Abstract
NADPH oxidase (NOX)-derived reactive oxygen species (ROS) act as signaling determinants that induce different cellular processes. To characterize NOX function during fungal development, we utilized the genetically tractable ascomycete Sordaria macrospora. Genome sequencing of a sterile mutant led us to identify the NADPH oxidase encoding nox1 as a gene required for fruiting body formation, regular hyphal growth, and hyphal fusion. These phenotypes are shared by ∆nor1, lacking the NOX regulator NOR1. Further phenotypic analyses revealed a high correlation between increased ROS production and hyphal fusion deficiencies in ∆nox1 and other sterile mutants. A genome-wide transcriptional profiling analysis of mycelia and isolated protoperithecia from wild type and ∆nox1 revealed that nox1 inactivation affects the expression of genes related to cytoskeleton remodeling, hyphal fusion, metabolism, and mitochondrial respiration. Genetic analysis of ∆nox2, lacking the NADPH oxidase 2 gene, ∆nor1, and transcription factor deletion mutant ∆ste12, revealed a strict melanin-dependent ascospore germination defect, indicating a common genetic pathway for these three genes. We report that gsa3, encoding a G-protein α-subunit, and sac1, encoding cAMP-generating adenylate cyclase, act in a separate pathway during the germination process. The finding that cAMP inhibits ascospore germination in a melanin-dependent manner supports a model in which cAMP inhibits NOX2 activity, thus suggesting a link between both pathways. Our results expand the current knowledge on the role of NOX enzymes in fungal development and provide a frame to define upstream and downstream components of the NOX signaling pathways in fungi.
Keywords: NOX enzymes, fruiting body formation, RNA-seq, ascospore germination, STE12
DURING sexual reproduction, filamentous fungi generate complex fruiting bodies that contain and protect meiosporangia. We used the ascomycetous model fungus Sordaria macrospora to identify genes directly involved in fruiting body development (Kück et al. 2009; Engh et al. 2010; Kück et al. 2009). Due to its homothallic life style, S. macrospora is able to complete the sexual life cycle without the mating of strains with opposite sex, and therefore, fruiting body-deficient mutants can be recognized directly without the need for crossing experiments. In earlier work, we generated sterile mutants showing a developmental block after formation of young fruiting bodies (protoperithecia), but being unable to generate mature perithecia, and referred to these mutants as pro. Recently, we have applied next-generation genome re-sequencing to identify the genes affected in some of these mutants (Nowrousian et al. 2012). Based on this approach, we now have characterized mutant pro32 and show that it carries a mutation in the nox1 gene encoding NAPDH oxidase 1 (NOX1).
NADPH oxidase (NOX) enzymes are transmembrane proteins that are highly conserved among eukaryotes and produce reactive oxygen species (ROS) through the oxidation of NADPH (Lambeth 2004; Kawahara and Lambeth 2007). ROS have long been recognized as damaging agents due to uncontrolled oxidizing reactions with DNA, RNA, proteins, and lipids (Halliwell and Gutteridge 2007). However, there is increasing evidence that ROS act as signaling determinants that induce different cellular processes (Scott and Eaton 2008; Aguirre and Lambeth 2010; Heller and Tudzynski 2011).
In mammals, seven members of the NOX family (NOX1–5, DUOX1, and DUOX2) are known (Aguirre et al. 2005; Kawahara and Lambeth 2007). The activity of NOX2, the most intensively studied NOX, is regulated by a protein complex containing p22phox, p40phox, p47phox, p67phox, and the small GTPase RAC1. NOX1, NOX3, and NOX4 also require p22phox, whereas the activity of NOX5, DUOX1, and DUOX2 is independent of this regulator (Smith et al. 2012). To date, three members of the NOX family are known in fungi. NOX1 and NOX2 [synonymous (syn.) NOXA and NOXB] are homologs of mammalian NOX2 and have been found in most ascomycetes. In contrast, NOX3, the homolog of mammalian NOX5, has been detected only in some fungi like Aspergillus terreus, Magnaporthe grisea, Podospora anserina, and several Fusarium species (Aguirre et al. 2005; Scott and Eaton 2008; Brun et al. 2009). Fungal NOX1 and NOX2 enzymes are regulated by the p67phox homolog NOR1 (NOX regulating, syn. NOXR) and the small GTPase RAC1 (syn. RacA) (Kawahara and Lambeth 2007; Tanaka et al. 2008). In Epichloë festucae, the putative scaffold protein Bem1 was reported to associate with NOR1 and CDC24 (Takemoto et al. 2011). Another candidate NOX regulatory protein is tetraspanin Pls1. Strains from different ascomycetes, lacking the corresponding gene, have phenotypes similar to nox2 deletion strains (Lambou et al. 2008; Ryder et al. 2013; Siegmund et al. 2013). However, very little is known about the upstream and downstream components of the NOX signaling pathways in fungi.
In this study, we carried out a comprehensive genetic analysis using S. macrospora to elucidate the contribution of NOX enzymes to fungal sexual development. In addition to the characterization of sterile mutant pro32, we present a detailed functional analysis of deletion mutants nox1, nox2, and nor1 in S. macrospora, showing that NOX1 and NOR1 are required for fruiting body development and hyphal fusion. For the first time, we provide RNA-seq analysis of ∆nox1 protoperithecia and show NOX1-dependent gene expression compared to gene expression in protoperithecia of the sterile mutant pro1 and wild-type strains. Furthermore, phenotypic and genetic analysis led us to conclude that NOX2 and NOR1 contribute to a signaling pathway controlling ascospore germination and that transcription factor STE12 is part of this pathway. Finally, we relate cAMP levels to NOX2 function. Our analysis extends the current knowledge on the contribution of NOX enzymes to the regulation of two distinct fungal developmental processes and provides important hints to define the cellular routes regulated by these enyzmes.
Materials and Methods
Strains, media, and growth conditions
S. macrospora strains, listed in Table 1, were grown under standard laboratory conditions on complete medium (CM) or cornmeal malt fructification medium (BMM) media (Esser 1982; Nowrousian et al. 1999). Cultivation for ascospore germination assays and DNA extraction were performed as described previously (Nowrousian and Cebula 2005; Kamerewerd et al. 2008; Teichert et al. 2012). For rescue of fertility, strains were inoculated on filter paper covering solid BMM medium. Two consecutive transfers of the filter paper to fresh solid BMM medium were performed after 3 days. Continuous growth on solid BMM medium with or without filter paper served as control (adapted from Malagnac et al. 2004). Quantification of linear growth was performed using race tube assays. Recombinant plasmids were propagated in Escherichia coli XL1 Blue MRF′ (Stratagene, La Jolla, CA) under standard experimental conditions (Sambrook and Russell 2001).
Table 1. S. macrospora strains used in this study.
| Strain | Relevant genotype | Relevant phenotype | Reference source |
|---|---|---|---|
| S91327 | Wild type | F | Culture collection of the Department of General and Molecular Botany |
| S84595 | fus, spore color mutant | F | Culture collection of the Department of General and Molecular Botany |
| S96888 | ∆ku70::nat | F | Pöggeler and Kück (2006) |
| DD27, DD1 | ∆nox1::hph | S, DG, HFD | This study |
| DD194, DD299 | ∆nox2::hph/fus | F, GDB | This study |
| DD118-2 | ∆nox2::hph/∆ku70::nat | F | This study |
| DD492, DD574 | ∆nor1::hph/fus | S, GDB, DG, HFD | This study |
| S104701 | ∆gsa3::hph | F, GD, DG | Kamerewerd et al. (2008) |
| S114583, S114602 | ∆ste12::hph/fus | F, GDB | Kamerewerd et al. (2008) |
| S107115 | ∆gsa3::hph/∆nox2::hph/fus | F, GD | This study |
| S83812 | ∆gsa3::hph/∆ste12::hph/r2 | S, DG | Kamerewerd et al. (2008) |
| S144534, S114567 | ∆nox2::hph/∆ste12::hph/r2 | F, GDB | This study |
| DD1093, DD1161, DD1325 | ∆nox1::hph/nox1::nat | F, HFD | This study |
| S106371, S106375, S106755 | ∆nox2::hph/nox2::nat | F | This study |
| DD2843, DD2909, DD2958 | ∆nor1::hph/nor1::nat | F | This study |
| S69656 | ∆pro40::hph | S | Engh et al. (2007) |
| S109348 | pro32 | S, HFD | This study |
| DD291-3-1, DD290-4-3 | pro32/nox1::nat | F | This study |
F, fertile; S, sterile; HFD, hyphal fusion defect; GD, germination defect; GDB, germination defect in black ascospores; DG, decreased growth.
Preparation of nucleic acids
DNA and RNA were extracted using phenol/chloroform, and RNA was selectively precipitated (Pöggeler et al. 1997). For RNA-seq analysis, mycelia were grown in surface cultures for 4 days or directly on slides [Molecular Machines and Industries (MMI)] for fixation and dissection in situ (Teichert et al. 2012).
Genome sequencing of developmental mutant pro32
Mutant pro32 from our laboratory collection was backcrossed several times with wild type or red-spored fus mutant (Nowrousian et al. 2012) and finally crossed with fus. DNA was extracted from 40 sterile and 40 fertile progeny as described previously (Nowrousian et al. 2012). Five micrograms of pooled genomic DNA for pro32 and wild type, respectively, was subjected to 50-bp paired-end Illumina/Solexa sequencing with a HiSeq2000 at GATC Biotech (Constance, Germany). Cleaning of raw data, mapping to the S. macrospora reference genome, and analysis of sequence variants were performed as previously described (Nowrousian et al. 2012). The Burrows Wheeler Alignment tool (Li and Durban 2009) was used for mapping and SAMtools (Li et al. 2009) for SNP calling. Further bioinformatics analysis was done using custom-made Perl scripts.
