Significance
Bifunctional enzymes catalyze two sequential reactions of a metabolic pathway using spatially separated and covalently connected active sites. Combining two enzymes into one protein chain creates efficiency because the product of the first reaction can be shuttled directly to the active site, catalyzing the second reaction in a process called substrate channeling. Here, we use X-ray crystallography to elucidate the three-dimensional structural basis of substrate channeling in the proline catabolic pathway. Structures of the bifunctional enzyme proline utilization A (PutA) reveal an elaborate and dynamic tunnel system featuring a 75-Å-long substrate-channeling path that links the two active sites. Furthermore, we have identified the elusive quinone-binding site of PutA, which provides insight into how PutAs interact with the membrane.
Keywords: proline catabolism, X-ray crystallography, membrane association
Abstract
Proline utilization A (PutA) proteins are bifunctional peripheral membrane flavoenzymes that catalyze the oxidation of l-proline to l-glutamate by the sequential activities of proline dehydrogenase and aldehyde dehydrogenase domains. Located at the inner membrane of Gram-negative bacteria, PutAs play a major role in energy metabolism by coupling the oxidation of proline imported from the environment to the reduction of membrane-associated quinones. Here, we report seven crystal structures of the 1,004-residue PutA from Geobacter sulfurreducens, along with determination of the protein oligomeric state by small-angle X-ray scattering and kinetic characterization of substrate channeling and quinone reduction. The structures reveal an elaborate and dynamic tunnel system featuring a 75-Å-long tunnel that links the two active sites and six smaller tunnels that connect the main tunnel to the bulk medium. The locations of these tunnels and their responses to ligand binding and flavin reduction suggest hypotheses about how proline, water, and quinones enter the tunnel system and where l-glutamate exits. Kinetic measurements show that glutamate production from proline occurs without a lag phase, consistent with substrate channeling and implying that the observed tunnel is functionally relevant. Furthermore, the structure of reduced PutA complexed with menadione bisulfite reveals the elusive quinone-binding site. The benzoquinone binds within 4.0 Å of the flavin si face, consistent with direct electron transfer. The location of the quinone site implies that the concave surface of the PutA dimer approaches the membrane. Altogether, these results provide insight into how PutAs couple proline oxidation to quinone reduction.
Proline catabolism is an important pathway in bioenergetics and has been implicated in tumor suppression (1), lifespan extension (2), production of fungal virulence factors (3), and bacterial virulence (4, 5). The pathway (Fig. 1A) comprises the flavoenzyme proline dehydrogenase (PRODH) and the aldehyde dehydrogenase (ALDH) superfamily member Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (P5CDH, also known as ALDH4A1). PRODH catalyzes the FAD-dependent oxidation of proline to P5C. P5CDH is a misnomer, as the enzyme catalyzes the oxidization of l-glutamate-γ-semialdehyde (GSA), which is the hydrolysis product of P5C. The electrons abstracted from proline by PRODH flow into the electron transport chain whereas the carbon skeleton of l-proline ultimately enters the citric acid cycle via α-ketoglutarate.
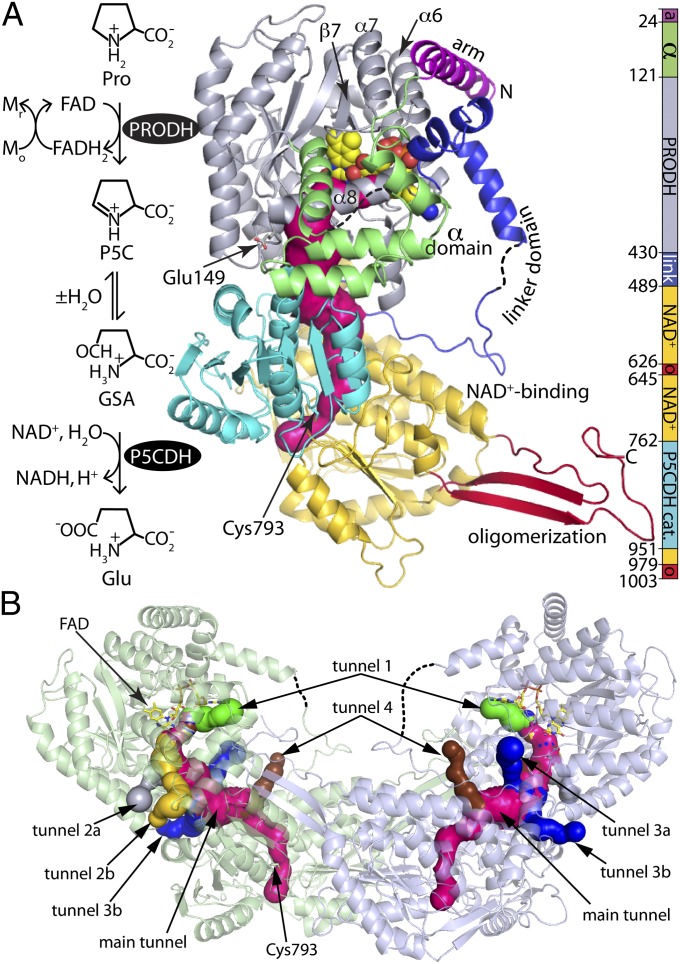
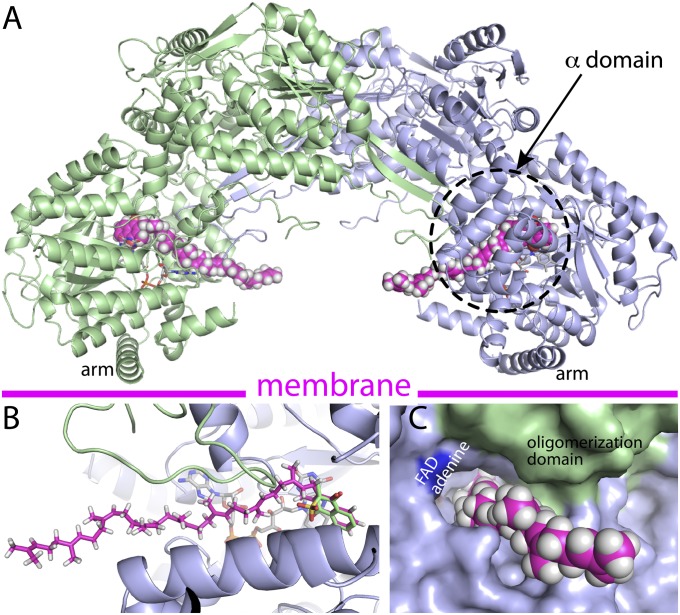
Fig. 1.
