50):34469–34489. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tasaki TS, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graciet E, Wellmer F. The plant N-end rule pathway: Structure and functions. Trends Plant Sci. 2010;15(8):447–453. doi: 10.1016/j.tplants.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Dougan DA, Micevski D, Truscott KN. The N-end rule pathway: From recognition by N-recognins, to destruction by AAA+proteases. Biochim Biophys Acta. 2012;1823(1):83–91. doi: 10.1016/j.bbamcr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Mogk A, Schmidt R, Bukau B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17(4):165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Kim H-K, et al. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 2014;156(1-2):158–169. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang C-S, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327(5968):973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106(20):8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starheim KK, Gevaert K, Arnesen T. Protein N-terminal acetyltransferases: When the start matters. Trends Biochem Sci. 2012;37(4):152–161. doi: 10.1016/j.tibs.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 28.Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56(1):2–5. doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttmann RP, et al. Specific proteolysis of the NR2 subunit at multiple sites by calpain. J Neurochem. 2001;78(5):1083–1093. doi: 10.1046/j.1471-4159.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- 30.Trajkovski M, et al. Nuclear translocation of an ICA512 cytosolic fragment couples granule exocytosis and insulin expression in beta-cells. J Cell Biol. 2004;167(6):1063–1074. doi: 10.1083/jcb.200408172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ort T, et al. Dephosphorylation of beta2-syntrophin and Ca2+/mu-calpain-mediated cleavage of ICA512 upon stimulation of insulin secretion. EMBO J. 2001;20(15):4013–4023. doi: 10.1093/emboj/20.15.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bean C, Facchinello N, Faulkner G, Lanfranchi G. The effects of Ankrd2 alteration indicate its involvement in cell cycle regulation during muscle differentiation. Biochim Biophys Acta. 2008;1783(6):1023–1035. doi: 10.1016/j.bbamcr.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi C, et al. Multiple molecular interactions implicate the connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J Biol Chem. 2008;283(21):14801–14814. doi: 10.1074/jbc.M708262200. [DOI] [PubMed] [Google Scholar]
- 34.Niswender CM, Conn PJ. Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu W, et al. Calpain-mediated mGluR1alpha truncation: A key step in excitotoxicity. Neuron. 2007;53(3):399–412. doi: 10.1016/j.neuron.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 36.Brini M, Carafoli E. The plasma membrane Ca²+ ATPase and the plasma membrane sodium calcium exchanger cooperate in the regulation of cell calcium. Cold Spring Harb Perspect Biol. 2011;3(2):a004168. doi: 10.1101/cshperspect.a004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James P, et al. Modulation of erythrocyte Ca2+-ATPase by selective calpain cleavage of the calmodulin-binding domain. J Biol Chem. 1989;264(14):8289–8296. [PubMed] [Google Scholar]
- 38.Green DR. Means to an End: Apoptosis and Other Cell Death Mechanisms. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 39.Moldoveanu T, et al. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24(5):677–688. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Hoeflich A, et al. Peri/nuclear localization of intact insulin-like growth factor binding protein-2 and a distinct carboxyl-terminal IGFBP-2 fragment in vivo. Biochem Biophys Res Commun. 2004;324(2):705–710. doi: 10.1016/j.bbrc.2004.09.111. [DOI] [PubMed] [Google Scholar]
- 41.Berg U, Bang P, Carlsson-Skwirut C. Calpain proteolysis of insulin-like growth factor binding protein (IGFBP) -2 and -3, but not of IGFBP-1. Biol Chem. 2007;388(8):859–863. doi: 10.1515/BC.2007.098. [DOI] [PubMed] [Google Scholar]
- 42.Hayden MS, Ghosh S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaecher K, Goust JM, Banik NL. The effects of calpain inhibition on IkB alpha degradation after activation of PBMCs: Identification of the calpain cleavage sites. Neurochem Res. 2004;29(7):1443–1451. doi: 10.1023/b:nere.0000026410.56000.dd. [DOI] [PubMed] [Google Scholar]
- 44.Adler J, Reuven N, Kahana C, Shaul Y. c-Fos proteasomal degradation is activated by a default mechanism, and its regulation by NAD(P)H:quinone oxidoreductase 1 determines c-Fos serum response kinetics. Mol Cell Biol. 2010;30(15):3767–3778. doi: 10.1128/MCB.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pariat M, et al. The sensitivity of c-Jun and c-Fos proteins to calpains depends on conformational determinants of the monomers and not on formation of dimers. Biochem J. 2000;345(Pt 1):129–138. [PMC free article] [PubMed] [Google Scholar]
- 46.Melloni E, Michetti M, Salamino F, Minafra R, Pontremoli S. Modulation of the calpain autoproteolysis by calpastatin and phospholipids. Biochem Biophys Res Commun. 1996;229(1):193–197. doi: 10.1006/bbrc.1996.1779. [DOI] [PubMed] [Google Scholar]
- 47.Elce JS, Hegadorn C, Arthur JS, Arthur C. Autolysis, Ca2+ requirement, and heterodimer stability in m-calpain. J Biol Chem. 1997;272(17):11268–11275. doi: 10.1074/jbc.272.17.11268. [DOI] [PubMed] [Google Scholar]
- 48.Farkas A, et al. Autolytic activation and localization in Schneider cells (S2) of calpain B from Drosophila. Biochem J. 2004;378(Pt 2):299–305. doi: 10.1042/BJ20031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baki A, Tompa P, Alexa A, Molnár O, Friedrich P. Autolysis parallels activation of mu-calpain. Biochem J. 1996;318(Pt 3):897–901. doi: 10.1042/bj3180897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy RM, Verburg E, Lamb GD. Ca2+ activation of diffusible and bound pools of mu-calpain in rat skeletal muscle. J Physiol. 2006;576(Pt 2):595–612. doi: 10.1113/jphysiol.2006.114090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chou JS, Impens F, Gevaert K, Davies PL. m-Calpain activation in vitro does not require autolysis or subunit dissociation. Biochim Biophys Acta. 2011;1814(7):864–872. doi: 10.1016/j.bbapap.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Minami Y, Emori Y, Kawasaki H, Suzuki K. E-F hand structure-domain of calcium-activated neutral protease (CANP) can bind Ca2+ ions. J Biochem. 1987;101(4):889–895. doi: 10.1093/oxfordjournals.jbchem.a121956. [DOI] [PubMed] [Google Scholar]
- 53.Johnson ES, Gonda DK, Varshavsky A. cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature. 1990;346(6281):287–291. doi: 10.1038/346287a0. [DOI] [PubMed] [Google Scholar]
- 54.Kampfl A, et al. mu-calpain activation and calpain-mediated cytoskeletal proteolysis following traumatic brain injury. J Neurochem. 1996;67(4):1575–1583. doi: 10.1046/j.1471-4159.1996.67041575.x. [DOI] [PubMed] [Google Scholar]
- 55.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8(4):339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 56.Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49(26):5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.