Generation of Δnox1, Δnox2, and Δnor1 deletion strains
Transformation of S. macrospora was performed as described previously with an enzyme mix of 1 g VinoTaste Pro (Novozymes, Blagsvaerd, Denmark), 0.3 g Caylase (Cayla, Toulouse, France), and 27 U Chitinase (ASA Spezialenzyme, Wolfenbüttel, Germany) (Walz and Kück 1995; Engh et al. 2007). For the generation of transgenic plasmids, homologous recombination in S. cerevisiae PJ69-4a and standard cloning procedures were used (James et al. 1996; Sambrook and Russell 2001; Colot et al. 2006; Bloemendal et al. 2012). To allow homologous recombination of nox knockout constructs in S. macrospora ∆ku70, the flanking regions of the corresponding nox genes together with the hph resistance cassette were inserted into pRS426 (Christianson et al. 1992). The 5′ and 3′ regions flanking the nox genes were amplified with specific oligonucleotides using PCR based on wild-type (S91327) genomic DNA [nox1: 5′ region 05007-5fw/05007-5rv (1039 bp), 3′ region 05007-3fw/05007-3rv (1035 bp); nox2: 5′ region 08741-5fw/08741-5rv (1065 bp), 3′ region 08741-3fw/08741-3rv (808 bp); nor1: 5′ region 02124-5fw/02124-5rv (1064 bp), 3′ region 02124-3fw/02124-3rv (1065 bp)]. The hph resistence cassette was obtained from vector pDrivehph (Nowrousian and Cebula 2005) by EcoRI restriction. Plasmids pKO-nox1, pKO-nox2, and pKO-nor1 were linearized with restriction enzymes (BamHI, pKO-nox1 and pKO-nor1; XhoI, pKO-nox2) and used to transform S. macrospora strain ∆ku70. To verify transformants, genomic DNA was isolated and tested for homologous recombination at the nox locus using PCR [∆nox1: 05007_vp1/d1 (1159 bp) and d2/05007_vp2 (2257 bp); ∆nox2: 08742_vp1/d1 (1111 bp) and d2/08742_vp2 (1144 bp); ∆nor1: 02124_vp1/d1 (1190 bp) and d2/02124_vp2 (1193 bp)]. To verify the deletion of nox genes, the following oligonucleotides were used: 05007-5′fw/05007_comp_5′rv (nox1 5′ fragment, 2017 bp), 05007_comp_3′fw/05007-3′rv (nox1 3′ fragment, 2248 bp), 08741-5′fw/08741_comp_5′rv (nox2 5′ fragment, 2053 bp), 08741_comp_3′fw/08741-3′rv (nox2 3′ fragment, 1910 bp), 02124-5′fw/02124_comp_5′rv (nor1 5′ fragment, 1979 bp), and 02124_comp_3′fw/02124-3′rv (nor1 3′ fragment, 1966 bp). Probes for Southern blot detection of nox genes were generated by restriction of complementing plasmids (nox1: pComp_nox1/XhoI; nox2: pComp_nox2/SphI; nor1: pComp_nor1/KspI). The hph probe was obtained from vector pDrivehph by EcoRI restriction (Nowrousian and Cebula 2005).
Plasmids for complementation of nox deletion strains were generated as follows: the nox ORFs and ∼1000 kb up- and downstream regions were amplified via PCR [nox1: 05007-5fw/05007-3rv (3889 bp); nox2: 08742-5fw/08742-3rv (3709 bp); nor1: 02124-5fw/02124-3rv (3842 bp)]. The amplicons were cloned in pRS426_nat (Klix et al. 2010) using homologous recombination in yeast. The complementation vectors pComp_nox1, pComp_nox2, and pComp_nor1 were transformed into the corresponding deletion strains. To verify ∆nox2 complementation, it was necessary to quantify the ascospore germination of black ascospores. For this, the deletion mutant ∆nox2/fus was transformed with pComp_nox2. Resulting strains carrying a nox2 deletion and an ectopical integration of nox2 were designated as ∆nox2/fus::nox2. To obtain black-spored ∆nox2::nox2, the ∆nox2/fus::nox2 was crossed with wild type, and black ascospores were isolated from recombinant asci. The black-spored ∆nox2::nox2 were verified by PCR. Double deletion strains were obtained via crossing of single deletion mutants followed by the isolation of single spores from recombinant asci as described previously (Esser and Straub 1958). All oligonucleotides and plasmids used in this study are listed in Table 2 and Table 3, respectively.
Table 2. Oligonucleotides used in this study.
| Oligonucleotide | Sequence (5′–3′) | Specificity |
|---|---|---|
| 05007-5fw | gtaacgccagggttttcccagtcacgacgggatccgaacaaacacaataacaccattgcc | 5′ nox1 with pRS426 overlap; BamHI |
| 05007-5rv | cgagggcaaaggaatagggttccgttgagggttggcgaccgctgaattcctcctc | 5′ nox1 with hph overhang |
| 05007-3fw | gcccaaaaatgctccttcaatatcagttgcgttatacttggcttataactataccc | 3′ nox1 with hph overlap |
| 05007-3rv | gcggataacaatttcacacaggaaacagcggatcctgattaggcggtattagttatggttg | 3′ nox1 with pRS426 overlap, BamHI |
| 08741-5fw | gtaacgccagggttttcccagtcacgacggtcgacgtccttcggtgatgtgccgagagtgc | 5′ nox2 with pRS426 overlap, SalI |
| 08741-5rv | cgagggcaaaggaatagggttccgttgaggcgtgtctgggttgcttctgttgtcgt | 5′ nox2 with hph overlap |
| 08741-3fw | gcccaaaaatgctccttcaatatcagttgcacgtctttgtcggaattcccgttta | 3′ nox2 with hph overlap |
| 08741-3rv | gcggataacaatttcacacaggaaacagcgtcgacctttgcggttgtcgctcatgcgatt | 3′ nox2 with pRS426 overlap; SalI |
| 02124-5fw | gtaacgccagggttttcccagtcacgacgggatccctggatacctctaggtcatcaattg | 5′ nor1 with pRS426 overlap, BamHI |
| 02124-5rv | cgagggcaaaggaatagggttccgttgagggttgaggtgttgttagacgtgcgta | 5′ nor1 with hph overlap |
| 02124-3fw | gcccaaaaatgctccttcaatatcagttgcaaccagctcgccgtctggttttgg | 3′ nor1 with hph overlap |
| 02124-3rv | gcggataacaatttcacacaggaaacagcggatcccgtgaggaagctggttgatcctgag | 3′ nor1 with pRS426 overlap; BamHI |
| 05007_vp1 | gccctgaggcgatttttgtttatc | ∆nox1 |
| 05007_vp2 | gcttttcgctctcacggtagattc | ∆nox1 |
| d1 | cgatggctgtgtagaagtactcgc | hph |
| d2 | atccgcctggacgactaaaccaa | hph |
| 08741_vp1 | ctaagcactttggtccttttcccc | ∆nox2 |
| 08741_vp2 | gattaggaagctgtagatgctcatggag | ∆nox2 |
| 02124_vp1 | ggacaatttccgaggagctggac | ∆nor1 |
| 02124_vp2 | gcttcatgtcagatcgcttgttcc | ∆nor1 |
| 05007_comp5′rv | gtgccactggtacttggagacttg | nox1 |
| 05007_comp3′fw | gtatcagacagcaatccttcgaaac | nox1 |
| 08741_comp5′rv | gttctccttcttgatctggatctcg | nox2 |
| 08741_comp3′fw | cacatgttcatcgtcttcttctttttc | nox2 |
| 02124_comp5′rv | ctatcatccttgacttccagtttcc | nor1 |
| 02124_comp3′fw | gtgctgaaatcaaaaaatgttagtcttg | nor1 |
| 5007 cDNA rev | gtgttccttccaaaacctgaaatc | nox1 |
| nox1_RT_fw | ggacatggataccacgcaga | nox1 (qRT-PCR) |
| nox1_RT_rv | ttccgcatgctctcaaagaa | nox1 (qRT-PCR) |
| nor1_RT_fw | ctggtatgcaggatttggca | nor1 (qRT-PCR) |
| nor1_RT_rv | gcctcgtttggtcggtagac | nor1 (qRT-PCR) |
| nox2_RT_fw_2 | ctggttcttttccccgtctg | nox2 (qRT-PCR) |
| nox2_RT_rv_2 | ggaccatgctgtcgtgatgt | nox2 (qRT-PCR) |
| ste12_RT_fw | gcctttcagtcccagtccac | ste12 (qRT-PCR) |
| ste12_RT_rv | ctgtcccatgttctgtccca | ste12 (qRT-PCR) |
| pro1_RT_fw | ttcgatcgattcgcattttg | pro1 (qRT-PCR) |
| pro1_RT_rv | tgatgaatatttgccgctcg | pro1 (qRT-PCR) |
| gsa3_RT_fw | tcgaccgaatgagatggatg | gsa3 (qRT-PCR) |
| gsa3_RT_rv | cacttcttgcgttcgctacg | gsa3 (qRT-PCR) |
| sac1_RT_fw | aggcttgcacttctcttcgg | sac1 (qRT-PCR) |
| sac1_RT_rv | ttgagcaggcccgttaatct | sac1 (qRT-PCR) |
| smta-1_RT_fw | catcgtcgccgaatacaaga | Smta-1 (qRT-PCR) |
| smta-1_RT_rv | aacgacgacactatcgggct | Smta-1 (qRT-PCR) |
| smtA-3_RT_fw | tcatgatgatggaatgggga | SmtA-3 (qRT-PCR) |
| smtA-3_RT_rv | ttgttttggcatccgtcttg | SmtA-3 (qRT-PCR) |
| smtA-2_RT_fw | agcatgctgcgtcattgagt | SmtA-2 (qRT-PCR) |
| smtA-2_RT_rv | cacccaacacatgcacctct | SmtA-2 (qRT-PCR) |
| smtA-1_RT_fw | cacgatccctttcacaacga | SmtA-1 (qRT-PCR) |
| smtA-1_RT_rv | ggcaagtagttttcgcgacc | SmtA-1 (qRT-PCR) |
| SMU6905for | ggcatcacggtcaatggtgt | teh (qRT-PCR) |
| SMU6905rev | tgctcagccatcatcctctca | teh (qRT-PCR) |
| pre1for | gcattcacgcccacatcaac | pre1 (qRT-PCR) |
| pre1rev | gttgtgccgaaggtgatgca | pre1 (qRT-PCR) |
| pre2for | tccacccgttccataccctg | pre2 (qRT-PCR) |
| pre2rev | tcgatgcaagctagttcgcg | pre2 (qRT-PCR) |
| ppg1-for | ctccgtgacaccaccttcag | ppg1 (qRT-PCR) |
| ppg1-rev | ggaggcatagcgcttcca | ppg1 (qRT-PCR) |
| ppg2for | cggtatctcgcctctcaacgt | ppg2 (qRT-PCR) |
| ppg2rev | gttgtgctcccattgtgcaga | ppg2 (qRT-PCR) |
| tap1_RT_fw | tgaccaagttgcatcccaag | tap1 (qRT-PCR) |
| tap1_RT_rv | caaccgtagccctcaacaca | tap1 (qRT-PCR) |
| SMU4533Ncofor | ccatggctccctcagtcgatcctaccacc | app (Northern blot) |
| SMU4533Ncorev | ccatggcctccgacggcttgttatcaaccaa | app (Northern blot) |
Table 3. Plasmids used in this study.
| Plasmid | Feature | Reference |
|---|---|---|
| pRS426 | URA3, lacZ_a, T7_promoter, T3_promoter, bla, FRT, hph | Christianson et al. (1992) |
| pDrivehph | pDrive with hph | Nowrousian and Cebula (2005) |
| pKO-nox1 | pRS426 with 1000 kb 5′ and 3′ region of nox1, hph | This study |
| pKO-nox2 | pRS426 with kb 5′ and 3′ region of nox2, hph | This study |
| pKO-nor1 | pRS426 with 1000 kb 5′ and 3′ region of nor1, hph | This study |
| pComp-nox1 | pRS426 with nox1 gene and 1000 kb 5′ and 3′, nat | This study |
| pComp-nox2 | pRS426 with nox2 gene and 1000 kb 5′ and 3′, nat | This study |
| pComp-nor1 | pRS426 with nor1 gene and 1000 kb 5′ and 3′, nat | This study |
Microscopy
To investigate sexual propagation, S. macrospora was grown on slides with a thin layer of BMM medium at 27° in continuous light (Engh et al. 2007). Hyphal fusion assays were performed after 2 days of growth on Sordaria Westergaard's medium overlaid with a cellophane layer (Bio-Rad, München, Germany) (Bloemendal et al. 2012). Light microscopy was performed either with an AxioPhot microscope (Zeiss, Jena, Germany) capturing images with an AxioCam using the AxioVision digital system, or AxioImager microscope (Zeiss) capturing images with a Photometrix Cool SnapHQ camera (Roper Scientific). Processing of images was done with MetaMorph (version 7.7.5.0, Universal Imaging) and Adobe Photoshop (Adobe Systems, Dublin, Ireland). The documentation of sexual propagation of complemented strains was done on BMM medium in Petri dishes using a Stemi 2000-C binocular (Zeiss) capturing images with AxioCam ERc5s.