Structure of GsPutA. (A) Reactions catalyzed by PutA and a ribbon drawing of the protomer of the resting enzyme (state I in Fig. S1). The pink surface represents the substrate-channeling tunnel. The domains are colored according to the legend on the right. The dashes denote disordered sections of the α and linker domains. (B) Ribbon drawing of the dimer, with the two protomers colored green and blue. The surfaces represent the substrate-channeling tunnel system. The main substrate-channeling tunnel is colored pink. Smaller tunnels that connect the main tunnel to the bulk medium are colored green (tunnel 1), silver (tunnel 2a), yellow (tunnel 2b), blue (tunnels 3a and 3b), and brown (tunnel 4).
Curiously, in Gram-negative bacteria, PRODH and P5CDH are combined into a single polypeptide, which is known as proline utilization A (PutA). Two functional classes of PutA exist: bifunctional and trifunctional (6). Bifunctional PutAs, which are the focus of this work, are peripheral membrane-associated enzymes that exhibit both PRODH and P5CDH catalytic activities. Localization of PutA at the inner bacterial membrane facilitates the efficient transfer of electrons from proline, through the FAD, to quinone electron acceptors in the membrane. Trifunctional PutAs additionally serve as the transcriptional repressor of the put regulon and have a ribbon–helix–helix DNA-binding domain in the N terminus of the polypeptide chain. Trifunctional PutAs are unique in that proline reduction of the flavin induces a shift in the subcellular location from DNA-bound to membrane-associated. PutAs thus serve as interesting systems for studying substrate channeling, membrane association, and, in the case of trifunctional PutAs, redox regulation of protein location and function.
PutAs are challenging structural biology targets, presumably because of their long polypeptide length (typically >1,000 residues), multidomain architecture, inherent flexibility, and membrane localization. Since the discovery of PutA in the 1970s, just one structure of the full-length enzyme has been determined, that of the 999-residue bifunctional PutA from Bradyrhizobium japonicum (BjPutA) (7). The structure revealed the basic PutA blueprint, including the spatial disposition of the two catalytic domains and a buried tunnel connecting the two active sites that functions in substrate channeling. However, little is known about how the substrate-channeling tunnel responds to ligand binding and flavin reduction, where substrates and water enter the tunnel, and where l-glutamate exits the tunnel. Furthermore, the location of the quinone-binding site and the structural basis of PutA membrane association have remained elusive.
To address this knowledge gap, we have determined crystal structures (Tables S1–S3) of PutA from Geobacter sulfurreducens PCA (GsPutA; 1,004 residues) corresponding to several of the conformations populated during the catalytic cycle (Fig. S1). The structures reveal a dynamic substrate-channeling tunnel system that is remodeled by proline binding. Furthermore, we report previously unidentified structures of PutA-quinone and PutA-detergent complexes, which provide insight into the mechanism of quinone reduction and the structural basis of membrane association.
Results
Tertiary and Quaternary Structure.
The phase problem for GsPutA was solved using experimental phasing because of the low (27%) global sequence identity to BjPutA (Table S1). Crystal structures representing states I, II, VIII, and IX of the catalytic cycle (Fig. S1) were determined (Tables S2 and S3). The high resolution limits of the structures range from 1.9 Å to 2.2 Å. These are the highest resolution PutA structures to date and the only PutA structures that have bona fide active site ligands bound.
The protomer consists of seven domains: arm, α domain, PRODH (βα)8 barrel, linker, Rossmann NAD+-binding domain, P5CDH catalytic domain, and oligomerization domain (Fig. 1A). This domain architecture is also present in BjPutA despite the low amino acid sequence identity with GsPutA. The two structures are related by a root mean square deviation of 2.0 Å for 800 aligned residues, implying high overall structural similarity.
