Abstract
Orotidine 5'-monophosphate decarboxylase (ODCase) accelerates the decarboxylation of orotidine 5'-monophosphate (OMP) to uridine 5'-monophosphate (UMP) by 17 orders of magnitude. Eight new crystal structures with ligand analogues combined with computational analyses of the enzyme’s short-lived intermediates and the intrinsic electronic energies to distort the substrate and other ligands improve our understanding of the still controversially discussed reaction mechanism. In their respective complexes, 6-methyl-UMP displays significant distortion of its methyl substituent bond, 6-amino-UMP shows the competition between the K72 and C6 substituents for a position close to D70, and the methyl- and ethyl-ester of OMP both induce rotation of the carboxylate group substituent out of the plane of the pyrimidine ring. MD and QM/MM computations of the enzyme-substrate (ES) complex also show the bond between the carboxylate group and the pyrimidine ring to be distorted with the distortion contributing a 10–15% decrease of the ΔΔG‡ value. These results are consistent with ODCase using both substrate distortion as well as transition state stabilization, primarily exerted by K72, in its catalysis of the OMP decarboxylation reaction.
Keywords: ODCase, OMPDC, OMPDCase, crystal structure, substrate distortion, ligand distortion
Introduction
The most-widely accepted general mechanistic feature of reaction acceleration by enzymes is transition state (TS) stabilization.1,2 A more rarely put forward and occasionally controversially discussed concept is substrate distortion.3 One enzyme whose mechanism has recently been the subject of interest-generating diverging points of view but for which no general agreement about the overall mechanism has been reached is orotidine-5'-monophosphate decarboxylase (ODCase).4–6 This enzyme catalyzes the conversion of orotidine-5’-monophosphate (OMP) into uridine-5’-monophosphate (UMP) (Fig. 1A), the last step of de novo pyrimidine biosynthesis. ODCase is also one of the most proficient enzymes known. The t1/2 of the decarboxylation reaction in water at neutral pH is estimated at about 78 million years.7 In contrast, the enzyme catalyzes dozens of decarboxylation reactions in a second, accelerating the reaction by 17 orders of magnitude.7 It achieves this enormous catalytic power without employing cofactors or metal ions.6,8–10
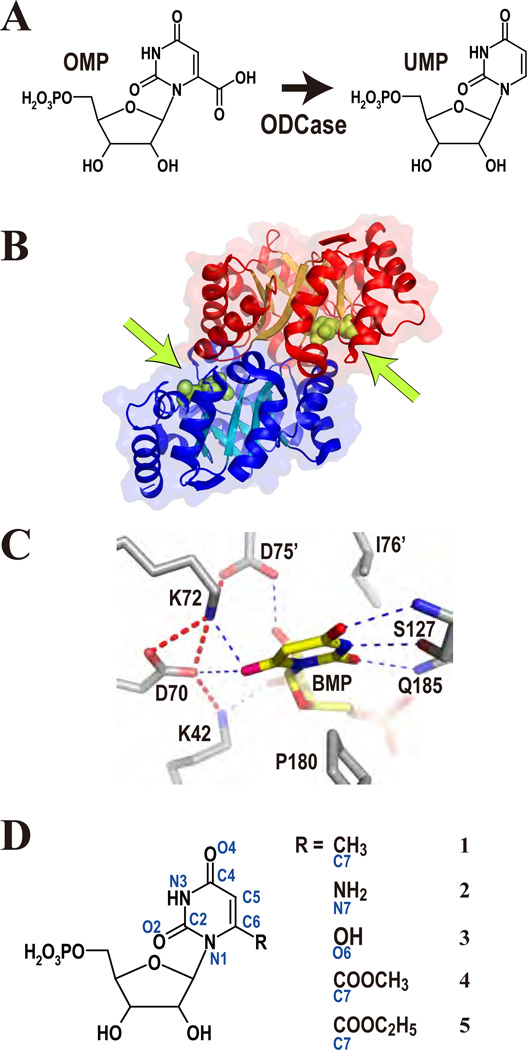
Figure 1.