Detection and quantification of ROS
The detection of ROS by nitroblue tetrazolium (NBT) staining was performed as described previously (Malagnac et al. 2004) using cultures with mycelia covering the whole plate (grown for 4 days on solid BMM media). This assay was performed with at least three independent biological replicates per strain. Before and 30 min after NBT addition, pictures were taken of all plates with defined camera settings to determine the mean tonal range (Adobe Photoshop, histogram). For every plate the mean tonal range values after the NBT assay were normalized to the values before NBT addition. Resulting values for the mutant strains were then normalized to the corresponding value for the wild type.
Ascospore germination assays
In this study, we used three different ascosopore germination assays. In the first assay, we quantified germination of ascospores discharged from perithecia generated by selfing (ascospore germination assay of selfing strains). For this, fertile strains were cultivated on BMM medium for 5–10 days at 27° in continuous light. When the first discharged ascospores were observed, a Petri dish with thin BMM medium and 0.5% sodium acetate (BMM-Ac) was put upside down on the S. macrospora culture to directly catch discharged ascospores. After 5 hr incubation at 27°, a minimum of 500 ascospores were microscopically analyzed for germination.
In our second ascospore germination assay, we investigated the effect of different concentrations of ascorbate (antioxidant) and cAMP on germinating ascospores using cell permeable N6, 2′-O-dibutyryl-cAMP (db-cAMP, BioLog). In this assay, we used the fertile wild-type and fus strains, which were cultivated and analyzed as described for the ascospore germination assay of selfing strains, except that ascospores were discharged on BMM-Ac medium supplemented with different concentrations of db-cAMP or ascorbate.
To investigate the influence of a certain gene deletion on ascospore germination in sterile mutants, we performed an ascospore germination assay as a crossing experiment (crossing ascospore germination assay). All the investigated strains carried the fus mutation and were crossed with wild type (black ascospores), leading to recombinant asci containing black and red ascospores. After 11 days, wild type (black) and fus (red) ascospores were isolated from recombinant perithecia and recovered on BMM-Ac medium. Colonies from at least 100 germinated black (wild type) and 100 germinated red (fus) spores were tested for hygromycin B resistance to detect those carrying a gene deletion.
RNA-seq analysis
For RNA-seq analysis, RNA isolated from wild-type and ∆nox1 mycelia and protoperithecia was used. For protoperithecia isolation by laser microdissection (LM), the ∆nox1 mutant was grown directly on slides for fixation and dissection in situ (Teichert et al. 2012). RNA was isolated from protoperithecia with the Arcturus PicoPure kit (Applied Biosystems, Carlsbad, CA) and amplified in two linear amplification rounds using the TargetAmp 2-Round aRNA Amplification kit 2.0 (Epicentre Biotechnologies, Madison, WI) with modifications as described in Teichert et al. (2012). For RNA isolation of total mycelia, strains were precultured for 2 days on solid BMM. From these plates, three 20-ml BMM liquid cultures were inoculated and incubated for an additional 4 days. RNA from mycelia was isolated using phenol/chloroform extraction. RNA from protoperithecia (3.5 µg) and total mycelia (400 µg) was used for library preparation and Illumina/Solexa sequencing at GATC Biotech. cDNA libraries were prepared with the TrueSeq RNA sample preparation kit (Illumina, San Diego, CA) and sequenced with a HiSeq2000. For wild-type total mycelia, RNA from one sample was sequenced, for ∆nox1 mycelia and ∆nox1 protoperithecia, RNA from two independent biological replicates was used for sequencing. Resulting reads were cleaned and mapped to the reference S. macrospora genome v2 (Teichert et al. 2012). Differential expression was evaluated using DESeq (Anders and Huber 2010) and a method called “classical analysis.” In this analysis, genes were grouped into five groups (0 to 4) containing genes that are not differentially expressed (group 0) to genes that are strongly and significantly differentially expressed (group 4) as described in Teichert et al. (2012). Based on the DESseq and classical analysis, a consensus was calculated according to the following criteria for differential expression similar to what was described before (Teichert et al. 2012): a gene is described as differentially regulated if ratios in both DESeq and classical analysis are >4 or <0.25, DESeq adjusted P-value ≤ 0.1, and a gene in groups 1–4 in the classical analysis. For comparisons of mycelia vs. protoperithecia samples, ratio thresholds were set to >8 and <0.125 (Supporting Information, File S1).
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed as described previously (Nowrousian et al. 2005) using the master mix from the Promega GoTaq qPCR kit for SybrGreen. qRT-PCR was performed in a StepOnePlus (Applied Biosystems) using StepOne software v2.2. Sequences of oligonucleotides used are given in Table 2.
Accession numbers
Raw sequence data from sequencing mutant pro32 (pro32/fus) and wild type (wt_3) were submitted to the National Center for Biotechnology Information (NCBI) sequence read archive (accession no. SRP033637). The RNA-seq reads and derived expression ratios were submitted to the Gene Expression Omnibus (GEO) database (accession no. GSE49363).
Results
Genome sequencing of mutant pro32 identifies a point mutation in nox1 encoding NADPH oxidase 1
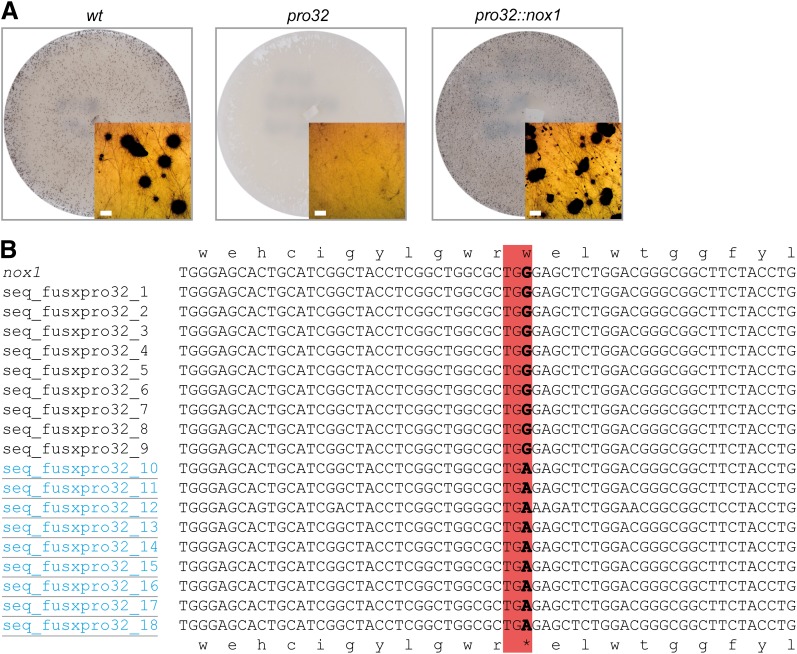
We have previously generated a collection of sterile pro mutants with a developmental block after protoperithecia formation (Kück et al. 2009). Using next-generation sequencing for efficient and time-saving identification of mutations (Nowrousian et al. 2012), we sequenced the genome of sterile pro32 (Figure 1A). After cleaning of raw sequence data from pro32 and a wild-type reference strain, the majority of reads (>95%) mapped to the reference genome (Table S1). We identified one mutation with 100% penetrance at position 810 of SMAC_05007 encoding a putative NADPH oxidase 1 (NOX1) in pro32, but not in the wild-type sample (Table 4). The point mutation in pro32 results in a transition from G to A at position 810 of the SMAC_05007 ORF, creating an early stop codon. As a consequence, the highly conserved NOX1 ferredoxin reductase-like C-terminal domain is missing from the NOX1 protein of the mutant strain (Figure S1). After crossing of pro32 to spore color mutant fus that carries a mutation in the tih melanin biosynthesis gene (Nowrousian et al. 2012), PCR fragments covering the nox1 gene were obtained from nine fertile and nine sterile ascospore progeny. Sequencing of PCR fragments confirmed the nox1 base pair substitution in all strains with the sterile pro32 phenotype (Figure 1B). NOX1 function in fruiting body development was further verified by transformation of pro32 with a wild-type nox1 gene including 5′ and 3′ flanking regions. The resulting transformants showed a restoration of fertility, as indicated by the formation of mature fruiting bodies (Figure 1A).
Figure 1.
Genome sequencing of pro32 mutants reveals a mutation in the nox1 gene. (A) Sexual phenotypes of indicated strains. Insets show a detailed view of either wild-type black perithecia or mutant light brown protoperithecia. Bar, 100 µm. (B) Sequence comparison of the nox1 gene from wild type and 18 ascospore isolates from a cross between pro32 and fus strains. Sterile strains (underlined and blue) show a G to A transition, which changes nox1 W222 codon to a TGA stop codon (boxed).
Table 4. Summary of small sequence variants detected in the pro32 genome when compared to the reference genome.
| Genotype | Sequence sample | No. of small variants with coverage >40% | No. of mutations with 100% penetrance | Location of putative mutations |
|---|---|---|---|---|
| wt | wt_3 | 146 | — | — |
| pro32 | pro32/fus | 142 | 3 | G810A in SMAC_05007 results in stop codon at W222 |
| C926T in SMAC_05015 does not result in change of amino acid sequence | ||||
| C2620T in SMAC_05015 results in V310I |
Mutations caused by small sequence variants were identified by screening the sequence data for SNPs and indels (insertions/deletions) of <4 bases with a coverage of at least 40% of the average coverage for that sample. For these putative mutations, it was subsequently checked whether they had 100% penetrance, i.e., all the reads in the strain had the SNP/indel and none of the reads in the sequenced wild-type sample carried this specific mutation.
Deletion mutants lacking nox1 or nor1 show developmental defects
To further investigate NOX1 function in S. macrospora, we generated deletion mutants of nox1 as well as nor1 (SMAC_02124), which encodes the NOX regulator NOR1. These genes show 89 and 94% similarity to the corresponding genes from Neurospora crassa (accession nos. XP_964104 and XM_958018, respectively). The nox1 and nor1 genes were replaced by the hygromycin B resistance cassette through homologous recombination at the 5′ and 3′ regions flanking the target genes in a Δku70 host (Figure S2). The transformants were crossed with fus, a mutant generating red ascospores, to obtain homokaryotic strains carrying the nox1 or nor1 deletions without the ku70 deletion. Correct replacement of nox1 or nor1 with the hph cassette was verified by Southern hybridization using hph-, nox1-, or nor1-specific probes (Figure S3).