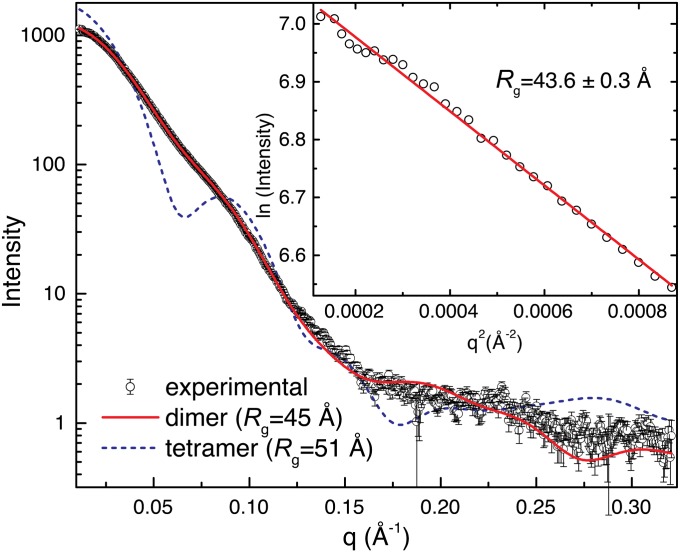
The oligomeric state and quaternary structure of GsPutA in solution were determined using small-angle X-ray scattering (SAXS) (8). Guinier analysis yields a radius of gyration (Rg) of 43.6 ± 0.3 Å (Fig. 2). This value is close to the Rg of the dimer in the crystallographic asymmetric unit (44.6 Å) (Fig. 1B). In the dimer, the oligomerization domain of one protomer forms an intermolecular β-sheet with the catalytic domain of the other protomer; this mode of dimerization is typical for ALDHs. The scattering profile calculated from the dimer agrees well with the experimental one (Fig. 2), and the envelope from shape reconstruction calculations is consistent with the dimer (Fig. S2).
Fig. 2.
SAXS analysis of GsPutA. (Inset) A Guinier plot spanning the range of qRg 0.490–1.29; the linear fit has R2 of 0.995. Theoretical curves were calculated from the GsPutA dimer in the asymmetric unit (red) and the BjPutA tetramer (blue dashes).
That GsPutA is dimeric is unexpected because BjPutA, which is also a bifunctional PutA, forms a ring-shaped tetramer containing two dimers and having Rg of 51 Å (7). Nevertheless, the scattering profile of the BjPutA tetramer deviates substantially from that of GsPutA (Fig. 2), confirming that GsPutA and BjPutA indeed have different oligomeric states.
The Tunnel System of the Resting Enzyme.
The structure of GsPutA, with the flavin oxidized and the substrate pockets unoccupied, reveals the previously unidentified substrate-channeling tunnel system of the resting enzyme (state I) (Fig. S1). We note that the BjPutA structure has adventitious sulfate ions from the crystallization solution bound in both active sites (7).
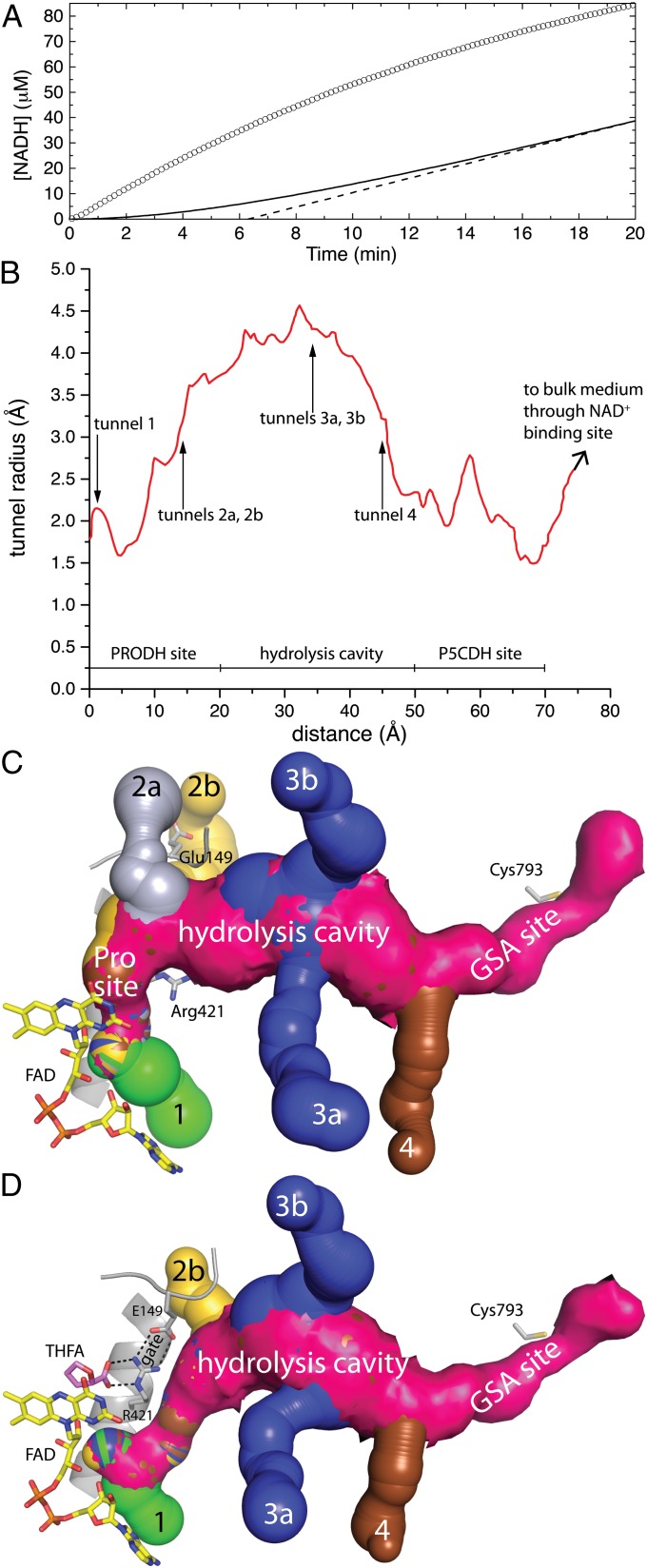
The PRODH and P5CDH active sites are connected by a tunnel, which presumably enables substrate channeling (Fig. 1). Consistent with this idea, steady-state kinetic measurements show that NADH is generated from proline without a perceptible time lag (Fig. 3A). In contrast, a free-diffusion model predicts a substantial lag in the production of NADH and a transient time of 6.3 min. (Fig. 3A). The large discrepancy between the experimental progress curve and the one simulated from the free-diffusion model is consistent with a substrate-channeling mechanism.