(A) The reaction catalyzed by ODCase. (B) The physiological dimer of MtODCase. The green arrows indicate the binding positions of the ligands, which are drawn as green spheres. (C) The electrostatic and hydrogen bond network in the active site of ODCase. WT-MtODCase complexed with BMP (PDB: 1x1z) is presented as an example. (D) The substrate analogues described in this paper. Atom names are in blue. 1: 6-methyl-UMP, 2: 6-amino-UMP, 3: BMP, 4: OMP-methyl-ester, 5: OMP-ethyl-ester
More than 180 crystal structures from 18 sources have been determined thus far. They all show a dimer of TIM-barrel fold subunits with their active sites deeply buried and located at the dimer interface (Fig 1B). The amino acids making up the active sites and their three-dimensional structures are also very similar in all structurally known ODCases (Fig 1C). The electrostatic network of K42-D70-K72 from one subunit and D75’† from the other subunit (numbering based on ODCase from Methanothermobacter thermoautotrophicus (MtODCase)) is the most characteristic feature in the catalytic site; these residues are completely conserved and face the carboxylate of OMP, which is released during the reaction. Mutation of any of D70, K72 or D75’ into alanine reduces the enzyme’s activity by more than 5 orders of magnitude (Supporting Fig. S1).11,12 Mutation of the remaining member of the network, K42, decreases the kcat value a hundredfold.11,12 Random mutagenesis of E. coli chromosomal ODCase confirms that these four residues do not tolerate any substitutions.13 In addition, quite a number of residues surrounding the substrate-binding site have also been mutated to estimate their contribution to catalysis (Supporting Fig. S1), including the substrate destabilizing effect of a conserved hydrophobic patch.14
We have extensively investigated the reaction mechanism of this enzyme by crystallographic and kinetic means. We proposed that the distortion of the reacting group plays a considerable role in catalysis.15,16 The crystallographic results that support such an assignment are an OMP carboxyl group slightly rotated and tilted from the pyrimidine plane when complexed with the D70A/K72A mutant of MtODCase16 and a substituent significantly bent from the pyrimidine plane despite the resonance of the pyrimidine ring15 in 6-cyano-UMP complexes, a compound which is very slowly converted into 6-hydroxyl-UMP (BMP) by MtODCase (t1/2 = 5 hours).17,18 Similarly distorted ligands were found in the structures of several compounds bound to human-ODCase (HsODCase)19,20 and it has been suggested that the size of the C6-substituents of the pyrimidine ring is the dominant factor in determining the extent of the observed deviation from low energy conformations.20 As the stable existence of such distorted structures strongly argues for the involvement of substrate distortion in ODCase catalysis we engaged in further analysis and investigated the crystal structures of four more complexes, i.e. the structures of 6-methyl-UMP, 6-amino-UMP, OMP-methyl-ester, and OMP-ethyl-ester (Fig. 1D) in complex with MtODCase. 6-amino-UMP is a very good inhibitor of MtODCase (Ki = 840 ± 25 nM), while 6-methyl-UMP is only a moderate one (Ki = 134 ± 5 µM).21 The small amino and methyl substituents are good probes to elucidate the effect of the size of a C6-substituent on the distortion of its bond to the pyrimidine ring when bound in the ODCase active site. The two esters are structurally similar to but a bit larger than OMP; they can be regarded as good mimics of the carboxylate group without being subject to catalytic transformation. To complement the rather static nature of crystallographic experiments, we also performed computational simulations to better characterize the structures of short-lived reaction intermediates. Our present interpretation of the ODCase reaction mechanism is based on the results of our structural analyses as well as the computational simulations, and takes into account the existing relevant literature.
Materials and Methods
6-methyl-UMP and 6-amino-UMP were synthesized as previously described.21 OMP-methyl-ester and OMP-ethyl-ester were synthesized from uridine. The introduction of methoxy or ethoxycarbonyl moieties at the C-6 position was achieved via a lithium diisopropylamide-mediated reaction with the appropriate alkyl chloroformate. Deprotection of the protecting groups with trifluoroacetic acid22 followed by reaction with phosphorus oxychloride afforded the mono-phosphorylated OMP-methyl- or ethyl-ester (Supporting Scheme S1).23,24 Finally, OMP-ester monophosphates were neutralized with 0.5 M NH4OH solution at 0 °C and lyophilized to obtain the ammonium salts of the corresponding nucleotides. Details of the syntheses are described in the Materials and Methods section of the Supporting Information.
Wild-type orotidine 5’-monophosphate decarboxylase from Methanothermobacter thermoautotrophicus (MtODCase) and its mutants were expressed and purified as described using Ni-NTA and gel-filtration chromatography.16,25 All proteins were dialyzed against crystallization buffer composed of 20 mM HEPES-NaOH pH 7.5, 150 mM NaCl and 5 mM DTT. At room temperature, MtODCase (10 mg/mL) in crystallization buffer and incubated with the respective OMP derivative ligand (5–10 mM final concentration) was mixed with equal amounts of various precipitant solutions for crystallization using the hanging-drop vapor diffusion method. Initially, clusters of crystals grew when 1.1–1.36 M sodium citrate and 5% (v/v) dioxane at pH 6–9 were used as precipitant. Several cycles of microseeding were necessary to obtain single, diffraction-quality crystals. Crystals were dipped in a cryo-protectant buffer consisting of 1.2 M sodium citrate, 15% glycerol and 0.1 M MES-Na at pH 6.5 before being flash-frozen in a nitrogen stream at 95 – 100 K.