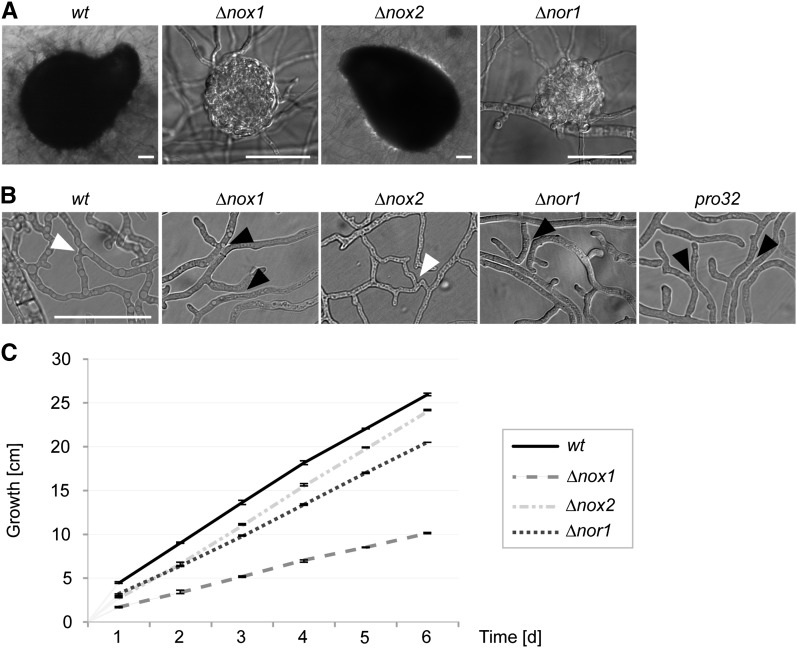
∆nox1 and ∆nor1 mutants showed distinctive similar phenotypes when compared to the wild-type strain. Both mutants have a sterile phenotype, being able to generate protoperithecia, but no perithecia (Figure 2A). As shown in Figure 2B, microscopic investigations revealed that both strains, as well as the pro32 mutant, are defective in vegetative cell fusion. In addition, ∆nox1 and ∆nor1 mutants were characterized by a significant reduction in hyphal growth by ∼61 and 21%, respectively (Figure 2C). With these phenotypes, pro32, ∆nox1, and ∆nor1 resemble other pro mutants found in a screen to detect strains having a developmental block after protoperithecia formation (Kück et al. 2009). To complement ∆nox1 and ∆nor1 mutants, we transformed them with full-length copies of nox1 or nor1 genes. In all cases, the reduced growth and sterility phenotypes were rescued in the transformants. However, hyphal fusion was only observed in the ∆nor1::nor1 and pro32::nox1 strains, but not in ∆nox1::nox1 (Figure S4). Further, the ectopic integration of nox1 in the corresponding deletion mutant leads only to a reduced number of perithecia, suggesting dose-dependent effects in the ∆nox1::nox1 complemented strain. Nevertheless, complementation analysis of ∆nor1 and pro32 prove that functional nox1 and nor1 genes are required for vegetative cell fusion, hyphal growth, and proper fruiting body formation.
Figure 2.
Phenotype of nox and nor1 deletion mutants. (A) Strains ∆nox1 and ∆nor1 are unable to develop black perithecia, and thus produce only slightly pigmented protoperithecia. Mutant ∆nox2 shows wild-type-like sexual development. Pictures were taken after 7 days of growth on BMM medium and incubation at 27° in constant light. Bar, 50 µm. (B) ∆nox1, pro32, and ∆nor1, but not ∆nox2, are affected in hyphal fusion. Strains were grown for 2 days on minimal medium on a cellophane layer in 27° in constant light. The assay was performed at least three times for every mutant. Hyphal fusion is indicated by white arrowheads; the lack of hyphal fusion between hyphae in close contact is marked by black arrowheads. Bar, 50 µm. (C) ∆nox1 and ∆nor1, but not ∆nox2 mutants, show decreased linear growth. Growth was followed in race tubes for 6 days in three replicates.
For P. anserina, it has been shown that the fertility defect of a PanoxA deletion mutant is rescued by serial transfers of the fungus to nutrient-rich medium (Malagnac et al. 2004). We observed a similar rescue of the S. macrospora nox1 deletion mutant (Figure S5). After germination of ∆nox1 ascospores from the rescued strain, we again obtained sterile strains that showed only protoperithecia formation. To determine whether this rescue phenotype is a general feature of sterile pro mutants, we performed serial media shifts using ∆nor1 and the unrelated ∆pro40 mutant (Figure S5). In contrast to ∆nox1, these mutants remained sterile even after several transfers.
Hyphal fusion mutants show elevated levels of ROS
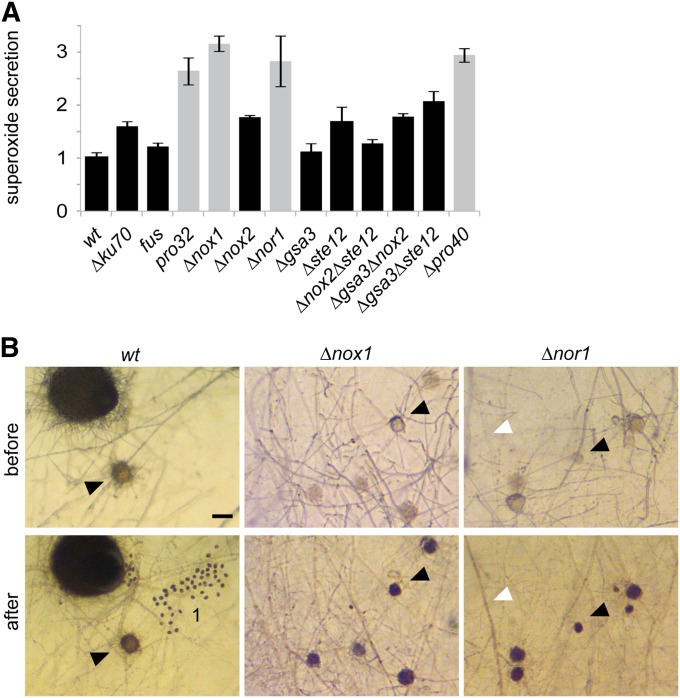
NADPH oxidases are known to produce ROS in a highly spatiotemporally regulated fashion (Aguirre and Lambeth 2010). Using the NBT assay for the detection of superoxide, we quantified the level of ROS in pro32, ∆nox1, and ∆nor1, as well as in other mutant strains affected in perithecia development and/or defective in hyphal fusion. Compared to wild type, sterile and hyphal fusion defective pro32, ∆nox1, and ∆nor1 showed enhanced ROS levels in vegetative hyphae (Figure 3A) and protoperithecia (Figure 3B). This enhancement of ROS is also shared by the hyphal fusion mutant ∆pro40 (Engh et al. 2007). In contrast, all investigated mutants with wild-type-like cell fusion showed wild-type-like levels of ROS. As a consequence of the hyphal fusion defect in ∆nox1 and other developmental mutants, we propose conditional nutrient starvation, and thus sterility in these strains.
Figure 3.
Mutants affected in hyphal fusion show increased levels of NBT reduction. (A) Quantitative measurement (fold change) of NBT precipitates after 30 min of incubation in at least three replicates. Gray bars indicate hyphal fusion-deficient strains. Hyphal fusion defect correlates with sterility, except for sterile mutant ∆gsa3∆ste12, which can undergo normal hyphal fusion. Normalization was done in reference to wild type. (B) Detailed view of sexual (black arrowheads) and vegetative (white arrowheads) structures stained by NBT. Bar, 50 µm, 1 = ascospores.
In protoperithecia, NOX1 regulates transcription of genes for hyphal fusion and cytoskeleton remodeling
To obtain information about differentially regulated genes that are dependent on NOX1 activity, we performed RNA-seq analysis using RNA samples obtained from wild-type and ∆nox1 mycelia and protoperithecia (Table S2). Mycelia were isolated from surface liquid cultures, while protoperithecia samples were obtained using a recently developed laser microdissection technique that allows protoperithecia isolation with minimal mycelial contamination (Teichert et al. 2012).
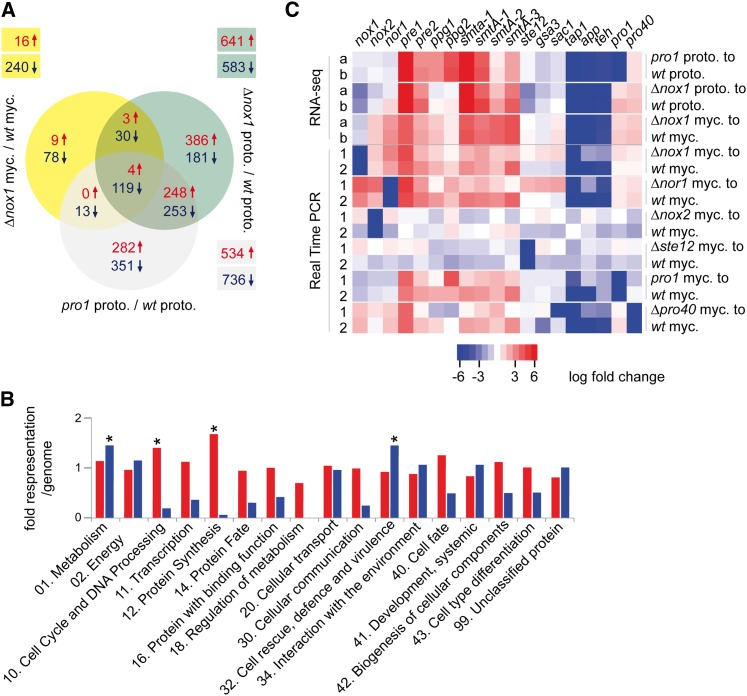
To allow conclusions about gene expression in protoperithecia of different developmental mutants, we included RNA-seq data obtained in a previous study for protoperithecia of the wild-type and the sterile mutant pro1 (File S1) (Teichert et al. 2012). pro1 encodes C6 zinc finger transcription factor PRO1 (Masloff et al. 1999) and shares the sterile phenotype with ∆nox1 strains. Comparison of genome-wide expression patterns between the different samples (total mycelia from wild type and ∆nox1 as well as protoperithecia from wild type, ∆nox1, and pro1) showed that the protoperithecial samples cluster apart from the mycelial samples, and that ∆nox1 protoperithecia are more similar to pro1 protoperithecia than to wild-type protoperithecia (Figure S6A). This reinforces a previous finding that expression patterns of protoperithecia are distinct from those of nonreproductive mycelia (Teichert et al. 2012). Furthermore, the fact that pro1 and ∆nox1 protoperithecia cluster together indicates that there is a common mutant-specific expression pattern in young fruiting bodies blocked at a similar developmental stage. These findings were confirmed by a comparison of differentially expressed genes in protoperithecia and mycelial samples (Figure 4A) and an analysis of the 500 most strongly expressed genes in each of the samples (“top500” analysis, Figure S6B). A total of 501 genes are differentially expressed in both, ∆nox1 and pro1 protoperithecia, compared to wild-type protoperithecia, whereas only 33 genes are differentially expressed specifically in the comparison of ∆nox1 and wild-type protoperithecia as well as ∆nox1 and wild-type mycelia (Figure 4A). The top500 analysis showed that intersections between wild-type and ∆nox1 mycelia and intersections between the protoperithecial samples contained more genes than intersections between protoperithecial and mycelial samples (Figure S6B).