Fig. 3.
Kinetic evidence of substrate channeling and the dynamic tunnel system of GsPutA. (A) Steady-state progress curve of NADH production from proline by GsPutA. The circles represent the experimental curve for the GsPutA PRODH-P5CDH coupled reaction using 0.75 μM GsPutA, 40 mM proline, 150 μM menadione, and 0.2 mM NAD+ (pH 7.5). The solid curve represents the nonchanneling PRODH and P5CDH reaction calculated from a free diffusion model (Eq. S1). The dashed line shows the extrapolation of the nonchanneling model, which yields a transient time of 6.3 min. (B) Plot of the radius of the main tunnel of the resting enzyme as a function of the distance from the flavin calculated using Mole (10). The locations of the ancillary tunnels are indicated. (C) Surface rendering of the tunnel system of the resting enzyme. Note that the Glu149-Arg421 ion pair gate is open. (D) Surface rendering of the tunnel system of the GsPutA–THFA complex. Note that the Glu149-Arg421 ion pair gate is closed, which eliminates tunnel 2a.
Although the two active sites are separated by a linear distance of 45 Å (measured from the FAD N5 to Cys793), the actual distance traveled by the reaction intermediate(s) is closer to 75 Å because the tunnel traces out a curved path (Fig. 3 B and C). For reference, the tunnels of tryptophan synthase and carbamoyl phosphate synthetase are 25 Å and 80 Å in length, respectively (9). The middle section of the GsPutA tunnel has a length of ∼30 Å and, with a radius of 3.5–4.5 Å, is the widest part of the tunnel (Fig. 3B). Because of its location midway between the active sites, large radius, and hydrophilic character (Fig. S3A), the middle section seems to be a suitable location for the P5C hydrolysis reaction. We thus refer to this region as the hydrolysis cavity.
The tunnel walls are lined almost exclusively with charged and polar side chains, which impart a highly hydrophilic character to the tunnel interior (Fig. S3A). In contrast, the tunnel of tryptophan synthase is mostly hydrophobic, which is appropriate for channeling indole (9). The ammonia section of the tunnel in carbamoyl phosphate synthetase features unreactive side chains and backbone atoms whereas the carbamate section is more hydrophilic (9).
The conservation of residues lining the tunnel was determined. Analysis of diverse PutA sequences reveals 67 identical residues. Many of these residues line the tunnel near the two active sites (Fig. S3A). Although none of the identities map to the middle section of the tunnel, the predominance of charged and polar side chains lining the middle section is also observed in BjPutA (7). Thus, the hydrophilic character of the tunnel appears to be conserved.
Analysis of the resting GsPutA structure with Mole (10) reveals six ancillary tunnels that connect the main substrate-channeling tunnel to the bulk medium (Figs. 1B and 3C). Tunnels 1, 2a, and 2b intersect the main tunnel near the PRODH site, connecting the si face of the FAD isoalloxazine to the bulk medium. Tunnels 3a and 3b flow into the hydrolysis cavity. Tunnel 4 intersects the main tunnel near the base of the GSA-binding site.
Proline Analogs Remodel the PRODH Half of the Tunnel System.
The structures of GsPutA complexed with the proline analogs l-tetrahydrofuroic acid (THFA) and l-lactate were determined (Table S2). These structures are the first of a full-length PutA complexed with a bona fide proline analog and represent state II of the kinetic scheme (Fig. S1). The PRODH sites of the two complexes are very similar, and we will focus on the THFA complex because THFA is isostructural to proline.
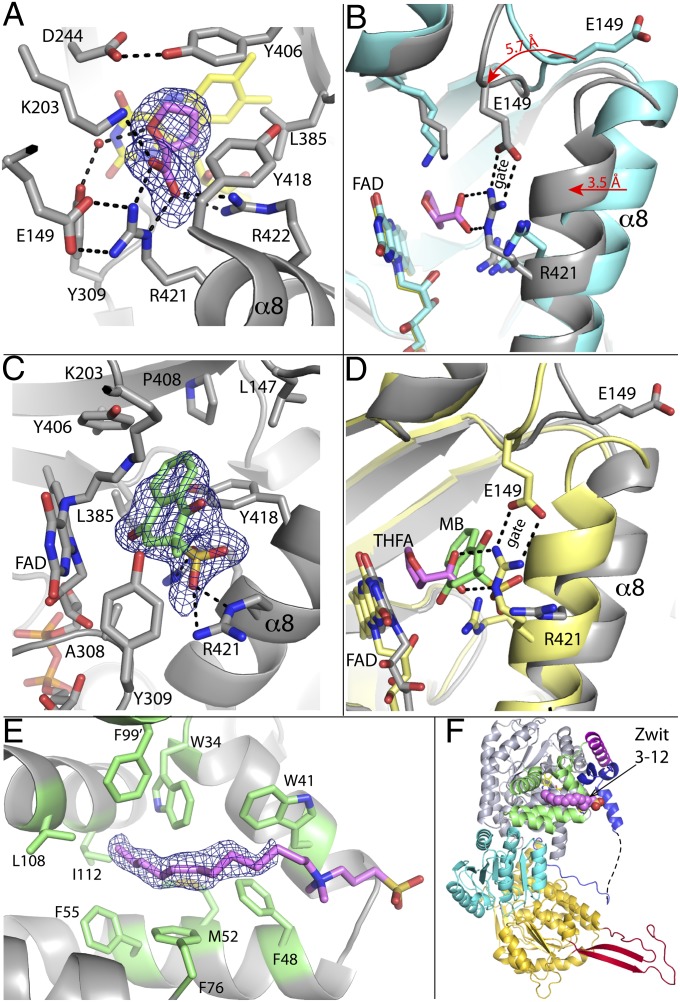
THFA binds between the si face of the isoalloxazine and α8 of the PRODH barrel (Fig. 4A). The inhibitor carboxylate forms ionic interactions with Arg421, Arg422, and Lys203 whereas the methylene groups of the ring contact Tyr406, Tyr418, and Leu385. The plethora of interactions results in the complete burial of the inhibitor and the si face of the isoalloxazine. The C5 atom of THFA, which represents the hydride donor of proline, is poised 3.5 Å from the flavin N5 atom. This arrangement is consistent with a direct hydride transfer mechanism, which has been shown for PutAs (11) and monofunctional PRODHs (12). All of the interacting residues in the GsPutA–THFA and GsPutA–l-lactate complexes are conserved in PutAs and monofunctional PRODHs, and analogous interactions are observed in the THFA complexes of an Escherichia coli PutA PRODH domain construct (13) and a monofunctional PRODH (14). Thus, substrate recognition appears to be conserved in the PutA/PRODH family.