Figure 4.
Results of RNA-seq-based expression analysis in developmental mutants. (A) Consensus analysis of differentially regulated genes identified by DESeq and classical statistical methods. Differentially regulated genes in relation to the corresponding wild-type tissue are indicated in yellow (∆nox1 mycelia), green (∆nox1 protoperithecia), and gray (pro1 protoperithecia). The numbers of up- (↑, red) and down- (↓, blue) regulated genes are indicated. (B) Illustration of functional categories (FunCat) (Ruepp et al. 2004) of up- (red) and down- (blue) regulated genes in protoperithecia from ∆nox1 mutant compared to wild-type protoperithecia. For each FunCat category, the fold representation compared to its representation among all predicted proteins is given. Asterisks indicate statistically overrepresented functional groups (P-value ≤ 0.05). (C) Heatmap of regulated genes in RNA-seq and qRT-PCR experiments using RNA from protoperithecia (proto.) and mycelium (myc.). Different statistical analyses are indicated by “a” (DESeq) and “b” (classical). Numbers (1 and 2) indicate biological replicates. Up- (red) and down- (blue) regulated genes are indicated.
To identify the possible functions of genes differentially regulated in ∆nox1 compared to wild-type protoperithecia, we performed functional categories (FunCat, P < 0.05) and BLAST analyses, using the N. crassa orthologs for FunCat analysis. Figure 4B illustrates functional categories (FunCat) for all N. crassa orthologs, in total 554 up- (red) and 432 down-regulated (blue) genes. Significantly overrepresented categories correspond to metabolism and cell rescue (down-regulated compared to wild type), as well as cell cycle and protein synthesis (up-regulated compared to wild type).
Furthermore, BLAST analysis of differentially regulated genes in ∆nox1 protoperithecia revealed that several of these genes encode proteins important for cytoskeleton remodeling, like CDC42, profilin, cofilin, and coronin-1 (Table S3). Coronin-1 has been shown to have a major role in actin organization and dynamics in N. crassa (Echauri-Espinosa et al. 2012). In a second group, genes important for hyphal fusion are differentially regulated in ∆nox1 protoperithecia compared to wild-type protoperithecia. Among these is the ham-10 gene, which is essential for hyphal fusion and perithecia formation in N. crassa (Fu et al. 2011) (Table S3, Table S4). The third group contains genes encoding subunits of NADH:oxidoreductases of the respiratory chain and of mitochondrial ATPase. Consistent with this, mutants unable to assemble the respiratory chain complex I are female sterile in N. crassa (Duarte and Videira 2000) (Table S3, Table S4). The enrichment of these three groups together with the FunCat analysis suggests that NOX1 has regulatory functions, e.g., in cytoskeleton remodeling, hyphal fusion, metabolism, and mitochondrial respiration.
Extensive genetic analysis has identified a large number of developmental genes involved in fruiting body formation (Pöggeler et al. 2006; Kück et al. 2009; Engh et al. 2010). Results from the RNA-seq-based differential expression analysis of these genes are given in Table 5. In Figure 4C results from qRT-PCR analysis are shown to verify some of the results of the RNA-seq analysis, and to further compare gene expression patterns in mycelia and protoperithecia from ∆nox1 and other developmental mutants. RNA-seq data show that both pheromone receptor genes pre1 and pre2 and the two mating type genes SmtA-1 and SmtA-3 are up-regulated in ∆nox1 mycelia and protoperithecia. Similarly, genes for melanin biosynthesis (pks, teh, sdh, and tih) and two genes associated with fruiting body maturation, app and tap1, are down-regulated in ∆nox1 mycelia and protoperithecia, similar to the situation in other pro mutants (Nowrousian et al. 2005, 2007; Teichert et al. 2012), whereas they only show minor expression changes in fertile mutants ∆nox2 and ∆ste12 compared to wild type (Figure 4C). These findings are in line with the proposed hypothesis of a common pro mutant-specific expression pattern.
Table 5. Log2 ratios of relative expression of known developmental genes in Δnox1 mycelium and protoperithecia.
| S. macrospora locus tag | Gene | Log2 ratios of gene expression in mycelium of Δnox1 vs. wt | Log2 ratios of gene expression in protoperithecia of Δnox1 vs. wt | ||
|---|---|---|---|---|---|
| DESeq analysis | Classical analysis | DESeq analysis | Classical analysis | ||
| Transcription factor genes | |||||
| SMAC_00338 | pro1 | 1.011 | 1.295 | 0.955 | 1.787 |
| SMAC_03223 | pro44 | −0.358 | −0.140 | −2.059 | −1.380 |
| SMAC_05219 | mcm1 | −0.041 | 0.081 | −1.296 | −0.565 |
| SMAC_06479 | ste12 | 0.050 | 0.330 | −2.836 | −2.196 |
| Pheromone pathway genes | |||||
| SMAC_02283 | pre1 | 3.907 | 4.041 | 7.720 | 7.381 |
| SMAC_08994 | pre2 | 2.104 | 2.231 | 2.867 | 3.631 |
| SMAC_05970 | ppg1 | 1.434 | 1.446 | −0.365 | 0.320 |
| SMAC_12697 | ppg2 | 0.496 | 0.540 | −0.166 | 0.583 |
| Mating type genes | |||||
| SMAC_05404 | Smta-1 | 4.052 | 4.188 | a | a |
| SMAC_05401 | SmtA-1 | 3.331 | 3.366 | 3.750 | 4.562 |
| SMAC_05402 | SmtA-2 | 3.836 | 3.827 | 1.440 | 2.207 |
| SMAC_05403 | SmtA-3 | 4.295 | 4.303 | 3.162 | 3.862 |
| Nox genes | |||||
| SMAC_05007 | nox1 | −1.109b | −0.913b | −3.076b | −2.404b |
| SMAC_08741 | nox2 | 1.210 | 1.430 | −1.006 | −0.299 |
| SMAC_02124 | nor1 | 2.532 | 2.718 | −0.376 | 1.025 |
| MAP kinase cell integrity pathway genes | |||||
| SMAC_03673 | mik1 | 0.479 | 0.717 | 1.284 | 1.939 |
| SMAC_02183 | mek1 | 0.813 | 0.967 | 0.401 | 1.033 |
| SMAC_05504 | mak1 | 0.546 | 0.743 | −1.545 | −0.870 |
| STRIPAK complex genes | |||||
| SMAC_08794 | pro11 | 0.255 | 0.474 | 1.400 | 2.050 |
| SMAC_02580 | pro22 | 0.230 | 0.417 | −0.985 | −0.273 |
| SMAC_00877 | mob3 | −0.245 | −0.084 | 0.311 | 0.988 |
| SMAC_01224 | pro45 | 0.372 | 0.601 | −0.210 | 0.438 |
| SMAC_01919 | pp2AA | 0.876 | 0.95 | 1.152 | 1.841 |
| SMAC_04678 | pp2Ac1 | 1.234 | 1.305 | 1.173 | 1.827 |
| Melanin biosynthesis genes | |||||
| SMAC_03130 | pks | −4.541 | −4.251 | −6.916 | −6.057 |
| SMAC_05880 | teh | −4.829 | −4.679 | −6.227 | −5.523 |
| SMAC_02101 | sdh | −3.656 | −3.603 | −4.134 | −2.870 |
| SMAC_05650 | tih | −5.819 | −5.601 | −7.691 | −6.990 |
| G-protein alpha subunit genes | |||||
| SMAC_05328 | gsa1 | −0.094 | 0.120 | −0.360 | 0.311 |
| SMAC_06605 | gsa2 | 0.235 | 0.386 | −0.422 | 0.220 |
| SMAC_07195 | gsa3 | −0.453 | −0.311 | −1.325 | −0.682 |
| Genes associated with ascospore germination | |||||
| SMAC_01638 | sac1 | 0.714 | 0.825 | −0.619 | 0.037 |
| SMAC_09071 | rac1 | 0.235 | 0.393 | 0.875 | 1.729 |
| Genes associated with fruiting body maturation | |||||
| SMAC_06095 | app | −8.505 | −8.274 | −7.069 | −6.419 |
| SMAC_03372 | tap1 | −8.984 | −8.722 | −9.364 | −8.716 |
| SMAC_00522 | fbm1 | 1.470 | 1.810 | 0.181 | 1.150 |
| Other essential genes for fruiting body development | |||||
| SMAC_07802 | pro4 | 2.579 | 2.901 | 0.536 | 1.347 |
| SMAC_04848 | pro41 | 2.354 | 2.511 | 0.406 | 1.110 |
| SMAC_06775 | acl1 | −1.353 | −1.174 | −0.734 | 0.100 |
| SMAC_08608 | asf1 | 0.879 | 0.988 | 1.141 | 1.809 |
| SMAC_06539 | atg7 | −0.355 | −0.233 | −0.643 | 0.041 |
| SMAC_04815 | pro40 | 1.225 | 1.468 | 1.225 | 1.468 |
No reads mapped in wild-type protoperithecia. Therefore no ratios can be calculated.
Counted reads map to 5′ and 3′ UTRs of nox1, which were not deleted in the ∆nox1 mutant.