Fig. 4.
Ligand binding to GsPutA. (A) Electron density and interactions for THFA bound to oxidized GsPutA. The cage represents a simulated annealing σA-weighted Fo − Fc omit map (2.5σ). (B) Comparison of the open (cyan) and THFA-bound closed (gray) PRODH active sites. The arrows show the directions of conformational changes induced by THFA binding. (C) Electron density and interactions for MB bound to NPPG-inactivated GsPutA. The cage represents a simulated annealing σA-weighted Fo − Fc omit map (2.5σ). (D) Comparison of the PRODH active sites of GsPutA-THFA (pink THFA, yellow protein) and inactivated GsPutA-MB (green MB, gray protein), highlighting the proximity of the proline and quinone sites and the structural differences involving α8 and Glu149. (E) Electron density for Zwittergent 3-12. The cage represents a simulated annealing σA-weighted Fo − Fc omit map (2σ). The head group is included in this figure to guide the eye but is omitted in the deposited structure. Phe99 is contributed by the α domain of a symmetry-related molecule. (F) The location of Zwittergent 3-12 (pink spheres) in the protomer, which is oriented and colored as in Fig. 1A.
The binding of THFA induces profound conformational changes, which were heretofore unknown because the structure of the resting state of PutA was unknown. Upon binding, the α8 helix (residues 415–427) of the PRODH barrel shifts 3.5 Å toward the isoalloxazine, and the β1-α1 loop (residues 147–151) of the barrel moves by 5 Å toward the active site (Fig. 4B). The latter movement draws conserved Glu149 from the surface of the enzyme (Fig. 1A) into the active site where it forms an ion pair with Arg421 of α8 (Figs. 4 A and B). Arg421 also forms a key ionic interaction with the THFA carboxylate (Fig. 4B).
These conformational changes close the PRODH active site and remodel the PRODH half of the tunnel system. Formation of the Glu149-Arg421 ion pair eliminates tunnel 2a and shrinks tunnel 2b (Fig. 3D). Furthermore, the main tunnel near the entrance to the hydrolysis cavity (15 Å from the flavin) narrows by 1 Å in diameter. Also, the main tunnel is diverted below the newly formed Glu149-Arg421 ion pair where it merges with tunnel 1. Thus, the Glu149-Arg421 ion pair appears to be a gate that senses the occupancy of the proline-binding site and controls traffic between the PRODH active site and the main tunnel.
Reduction Reconfigures the Flavin.
The structure of GsPutA with the flavin reduced was determined from crystals of the oxidized enzyme that had been soaked in dithionite followed by cryo-trapping (Table S3). This structure provides insight into the conformation of the PRODH site after release of P5C into the hydrolysis cavity and before the binding of the quinone, which is relevant to states V–VIII (Fig. S1).
Reduction induces substantial conformational changes in the flavin. Whereas the oxidized isoalloxazine is planar (Fig. S4A), the reduced isoalloxazine has a 23° butterfly bend (Fig. S4B). Furthermore, reduction induces a crankshaft rotation of the ribityl chain that affects all three hydroxyl groups, shifting the 2′- and 3′-hydroxyl groups to the pyrimidine side of the isoalloxazine and the 4′-hydroxyl to the dimethylbenzene side. Similar conformational changes in the reduced FAD have been observed for a PutA PRODH domain construct (15).
Structural and Kinetic Characterization of the Quinone-Binding Site.
The mechanism-based inactivator N-propargylglycine (NPPG) was essential for identification of the quinone site. We previously showed that NPPG is an irreversible inactivator of PutAs and PRODHs (16, 17). Briefly, NPPG is initially oxidized to N-propargyliminoglycine with concomitant reduction of the FAD, which is analogous to the oxidation of proline to P5C. Subsequent steps in the mechanism lead to imine linkage between an active site Lys and the N5 atom of the reduced flavin. Biochemical studies have shown that inactivation by NPPG locks PutA into a conformation that mimics the proline-reduced enzyme (16). Thus, NPPG-inactivated PutA is a surrogate for the reduced enzyme that can be studied under normal (aerobic) laboratory conditions.
The crystal structure of NPPG-inactivated GsPutA was determined for comparison with the dithionite-reduced enzyme (Table S3). Consistent with previous structures of NPPG-inactivated PRODHs (16, 17), electron density maps show that Lys203 is covalently attached to the flavin N5 via a 3-carbon linker (Fig. S4C). The inactivated flavin is very similar in structure to the dithionite-reduced flavin. In particular, the isoalloxazine has a large butterfly bend angle (27°), and the ribityl chain is reorganized (Fig. S4C). The similarity between the dithionite-reduced and NPPG-modified flavins confirms that inactivated GsPutA is a good surrogate for the reduced enzyme.
The structure of NPPG-inactivated GsPutA complexed with menadione bisulfite (MB) was determined at 1.95 Å resolution (Table S3). MB was used because menaquinones are the predominant respiratory quinones in Geobacter (18), and the sodium salt of MB has high aqueous solubility. Electron density in one of the protomers is consistent with a single conformation (conformation A) of the (R) enantiomer of MB (Fig. 4C). Density in the other protomer suggests conformational disorder and was modeled as the (R) enantiomer in conformation A with occupancy of 0.55 and the (S) enantiomer in a different conformation (conformation B) with occupancy of 0.45 (Fig. S4D). We focus here on conformation A as it is present in both active sites.
MB binds next to the middle ring of the isoalloxazine si face (Fig. 4C). The angle between the benzoquinone and isoalloxazine rings is ∼40°, and one of the carbonyl O atoms of MB is 3.2 Å and 4.0 Å from the flavin N10 and N5 atoms, respectively. This arrangement is consistent with direct electron transfer from the flavin to the quinone without an intermediary electron carrier (11). The 1,4-naphthoquinone is surrounded by several aromatic and nonpolar residues, including Leu147, Leu385, Tyr406, Pro408, Tyr309, and Tyr418. The amino group of modified Lys203 is 4.1 Å from a carbonyl of the benzoquinone, suggesting that Lys203 of the native enzyme could form a hydrogen bond with the quinone and possibly serve as a proton donor to the reduced quinone. The sulfate group interacts with Arg421 and Arg422; these interactions would be absent with biological menaquinones.
The steady-state kinetic parameters for GsPutA with MB and menadione were determined to test the efficacy of MB as a substrate and to verify that 1,4-naphthoquinones lacking an isoprenoid chain are electron acceptors of GsPutA (Table S4). Using saturating proline (350 mM), the Km and kcat values are 10.5 ± 1.2 μM and 0.66 ± 0.02 s−1 for MB and 8.2 ± 0.9 μM and 0.97 ± 0.02 s−1 for menadione. The catalytic efficiencies (kcat/Km) of the two quinones thus agree within a factor of two (63,000 ± 7,000 M−1⋅s−1 for MB and 118,000 ± 13,000 M−1⋅s−1 for menadione). The kinetic constants for proline at fixed quinone concentration are likewise similar for MB and menadione (Table S4). These results show that MB and menadione are efficient in vitro electron acceptors for GsPutA and suggest that the GsPutA–MB complex is functionally relevant to the GsPutA/PRODH catalytic mechanism.
Structure of GsPutA with Bound Detergent Molecule.
A structure of GsPutA was determined from a crystal that had been grown in the presence of Zwittergent 3–12 (N-dodecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate). Electron density consistent with the hydrocarbon tail of Zwittergent 3–12 was evident in one of the molecules in the asymmetric unit (Fig. 4E). Density for the head group is weak, implying conformational disorder. The density supported the modeling of the nine terminal C atoms of the tail at occupancy of 1.0. The refined B-factor of the modeled tail is 45 Å2. The head group could not be modeled with confidence but is included in Fig. 4E to guide the eye.
The hydrocarbon tail of Zwittergent 3–12 binds to an exposed hydrophobic patch of the α domain (Fig. 4F). The tail is surrounded exclusively by aromatic and nonpolar residues, including Trp, Phe, Leu, Ile, and Met. Phe99 is donated by the α domain of a neighboring protein in the crystal lattice. The observation of a detergent molecule bound in this location suggests that the α domain may be involved in membrane association.
Discussion
The GsPutA structures reveal a more complex and dynamic tunnel system than had been obtained from the lone BjPutA structure. In particular, the structures reveal six ancillary tunnels in addition to the main substrate-channeling pathway. The locations of the ancillary tunnels and their responses to ligand binding suggest hypotheses about their functions. Tunnels 1, 2a, and 2b connect the si face of the FAD isoalloxazine directly to the bulk medium, and, because proline and MB bind at the si face, these tunnels could be entryways for proline and quinone. In particular, the observation that tunnel 2a closes and tunnel 2b constricts upon THFA binding suggests that proline accesses the PRODH site via tunnels 2a/2b. Also, molecular modeling is consistent with tunnel 1 providing the entryway for quinones (Fig. 5). Tunnels 3a and 3b connect the middle section of the main substrate-channeling tunnel to the bulk medium. Because the hydrolysis of P5C likely occurs in the middle section, it is tempting to hypothesize that water for this reaction enters through these tunnels. Molecular dynamics simulations could be used to test this idea. Tunnel 4 connects to the base of the GSA-binding site, suggesting a possible escape route for glutamate. Interestingly, tunnel 4 is closed in some of the GsPutA structures due to variation of Asp321. For example, the carboxylate of Asp321 blocks tunnel 4 in the dithionite-reduced (Fig. S3B), NPPG-inactivated, and lactate-complexed enzymes. However, in the resting enzyme and THFA complex, Asp321 is disordered or rotated out into solvent, allowing tunnel 4 to reach the bulk solvent (Figs. 3 C and D). We note that the lactate complex has an adventitious 2-(N-morpholino)ethanesulfonic acid buffer molecule bound in the GSA site. Thus, the status of tunnel 4 may depend on the flavin redox state and occupancy of the GSA site, suggesting that tunnel 4 could be an additional dynamic feature of the tunnel system. These ideas about the functions of the ancillary tunnels could be tested by blocking one of more of them using site-directed mutagenesis. The GsPutA structures provide the basis for such studies.