NOX2 is required for the germination of melanized ascospores
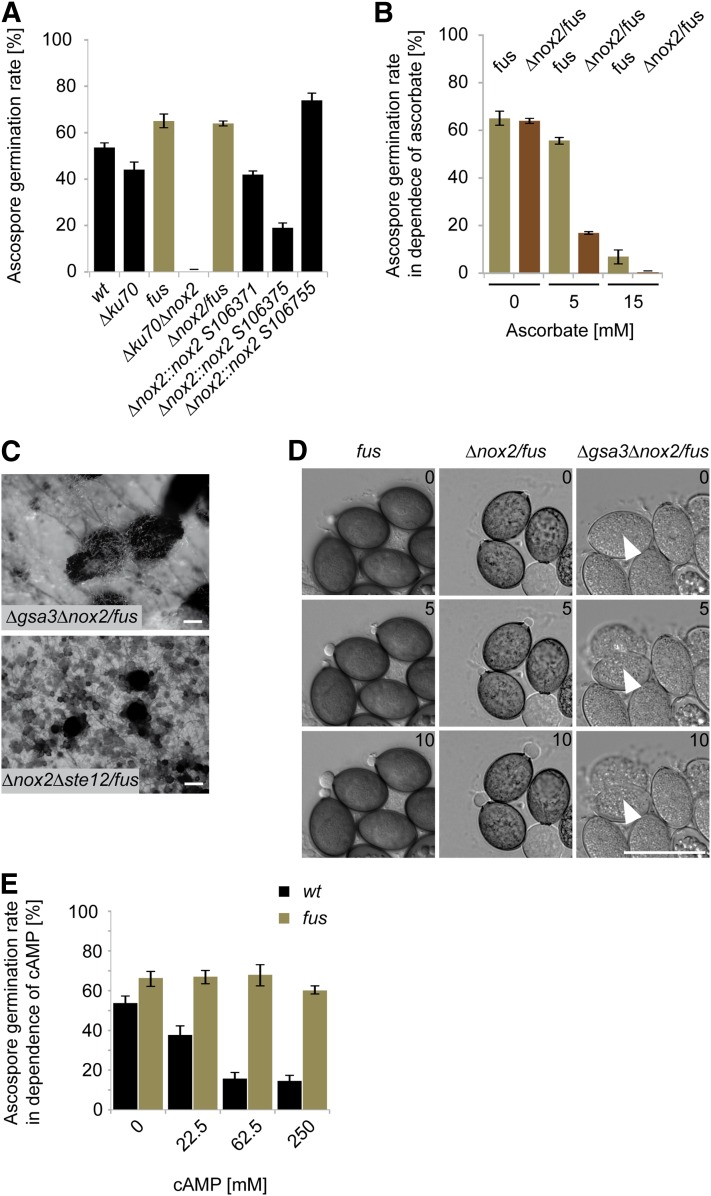
Like other ascomycetes, the genome of S. macrospora encodes a second NADPH oxidase, NOX2. The corresponding nox2 gene (SMAC_08741) shows 91% sequence similarity to its N. crassa homolog (accession no. XP_001728356). A homokaryotic ∆nox2/fus strain was generated with the strategy described in Materials and Methods. As can be seen in Figure 2, this strain has no defect in vegetative growth, cell fusion, or sexual development. However, when we performed random ascospore analysis from a cross of the primary transformant ∆ku70∆nox2 with the melanin-deficient fus mutant, we observed in a total of 390 analyzed viable ascospores a strict cosegregation of the hygromycin-B-resistant phenotype, corresponding to the nox2 deletion, with fus. To investigate ascospore germination further, we tested germination of diverse fertile strains generating either black or red spores. For sound statistical analysis, we investigated a minimum of 400 spores in each of three replicate experiments and tested their germination capacity on solid BMM-Ac medium. We analyzed wild type, fus, and the ∆ku70 strain, the latter of which was used as the recipient strain for generating the ∆nox2 mutant. As can be seen in Figure 5A, ascospores generally showed a germination rate between 40 and 70%. Similar results were obtained with the red-spored ∆nox2/fus strain. However, we were unable to isolate any viable (germinating) ascospores from black-spored ∆ku70/∆nox2 mutants, indicating that germination in ∆nox2 is suppressed by the presence of melanin.
Figure 5.
Phenotypes of ascospore germination-defective mutants. (A) Quantification of germination of ascospores discharged from selfed perithecia from wild type, different mutants, and ∆nox2::nox2 strains. A minimum of 400 ascospores per strain were tested in each of three biological replicates. Black bars represent strains with full melanized ascospores; brown bars represent strains with a block in melanin biosynthesis. (B) Ascospore germination under different ascorbate concentrations. A minimum of 400 ascospores from selfed perithecia were tested for each strain and ascorbate concentration. (C) Perithecia formation of ∆gsa3∆nox2/fus and ∆nox2∆ste12/fus mutants after 7 and 14 days incubation, respectively. Bar, 100 µm. (D) Time series of ascospore germination for the indicated mutants. The elapsed time in minutes is indicated. White arrowheads mark ascospores that burst instead of showing a germinaton vesicle. Bar, 50 µm. (E) Wild-type and fus mutant ascospore germination rates in the presence of db-cAMP. Discharged ascospores were collected and incubated on medium containing the indicated db-cAMP concentrations. A minimum of 400 ascospores per strain were tested for each of three biological replicates. Black bars represent strains with full melanized ascospores; brown bars represent strains with a block in melanin biosynthesis.
To verify the strict dependence of the germination defect on melanization, we generated a black-spored ∆nox2::nox2 complemented strain. Primary transformant ∆ku70∆nox2 was not appropriate for complementation experiments since it carries selection markers nat1 (noursethricin resistance) and hph (hygromycin B resistance) replacing ku70 and nox2, respectively. Therefore, we transformed hygromycin-resistant ∆nox2/fus with a wild-type copy of nox2. The red-spored ∆nox2/fus::nox2 was then crossed to wild type, and black ascospores were isolated carrying the ∆nox2 deletion and the ectopically integrated nox2 gene. Three randomly selected strains germinated at a frequency similar to wild type and ∆ku70 (Figure 5A), confirming that germination of black ascospores is strictly dependent on NOX2 function.
To assess whether the germination defect is restricted to ∆nox2, we tested further the ascospore germination phenotype of ∆nox1 and ∆nor1, both having a sterile phenotype. ∆nox1/fus and ∆nor1/fus double mutants were crossed to the wild type, and the germination assays of black and red spores were done as described in Materials and Methods. As can be seen from Table 6, ∆nor1, but not ∆nox1 ascospores, displayed the same melanin-dependent germination defect as ∆nox2 ascospores. This result indicates that NOR1 is required to regulate both NOX1 and NOX2.
Table 6. Frequency of hygromycin B resistance in black- and red-spored progeny from indicated crosses.
| Cross | 100 germinated black ascospores | 100 germinated red ascospores | ||
|---|---|---|---|---|
| HygS | HygR | HygS | HygR | |
| ∆nox1/fus × wt | 65 | 35 | 49 | 51 |
| ∆nox2/fus × wt | 100 | 0 | 43 | 57 |
| ∆nor1/fus × wt | 100 | 0 | 48 | 52 |
| ∆gsa3/fus × wt | 92 | 8 | 81 | 19 |
| ∆sac1/fus × wt | 80 | 20 | 81 | 19 |
| ∆ste12/fus × wt | 100 | 0 | 47 | 53 |
| ∆pro40/fus × wt | 45 | 55 | 43 | 57 |
From each cross, 100 black and 100 red colony forming ascospores were tested for hygromycin B resistance, indicating the deletion of the corresponding gene.
These data suggest that ROS is necessary for spore germination. It might be hypothesized that in the wild type, both NOX2 as well as other pathways contribute to ROS production allowing spore germination. The ∆nox2 mutant would then retain residual amounts of ROS generated independently of NOX2; however, the residual amount of ROS in ∆nox2 ascospores might be scavenged by melanin in black ascospores. To further test this hypothesis, we performed germination tests with different ascorbate concentrations in the germination media. Ascorbate is an antioxidant that scavenges ROS. As can be seen in Figure 5B, germination of ascospores on ascorbate-containing media is drastically reduced in strains lacking nox2 in the fus mutant background but less pronounced in the fus reference strain. Thus, the antioxidant ascorbate mimics the effect of melanin in ∆nox2/fus spores.
NOX2, NOR1, and transcription factor STE12 act in a genetic pathway determining ascospore germination
The melanin-dependent ascospore germination defect of ∆nox2 and ∆nor1 prompted us to look for other genes involved in this process. Previously, S. macrospora deletion mutants lacking the gsa3 (alpha subunit 3 of the heterotrimeric G-protein), sac1 (adenylate cyclase), and ste12 genes were shown to have an ascospore germination defect (Nolting and Pöggeler 2006; Kamerewerd et al. 2008). We performed further crossing experiments to test whether these genes act with nox2 and nor1 in the same genetic pathway. ∆gsa3/fus, ∆sac1/fus, and ∆ste12/fus were crossed to a wild-type strain and then 100 black and 100 red germinating ascospores from these crosses were tested for hygromycin B resistance. As can be seen in Table 6, the ste12 deletion, but not the gsa3 or sac1 deletions, strictly cosegregated with the fus mutation. Thus, the germination defect of ∆ste12 resembles the ∆nox2 and ∆nor1 ascospore germination defect. To further verify this result, we generated triple mutant ∆nox2∆ste12/fus, having a fertile phenotype (Figure 5C). From a cross of the triple mutant with wild type, we isolated a total of 146 ascospores. Of these, 68 were red colored and hygromycin B resistant, thus indicating the deletion of nox2 and/or ste12. In contrast, 78 black and red ascospores showed wild-type-like sensitivity to hygromycin B. These data thus verify the strict cosegregation of the nox2 or ste12 deletion with the fus mutation, indicating that NOX2 and STE12 act in the same pathway that controls ascospore germination.
In contrast to ∆nox2, ∆nor1, and ∆ste12, sac1 and gsa3 deletion mutants have a germination defect that is independent of melanin in ascospores (Table 6). To determine whether the gsa3 and the nox2 pathways overlap, we generated triple mutant ∆gsa3∆nox2/fus. Like the single deletion strains, this strain is fertile (Figure 5C); however, the ascospores are unable to generate germination vesicles in contrast to the control strains fus and ∆nox2/fus. Instead, they burst after ∼5 hr on germination medium (Figure 5D, File S2, File S3, File S4). None of the 4000 ascospores analyzed showed germination. Thus, the germination-related phenotypes of ∆nox2 and ∆gsa3 (Kamerewerd et al. 2008) are exacerbated in the ∆gsa3∆nox2/fus mutant. This result clearly shows that NOX2 and GSA3 have a different impact on ascospore germination. Therefore, we propose that ascospore germination is regulated by two pathways, namely the NOX2-NOR1-STE12 and the GSA3-SAC1 pathways (Figure 6).
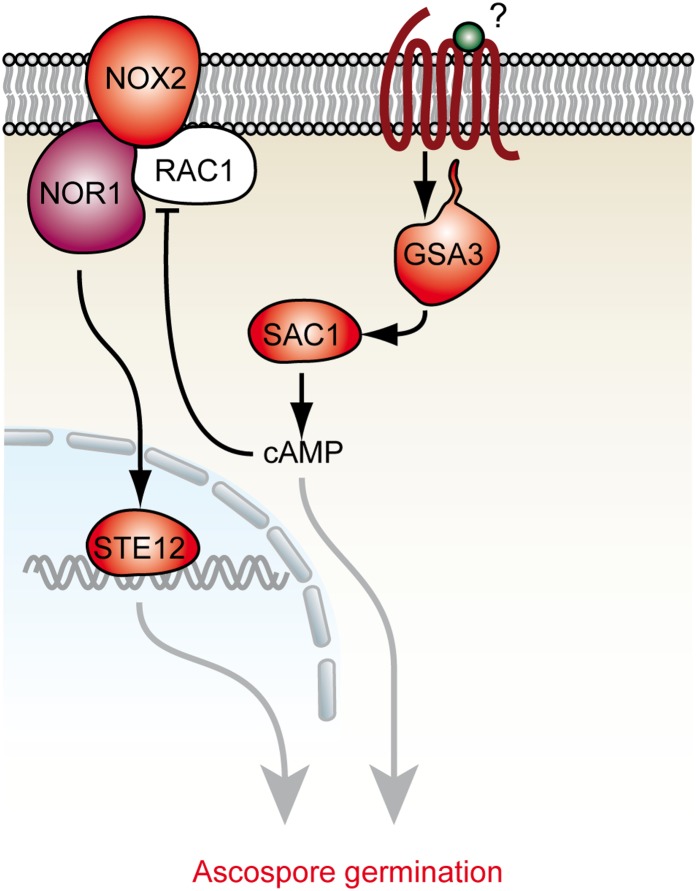
Figure 6.
Interplay of two pathways regulating ascospore germination in S. macrospora. During ascospore germination, two different signaling pathways, NOX2-NOR1-STE12 and GSA3-SAC1, exist and may be interconnected by cAMP. Work from other studies indicates that the GTPase RAC1 is inhibited by cAMP and thus negatively regulates the NOX2-NOR1 complex (Diebold et al. 2009).
cAMP inhibits germination of melanized ascospores
Recent reports indicate that cAMP, generated by an adenylate cyclase homologous to SAC1, inhibits the activity of a NOX2 homolog in human cells (Diebold et al. 2009; Li et al. 2012). At the molecular level, ROS production by the mammalian NOX2 complex is proposed to be reduced when the NOX2 activator RAC1 is directly inhibited by high cAMP concentrations. In analogy to this model, we hypothesized that increased cAMP levels could inhibit NOX2 activity in S. macrospora and reduce ascospore germination. To test this hypothesis, discharged ascospores from wild-type and fus strains were germinated on BMM-Ac medium supplemented with different concentrations of cell-permeable db-cAMP (Figure 5E). A concentration of 22.5 mM db-cAMP resulted in a reduction of ascospore germination of ∼30% in the wild type, but not in the red-spored fus mutant, which does not require NOX2 activity for germination. At higher db-cAMP concentrations of 62.5 mM and 250 mM, ascospore germination frequency of wild type, but not fus, dropped down by 70%. Thus, cAMP is able to mimic the ascospore germination defect of ∆nox2, ∆nor1, and ∆ste12 in a concentration-dependent manner. Based on these results, we hypothesize that cAMP generated by the GSA3-SAC1 pathway (Kamerewerd et al. 2008) exerts a negative regulatory role on NOX2 activity, which results in decreased production of ROS and failure to establish an ascospore germination vesicle, thus connecting the nox2/ste12 and gsa3/sac1 pathways (Figure 6).
Discussion
Fruiting body development is regulated by NOX1 and NOR1
The deletion of nox1 and nor1 genes led to a sterile phenotype that has been found previously in a set of S. macrospora developmental pro mutants, which can form protoperithecia, but are hampered in developing mature fruiting bodies (Kück et al. 2009). The deletion of nox1 or nor1 in other filamentous fungi leads to similar effects on sexual development. In Aspergillus nidulans, noxA deletion blocks the differentiation of cleistothecia (Lara-Ortiz et al. 2003), and similarly, P. anserina and N. crassa nox1 mutants are female sterile (Malagnac et al. 2004; Cano-Dominguez et al. 2008). Notably, in P. anserina (Malagnac et al. 2004) and S. macrospora, the sterile phenotype can be rescued by serially shifting the mutant to rich medium, indicating that sterility is related to the availability of nutritional factors, or ROS scavenging and signaling molecules. Alternatively, shifts to rich medium might induce the activity of NOX2, thereby bypassing the nox1 deletion.
A novel function of NOX1 and NOR1 was recently detected in N. crassa and Botrytis cinerea (Read et al. 2012; Roca et al. 2012), where a deletion of the corresponding genes abolishes fusion of conidial anastomosis tubes (CATs). S. macrospora does not form any conidia; however, hyphal fusion can be observed within the hyphal network (Rech et al. 2007). Our investigation showed that S. macrospora nox1 and nor1 mutants have a defect in hyphal fusion, as was also shown for corresponding mutants from E. festucae (Kayano et al. 2013). Both phenomena, sterility and hyphal fusion defect, are correlated but not strictly linked with each other. For example, the N. crassa ham-4 mutant is fertile but shows a hyphal fusion defect (Simonin et al. 2010) and the sterile Smatg8 mutant from S. macrospora still shows hyphal fusion (Voigt and Pöggeler 2013).
Since NOX enzymes are known to produce superoxide by the reduction of oxygen (Leto et al. 2009), we determined the levels of ROS using the NBT assay. Interestingly, we detected higher superoxide levels in all mutants tested that were defective in hyphal fusion, including ∆pro40. The superoxide level has been investigated in nox deletion mutants of several ascomycetes and diverging results have emerged. For example, while E. festucae nox1 mutants show decreased superoxide levels (Tanaka et al. 2008), increased levels of superoxide are detected in corresponding mutants of M. oryzae, P. anserina, and B. cinerea (Malagnac et al. 2004; Egan et al. 2007; Siegmund et al. 2013). The latter results are rather unexpected since NOX enzymes are known to generate superoxide. Nevertheless, our results highlight that not only a deletion of the nox1 gene leads to elevated superoxide levels, but also a deletion of pro40. Thus, our data support a general correlation between a hyphal fusion defect and enhanced superoxide levels generated by a yet unknown source in mutant strains, as was proposed previously (Malagnac et al. 2004; Egan et al. 2007; Siegmund et al. 2013).
Transcriptional profiling reveals differentially regulated genes in a nox1 mutant
Recently, transcriptional profiling of mycelia from a P. anserina Panox1 mutant was performed by microarray analysis and showed the deregulation of a large set of genes involved in carbohydrate degradation and secondary metabolism (Bidard et al. 2012). However, the authors analyzed total mycelia after 72 hr of growth, therefore expression patterns specific to sexual structures could not be elucidated in these experiments. Thus, our RNA-seq analysis of protoperithecia as well as mycelia significantly extends the current knowledge about NOX1-dependent gene expression by including sexual structures from a specific developmental stage. The increased levels of ROS in the ∆nox1 mutant might also be responsible for some of the gene expression changes observed.
One finding was that a large set of genes involved in cytoskeleton remodeling and hyphal fusion is differentially regulated in the ∆nox1 mutant. This finding is consistent with recent studies in Magnaporthe oryzae, where the NOX1 complex was shown to be important for the maintenance of the cortical F-actin network during penetration of the host plant (Ryder et al. 2013). These authors showed that ∆nox1 still initated penetration peg formation but was unable to proliferate in the infected plant tissue. Furthermore, Yno1p, the NOX enzyme from yeast, has a regulatory role in actin remodeling (Rinnerthaler et al. 2012). Interestingly, F-actin remodeling is also crucial for CAT fusion in N. crassa (Roca et al. 2010). Further, our RNA-seq analysis revealed that among the differentially expressed genes in ∆nox1 protoperithecia, there are some involved in the establishment of hyphal polarity. For example, cdc42, a gene central for the establishment of polarity, is up-regulated in ∆nox1 protoperithecia compared to wild-type protoperithecia. The activity of CDC42 is regulated by the guanine nucleotide exchange factor (GEF) CDC24 and the scaffold protein BEM1, both of which are able to interact with the NOR1 homolog of E. festucae in a yeast-two-hybrid assay (Takemoto et al. 2011). In N. crassa, Bem1 is actively recruited around the forming fusion pore of germlings, and bem1 deletion leads to a drastic reduction of germling fusion, indicating a cross-talk between polarity establishment and hyphal fusion (Schürg et al. 2012). In N. crassa it was further shown that ∆cdc24 and ∆cdc42 are fusion defective (Read et al. 2012). Taken together, our results support a model in which the differential expression of a set of genes involved in cytoskeleton remodeling and hyphal fusion is responsible for the inability of ∆nox1 mutants to undergo cell fusion, normal polar growth, and perithecia development. The regulated genes are prime candidates for further analysis of NOX1-dependent developmental functions.
Besides NOX enzymes, there are other ROS-producing enzymes and processes within a cell, among them complex I of the respiratory chain that includes NADH:oxidoreductases (Murphy 2009). Our RNA-seq data indicate the up-regulation of subunits of the respiratory chain NADH:oxidoreductase of complex I and Cytochrome c in ∆nox1, whereas two ATPase subunits are down-regulated. Thus, we propose that the enhanced ROS levels observed in ∆nox1 may be due to de-regulation of the mitochondrial respiratory chain, perhaps as a consequence of starvation of hyphae and protoperithecia. This would be consistent with the finding that a serial transfer to fresh media restores fertility of the ∆nox1 mutant strain (Figure S5). A similar phenomenon has been described for multiple human cell lines (Scherz-Shouval and Elazar 2007; Chen and Gibson 2008). There, starvation leads to an up-regulation of mitochondrial ROS, which in turn directly activates an autophagy-inducing pathway (Li et al. 2013).
NOX2, NOR1, and STE12 act in a genetic pathway controlling ascospore germination
Our results show that NOX2 and NOR1 are required for the germination of sexual spores. Similar defects have been observed in corresponding P. anserina (Malagnac et al. 2004) and N. crassa (Cano-Dominguez et al. 2008) mutants. We found that NOX2 is required for germination of melanized ascospores only, since melanin-deficient ascospores from ∆nox2 are able to germinate. Lambou et al. (2008) suggested for P. anserina that the weakened cell wall of pigment-deficient ascospores and the absence of melanin either triggered the germination process or prevented its inhibition. However, the ROS scavenging capacity of melanin (Riley 1997) might also play a role in this process. We propose that ROS is necessary for spore germination and a residual amount in ∆nox2 ascospores is scavenged by melanin, thus preventing germination. On the other hand, such residual ROS would not be scavenged in pigment-deficient ascospores, allowing ascospores to germinate.
The ascospore germination assays provided the novel finding that NOX2, NOR1, and the transcription factor STE12 might act in the same genetic pathway to regulate ascospore germination, while SAC1 and GSA3 act in a parallel but interconnected pathway. Indeed, the inhibitory effect of cAMP on germination of melanized ascospores suggests a link between both pathways. In human cells, an increased level of cAMP inhibits GTPase RAC1, which itself is required for NOX2 activity (Diebold et al. 2009). We propose that cAMP generated by adenylate cyclase SAC1 inhibits NOX2 activity via the RAC1 homolog (SMAC_09071) (Figure 6). The fact that gsa3 and sac1 deletion mutants are fertile suggests that cAMP might not play an important role in NOX1 regulation during growth and perithecia development. In summary, our analyses of ascospore germination in various mutants show that two genetic pathways, NOX2-NOR1-STE12 and GSA3-SAC1, are involved in the regulatory network governing this process.
Supplementary Material
Acknowledgments
We are grateful to Susanne Schlewinski, Swenja Ellßel, Ingeborg Godehardt, and Regina Ricke for their excellent technical assistance. We also thank Kathrin Grieß for help with some of the experiments. This work was funded by the Deutsche Forschungsgemeinschaft (Bonn-Bad Godesberg) through Forschergruppe FOR 1334, DFG Grant NO407/4-1, and DFG-CONACYT Germany–México Collaboration Grant 75306.