Fig. 5.
Model of GsPutA complexed with a biological quinone (MK-8) based on the GsPutA-MB crystal structure. (A) Model of the dimer with two MK-8 molecules bound. (B) Close-up view of the model showing the superposition of the naphthoquinones of MK-8 (magenta) and MB (green). (C) A view of the isoprenoid chain of MK-8 exiting through tunnel 1.
The GsPutA structures also reveal a dynamic aspect of PutA involving the Glu149-Arg421 ion pair. These residues are present in all PutA and PRODH sequences, and the formed ion pair has been observed in PutAs and PRODHs complexed with proline analogs (7, 13, 14, 19). The observation in these structures that the proline analog is completely buried raised the question of how proline enters the active site of the resting enzyme. We had hypothesized that the ion pair might be a gate that closes and opens upon substrate binding and product release, respectively (7). The GsPutA structures provide snapshots of the gate in action, validating the ion pair gate hypothesis.
Conformational changes, such as gating, occur in other bifunctional enzymes, including tryptophan synthase (20) and glutamine amidotransferases (21), two well-studied bifunctional enzymes. Parallels between PutA and tryptophan synthase are especially apparent. Tryptophan synthase exhibits an elegant mechanism of allosteric control in which chemical events in the α- and β-catalytic sites induce conformational changes that synchronize catalysis at the two sites and prevent escape of the intermediate indole (20). The closure of the PRODH site upon proline binding is analogous to the closure of the α-subunit of tryptophan synthase by 3-indole-d-glycerol 3′-phosphate in that the inducing ligand is the first substrate of the pathway. The binding of 3-indole-d-glycerol 3′-phosphate also induces partial closure of the β-subunit. Similarly, accessibility of the P5CDH site via tunnel 4 may depend on the flavin redox state and occupancy of the GSA site. Whether the active sites of PutA are synchronized to the extent that they are in tryptophan synthase remains to be seen. Nevertheless, PutA provides another example of large conformational changes playing a central role in substrate channeling.
The GsPutA–MB complex provides previously unidentified structural information about the quinone-binding site of PutA. The quinone ring binds within 4 Å of the isoalloxazine, which is consistent with direct electron transfer and reminiscent of other flavoenzyme–quinone complexes (22–24), especially NAD(P)H/quinone acceptor oxidoreductase (25, 26). Although both MB and THFA bind at the si face of the isoalloxazine, the two ligands associate with different conformations of the active site and provoke different protein conformational changes (Fig. 4D). In particular, whereas closure of the ion pair gate accompanies THFA binding, the PRODH site remains open when MB is bound (Fig. 4D). The open active site presumably is needed to accommodate the larger bulk of MB compared with proline. The unique binding conformation found with MB is consistent with a two-site ping-pong mechanism for proline:quinone oxidoreductase activity as observed previously for E. coli PutA (27).
Finally, the GsPutA–MB complex provides clues about the structural basis of PutA membrane association. The GsPutA–MB structure was used to model the enzyme complexed with menaquinone-8 (MK-8) (Fig. 5), the most abundant menaquinone in Geobacter (18). A model of MK-8 was manually fitted to the GsPutA–MB crystal structure by overlapping the naphthoquinones (Fig. 5B), and the dihedral angles of the isoprenoid chain were adjusted to avoid steric clash with the protein. Two cavities for the isoprenoid chain are evident: tunnel 1 and the main substrate-channeling tunnel. The former was selected because it allows the isoprenoid chain to exit the protein (Figs. 5 A and C) rather than being sequestered in the middle of the protein. In the resulting model, the isoprenoid chain exits the protein at the concave face of the dimer (Fig. 5A), implying that this surface is oriented toward the membrane.
The idea that the concave face of GsPutA approaches the membrane was explored with electrostatic potential calculations. This face of GsPutA presents adjacent regions of neutral and positive electrostatic potential (Fig. S5A), which is the expected pattern for a peripheral membrane association surface (23, 28–30). In contrast, the convex face (Fig. S5B) and sides (Fig. S5C) of GsPutA feature regions of strong negative potential. The prominent neutral portions of the concave face correspond to the arm and α domains whereas the strong positive patch corresponds to α6, β7, and α7 of the PRODH barrel (Fig. S5A). The arm is reminiscent of the hydrophobic ridge of yeast NADH dehydrogenase that is proposed to contact the membrane (23), and involvement of the α domain in membrane association is consistent with the observation of a detergent molecule bound to this region (Fig. 4E) and fluorescence quenching studies of E. coli PutA (31). The GsPutA structures provide a foundation for designing experiments to test this hypothesis about PutA-membrane association.
Materials and Methods
The initial structure of GsPutA was determined with single-wavelength anomalous diffraction phasing based on a mercury derivative (Table S1). This structure was used to calculate initial phases for the other structures using molecular replacement (Tables S2 and S3). See SI Materials and Methods for detailed information regarding the preparation and crystallization of GsPutA, phasing and refinement calculations, SAXS experiments, and kinetics measurements.