Footnotes
Communicating editor: J. Heitman
Literature Cited
- Aguirre J., Rios-Momberg M., Hewitt D., Hansberg W., 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13: 111–118. [DOI] [PubMed] [Google Scholar]
- Aguirre J., Lambeth J. D., 2010. Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free Radic. Biol. Med. 49: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W., 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard F., Coppin E., Silar P., 2012. The transcriptional response to the inactivation of the PaMpk1 and PaMpk2 MAP kinase pathways in Podospora anserina. Fungal Genet. Biol. 49: 643–652. [DOI] [PubMed] [Google Scholar]
- Bloemendal S., Bernhards Y., Bartho K., Dettmann A., Voigt O., et al. , 2012. A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84: 310–323. [DOI] [PubMed] [Google Scholar]
- Brun S., Malagnac F., Bidard F., Lalucque H., Silar P., 2009. Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Mol. Microbiol. 74: 480–496. [DOI] [PubMed] [Google Scholar]
- Cano-Dominguez N., Alvarez-Delfin K., Hansberg W., Aguirre J., 2008. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot. Cell 7: 1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Gibson S. B., 2008. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy 4: 246–248. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold I., Djordjevic T., Petry A., Hatzelmann A., Tenor H., et al. , 2009. Phosphodiesterase 2 mediates redox-sensitive endothelial cell proliferation and angiogenesis by thrombin via Rac1 and NADPH oxidase 2. Circ. Res. 104: 1169–1177. [DOI] [PubMed] [Google Scholar]
- Duarte M., Videira A., 2000. Respiratory chain complex I is essential for sexual development in Neurospora and binding of iron sulfur clusters are required for enzyme assembly. Genetics 156: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echauri-Espinosa R. O., Callejas-Negrete O. A., Roberson R. W., Bartnicki-Garcia S., Mourino-Perez R. R., 2012. Coronin is a component of the endocytic collar of hyphae of Neurospora crassa and is necessary for normal growth and morphogenesis. PLoS ONE 7: e38237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan M. J., Wang Z. Y., Jones M. A., Smirnoff N., Talbot N. J., 2007. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA 104: 11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh I., Würtz C., Witzel-Schlomp K., Zhang H. Y., Hoff B., et al. , 2007. The WW domain protein PRO40 is required for fungal fertility and associates with Woronin bodies. Eukaryot. Cell 6: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh I., Nowrousian M., Kück U., 2010. Sordaria macrospora, a model organism to study fungal cellular development. Eur. J. Cell Biol. 89: 864–872. [DOI] [PubMed] [Google Scholar]
- Esser K., 1982. Cryptogams-Cyanobacteria, Algae, Fungi, Lichens, Cambridge University Press, London. [Google Scholar]
- Esser K., Straub J., 1958. Genetic studies on Sordaria macrospora Auersw., compensation and induction in gene-dependent developmental defects. Z. Vererbungsl. 89: 729–746. [PubMed] [Google Scholar]
- Fu C., Iyer P., Herkal A., Abdullah J., Stout A., et al. , 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10: 1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J. M. C., 2007. Free Radicals in Biology and Medicine, Oxford University Press, Oxford. [Google Scholar]
- Heller J., Tudzynski P., 2011. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49: 369–390. [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A., 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerewerd J., Jansson M., Nowrousian M., Pöggeler S., Kück U., 2008. Three alpha-subunits of heterotrimeric G proteins and an adenylyl cyclase have distinct roles in fruiting body development in the homothallic fungus Sordaria macrospora. Genetics 180: 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T., Lambeth J. D., 2007. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol. Biol. 7: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayano Y., Tanaka A., Akano F., Scott B., Takemoto D., 2013. Differential roles of NADPH oxidases and associated regulators in polarized growth, conidiation and hyphal fusion in the symbiotic fungus Epichloë festucae. Fungal Genet. Biol. 56: 87–97. [DOI] [PubMed] [Google Scholar]
- Klix V., Nowrousian M., Ringelberg C., Loros J. J., Dunlap J. C., et al. , 2010. Functional characterization of MAT1–1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot. Cell 9: 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück U., Pöggeler S., Nowrousian M., Nolting N., Engh I., 2009. Sordaria macrospora, a model system for fungal development, pp. 17–39 in The Mycota XV: Physiology and Genetics, edited by Anke T., Weber D. Springer, Berlin. [Google Scholar]
- Lambeth J. D., 2004. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 4: 181–189. [DOI] [PubMed] [Google Scholar]
- Lambou K., Malagnac F., Barbisan C., Tharreau D., Lebrun M. H., et al. , 2008. The crucial role of the Pls1 tetraspanin during ascospore germination in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot. Cell 7: 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Ortiz T., Riveros-Rosas H., Aguirre J., 2003. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50: 1241–1255. [DOI] [PubMed] [Google Scholar]
- Leto T. L., Morand S., Hurt D., Ueyama T., 2009. Targeting and regulation of reactive oxygen species generation by Nox family NADPH oxidases. Antioxid. Redox Signal. 11: 2607–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durban R., 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Chen Y., Gibson S. B., 2013. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell. Signal. 25: 50–65. [DOI] [PubMed] [Google Scholar]
- Li N., Li B., Brun T., Deffert-Delbouille C., Mahiout Z., et al. , 2012. NADPH oxidase NOX2 defines a new antagonistic role for reactive oxygen species and cAMP/PKA in the regulation of insulin secretion. Diabetes 61: 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F., Lalucque H., Lepere G., Silar P., 2004. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 41: 982–997. [DOI] [PubMed] [Google Scholar]
- Masloff S., Pöggeler S., Kück U., 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P., 2009. How mitochondria produce reactive oxygen species. Biochem. J. 417: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolting N., Pöggeler S., 2006. A STE12 homologue of the homothallic ascomycete Sordaria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 62: 853–868. [DOI] [PubMed] [Google Scholar]
- Nowrousian M., Cebula P., 2005. The gene for a lectin-like protein is transcriptionally activated during sexual development, but is not essential for fruiting body formation in the filamentous fungus Sordaria macrospora. BMC Microbiol. 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Masloff S., Pöggeler S., Kück U., 1999. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol. Cell. Biol. 19: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M., Ringelberg C., Dunlap J. C., Loros J. J., Kück U., 2005. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genomics 273: 137–149. [DOI] [PubMed] [Google Scholar]
- Nowrousian M., Piotrowski M., Kück U., 2007. Multiple layers of temporal and spatial control regulate accumulation of the fruiting body-specific protein APP in Sordaria macrospora and Neurospora crassa. Fungal Genet. Biol. 44: 602–614. [DOI] [PubMed] [Google Scholar]
- Nowrousian, M., I. Teichert, S. Masloff, and U. Kück, 2012 Whole-genome sequencing of Sordaria macrospora mutants identifies developmental genes. G3 2: 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S., Kück U., 2006. Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378: 1–10. [DOI] [PubMed] [Google Scholar]
- Pöggeler S., Risch S., Kück U., Osiewacz H. D., 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147: 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S., Nowrousian M., Kück U., 2006. Fruiting body development in ascomycetes, pp. 325–355 in The Mycota I: Growth, Differentiation and Sexuality, edited by Kües U., Fischer R. Springer, Berlin. [Google Scholar]
- Read N. D., Goryachev A. B., Lichius A., 2012. The mechanisitc basic of self-fusion between conidial anastomosis tubes during fungal colony initiation. Fungal Biol. Rev. 26: 1–11. [Google Scholar]
- Rech C., Engh I., Kück U., 2007. Detection of hyphal fusion in filamentous fungi using differently fluorescence-labeled histones. Curr. Genet. 52: 259–266. [DOI] [PubMed] [Google Scholar]
- Riley P. A., 1997. Melanin. Int. J. Biochem. Cell Biol. 29: 1235–1239. [DOI] [PubMed] [Google Scholar]
- Rinnerthaler M., Buttner S., Laun P., Heeren G., Felder T. K., et al. , 2012. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. Proc. Natl. Acad. Sci. USA 109: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M. G., Kuo H. C., Lichius A., Freitag M., Read N. D., 2010. Nuclear dynamics, mitosis, and the cytoskeleton during the early stages of colony initiation in Neurospora crassa. Eukaryot. Cell 9: 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M. G., Weichert M., Siegmund U., Tudzynski P., Fleissner A., 2012. Germling fusion via conidial anastomosis tubes in the grey mould Botrytis cinerea requires NADPH oxidase activity. Fungal. Biol. 116: 379–387. [DOI] [PubMed] [Google Scholar]
- Ruepp A., Zollner A., Maier D., Albermann K., Hani J., et al. , 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32: 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder L. S., Dagdas Y. F., Mentlak T. A., Kershaw M. J., Thornton C. R., et al. , 2013. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. USA 110: 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W., 2001. Laboratory Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Scherz-Shouval R., Elazar Z., 2007. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 17: 422–427. [DOI] [PubMed] [Google Scholar]
- Schürg T., Brandt U., Adis C., Fleissner A., 2012. The Saccharomyces cerevisiae BEM1 homologue in Neurospora crassa promotes co-ordinated cell behaviour resulting in cell fusion. Mol. Microbiol. 86: 349–366. [DOI] [PubMed] [Google Scholar]
- Scott B., Eaton C. J., 2008. Role of reactive oxygen species in fungal cellular differentiations. Curr. Opin. Microbiol. 11: 488–493. [DOI] [PubMed] [Google Scholar]
- Siegmund U., Heller J., van Kan J. A. L., Tudzynski P., 2013. The NADPH oxidase complexes in Botrytis cinerea: evidence for a close association with the ER and the tetraspanin Pls1. PLoS ONE 8: e55879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonin A. R., Rasmussen C. G., Yang M., Glass N. L., 2010. Genes encoding a striatin-like protein (ham-3) and a forkhead associated protein (ham-4) are required for hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 47: 855–868. [DOI] [PubMed] [Google Scholar]
- Smith S. M., Min J., Ganesh T., Diebold B., Kawahara T., et al. , 2012. Ebselen and congeners inhibit NADPH oxidase 2-dependent superoxide generation by interrupting the binding of regulatory subunits. Chem. Biol. 19: 752–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D., Kamakura S., Saikia S., Becker Y., Wrenn R., et al. , 2011. Polarity proteins Bem1 and Cdc24 are components of the filamentous fungal NADPH oxidase complex. Proc. Natl. Acad. Sci. USA 108: 2861–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Takemoto D., Hyon G. S., Park P., Scott B., 2008. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol. Microbiol. 68: 1165–1178. [DOI] [PubMed] [Google Scholar]
- Teichert I., Wolff G., Kück U., Nowrousian M., 2012. Combining laser microdissection and RNA-seq to chart the transcriptional landscape of fungal development. BMC Genomics 13: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt O., Pöggeler S., 2013. Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy 9: 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz M., Kück U., 1995. Transformation of Sordaria macrospora to hygromycin B resistance: characterization of transformants by electrophoretic karyotyping and tetrad analysis. Curr. Genet. 29: 88–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.