Supplementary Material
Acknowledgments
We thank Prof. Chris Whitman and Dr. William Johnson, Jr., for providing NPPG. We thank Prof. Steve Almo and the New York Structural Genomics Research Consortium (NYSGRC) for clone 013560. We thank Drs. Jonathan Schuermann and Jay Nix for help with X-ray diffraction data collection and Kevin Dyer for collecting the SAXS data through the Structurally Integrated Biology for Life Sciences mail-in program. Research reported in this publication was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under Awards GM065546, GM061068, and P30GM103335. The NYSGRC is supported by NIGMS Grant U54GM094662. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by NIGMS Grant P41 GM103403 from the NIH. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357. Part of this work was conducted at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the DOE, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by the DOE Office of Biological and Environmental Research. Additional support comes from NIH project Macromolecular Insights on Nucleic Acids Optimized by Scattering (R01GM105404). The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the DOE under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4NM9 and 4NMA–4NMF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321621111/-/DCSupplemental.
References
- 1.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol. 2012;2:60. doi: 10.3389/fonc.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zarse K, et al. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15(4):451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee IR, et al. Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway. Genetics. 2013;194(2):421–433. doi: 10.1534/genetics.113.150326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnan N, Doster AR, Duhamel GE, Becker DF. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect Immun. 2008;76(7):3037–3044. doi: 10.1128/IAI.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakajima K, et al. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res. 2008;29(1):9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- 6.Singh RK, Tanner JJ. Unique structural features and sequence motifs of proline utilization A (PutA) Front Biosci (Landmark Ed) 2012;17:556–568. doi: 10.2741/3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srivastava D, et al. Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 2010;107(7):2878–2883. doi: 10.1073/pnas.0906101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 10.Berka K, et al. MOLEonline 2.0: Interactive web-based analysis of biomacromolecular channels. Nucleic Acids Res. 2012;40(Web Server issue):W222–W227. doi: 10.1093/nar/gks363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moxley MA, Becker DF. Rapid reaction kinetics of proline dehydrogenase in the multifunctional proline utilization A protein. Biochemistry. 2012;51(1):511–520. doi: 10.1021/bi201603f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrano H, Blanchard JS. Kinetic and isotopic characterization of L-proline dehydrogenase from Mycobacterium tuberculosis. Biochemistry. 2013;52(29):5009–5015. doi: 10.1021/bi400338f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, et al. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43(39):12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M, Arentson BW, Srivastava D, Becker DF, Tanner JJ. Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release. Biochemistry. 2012;51(50):10099–10108. doi: 10.1021/bi301312f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, et al. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA—membrane binding. Biochemistry. 2007;46(2):483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava D, et al. The structure of the proline utilization a proline dehydrogenase domain inactivated by N-propargylglycine provides insight into conformational changes induced by substrate binding and flavin reduction. Biochemistry. 2010;49(3):560–569. doi: 10.1021/bi901717s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White TA, Johnson WH, Jr, Whitman CP, Tanner JJ. Structural basis for the inactivation of Thermus thermophilus proline dehydrogenase by N-propargylglycine. Biochemistry. 2008;47(20):5573–5580. doi: 10.1021/bi800055w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedrick DB, et al. Polar lipid fatty acids, LPS-hydroxy fatty acids, and respiratory quinones of three Geobacter strains, and variation with electron acceptor. J Ind Microbiol Biotechnol. 2009;36(2):205–209. doi: 10.1007/s10295-008-0486-7. [DOI] [PubMed] [Google Scholar]
- 19.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10(2):109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn MF, Niks D, Ngo H, Barends TR, Schlichting I. Tryptophan synthase: The workings of a channeling nanomachine. Trends Biochem Sci. 2008;33(6):254–264. doi: 10.1016/j.tibs.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Mouilleron S, Golinelli-Pimpaneau B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr Opin Struct Biol. 2007;17(6):653–664. doi: 10.1016/j.sbi.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, et al. Structural insight into the type-II mitochondrial NADH dehydrogenases. Nature. 2012;491(7424):478–482. doi: 10.1038/nature11541. [DOI] [PubMed] [Google Scholar]
- 23.Iwata M, et al. The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proc Natl Acad Sci USA. 2012;109(38):15247–15252. doi: 10.1073/pnas.1210059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherney MM, Zhang Y, James MN, Weiner JH. Structure-activity characterization of sulfide:quinone oxidoreductase variants. J Struct Biol. 2012;178(3):319–328. doi: 10.1016/j.jsb.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Faig M, et al. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: Species comparison and structural changes with substrate binding and release. Proc Natl Acad Sci USA. 2000;97(7):3177–3182. doi: 10.1073/pnas.050585797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster CE, Bianchet MA, Talalay P, Zhao Q, Amzel LM. Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry. 1999;38(31):9881–9886. doi: 10.1021/bi990799v. [DOI] [PubMed] [Google Scholar]
- 27.Moxley MA, Tanner JJ, Becker DF. Steady-state kinetic mechanism of the proline:ubiquinone oxidoreductase activity of proline utilization A (PutA) from Escherichia coli. Arch Biochem Biophys. 2011;516(2):113–120. doi: 10.1016/j.abb.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt H, et al. Structural and mechanistic analysis of the membrane-embedded glycosyltransferase WaaA required for lipopolysaccharide synthesis. Proc Natl Acad Sci USA. 2012;109(16):6253–6258. doi: 10.1073/pnas.1119894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dym O, Pratt EA, Ho C, Eisenberg D. The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme. Proc Natl Acad Sci USA. 2000;97(17):9413–9418. doi: 10.1073/pnas.97.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106(24):9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W, Becker DF. Exploring the proline-dependent conformational change in the multifunctional PutA flavoprotein by tryptophan fluorescence spectroscopy. Biochemistry. 2005;44(37):12297–12306. doi: 10.1021/bi051026b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.