Diffraction datasets were collected at one of the following beamlines: 14ID-B, 14BM-C and 14BM-D at the Advanced Photon Source, Argonne, USA; 5A at the Photon Factory, Japan; or 41XU at SPring-8, Japan. Data were integrated, scaled, and truncated using HKL200026 and TRUNCATE.27 The dataset collected from a crystal of the K72A:6-amino-UMP complex was phased with the help of the molecular replacement program MOLREP using the wild-type:6-aza-UMP structure (PDB ID: 1DVJ) as the search model. All other data sets were phased directly from the refined model of the K72A:6-amino-UMP complex. Model-building and refinement were done using the program packages COOT28 and REFMAC.29 The parameter files for 6-amino-UMP, 6-methyl-UMP, OMP-methyl-ester and OMP-ethyl-ester were constructed modifying the file for uridine-5’-monophosphate (UMP). The bond between the C6 substituents and the pyrimidine rings in all four ligands were only restrained via bonding distance parameters; no planarity and angle restraint parameters regarding the bond were applied during refinement. The planarity restraints of the pyrimidine ring in 6-methyl-UMP and 6-amino-UMP (excluding the C6 substituents) are 10 times weaker than the default value in order to allow evaluation of their distorted structures. All refined structures were validated using MolProbity30 and deposited to Protein Data Bank. Statistics of all data collections and refinements are summarized in Supporting Table S1.
To simulate the decarboxylation reaction inside the active site of ODCase, we performed systematic ab initio QM (quantum mechanics)/MM (molecular mechanics) calculations combined with MD (molecular dynamics)-FEP (free energy perturbation) simulations and all-electron QM analyses for the entire enzyme complex. Technical issues of this modeling process are described elsewhere;31–33 the overall computational procedure is summarized as follows.
Initial coordinates of proteins were adopted from the X-ray geometry of wild-type MtODCase (1X1Z).17 The substrate (OMP) was modeled and placed at the original X-ray position of the substrate analog. Since ODCase works well around neutral pH,34,35 we assigned the standard protonation state to all polar residues. This assignment is consistent with the protonation state analysis of individual residues performed by the program PROPKA36,37 (Supporting Table S2). The resultant initial structure of the ES (Enzyme-substrate) complex contains the enzyme, the substrate OMP, crystal water molecules, and counterions to neutralize the total charge of the initial model. Next, we performed an MD simulation to reliably model the ES complex in the aqueous phase. After collecting trajectories for more than 2 ns periods, we randomly selected 10 representative protein structures for additional QM/MM structural refinements of the ES complex. In each QM/MM calculation, the QM region consists of the side chains of K42, D70, K72, D75’ and the pyrimidine ring of the substrate OMP. Considering the system size of the ODCase complex, we employed the restricted Hartree-Fock (RHF) method with the 6–31G(+)** basis set in all QM/MM reaction path optimizations. Then, by selecting one representative QM/MM optimized ES complex, we followed the direct decarboxylation path, a dissociation of carbon dioxide forming a vinyl anion intermediate.38–41 In this calculation, the reaction coordinate was defined as the bond distance between the C6 and C7 atoms. The strength of interaction energies between OMP and surrounding amino acid residues was estimated by all-electron QM computations for the entire protein complex using the Fragment Molecular Orbital method.42 For energetic analyses of the intrinsic nature of ligand distortion, we also performed ab initio QM calculations for the OMP analogs. To evaluate the energy cost of deforming the C6-C7 bond of the reactive substrate, we employed a computationally rather expensive method (MP2/aug-cc-pVDZ level) for two analog molecules (1-methyl-orotate methyl ester and 1-methyl-orotate). Further computational details are summarized in the Supporting Materials and Method section.
Results and Discussion
General Description
All of the eight complexes discussed in this paper (see Supporting Table S1) were crystallized under essentially the same crystallization conditions and resulted in equivalent crystal contacts. The crystallographic asymmetric unit contains only one subunit of the physiological dimer. The overall RMSD of Cα models superimposed on a reference structure, K72A-MtODCase complexed with BMP (PDB ID: 2ZZ7),15 range from 0.08 Å to 0.11 Å (all 215 Cα atoms, Supporting Table S1), i.e. the overall folds are practically identical.
For several residues/ligands in the active site, the electron density maps clearly indicate multiple conformations, which are accounted for in the refinement protocols. As sometimes neighboring residues or ligands both assume a number of orientations but only specific combinations of them can coexist we appended their respective names with labels A–C. For example, conformer A of a residue and conformer A of a close ligand exist in one asymmetric unit of the crystal, whereas conformer A of the amino acid and conformer B of the ligand can only be found in different asymmetric units of the crystal, e.g. because of physical overlap. Residues without an alternate conformation assignment, i.e. a single conformer, are present in all asymmetric units and potentially are in contact with all alternate conformations of neighboring side chains or ligand molecules. For instance, a single conformer residue might interact with all the multiple conformers of a ligand or vice versa. Unless otherwise specified, capitals in parenthesis following a residue or an atom name represent their conformation codes. For example, Nζ(A) and D75(B) indicate conformation A of the Nζ atom and conformation B of the D75 residue, respectively.
6-methyl-UMP and 6-amino-UMP
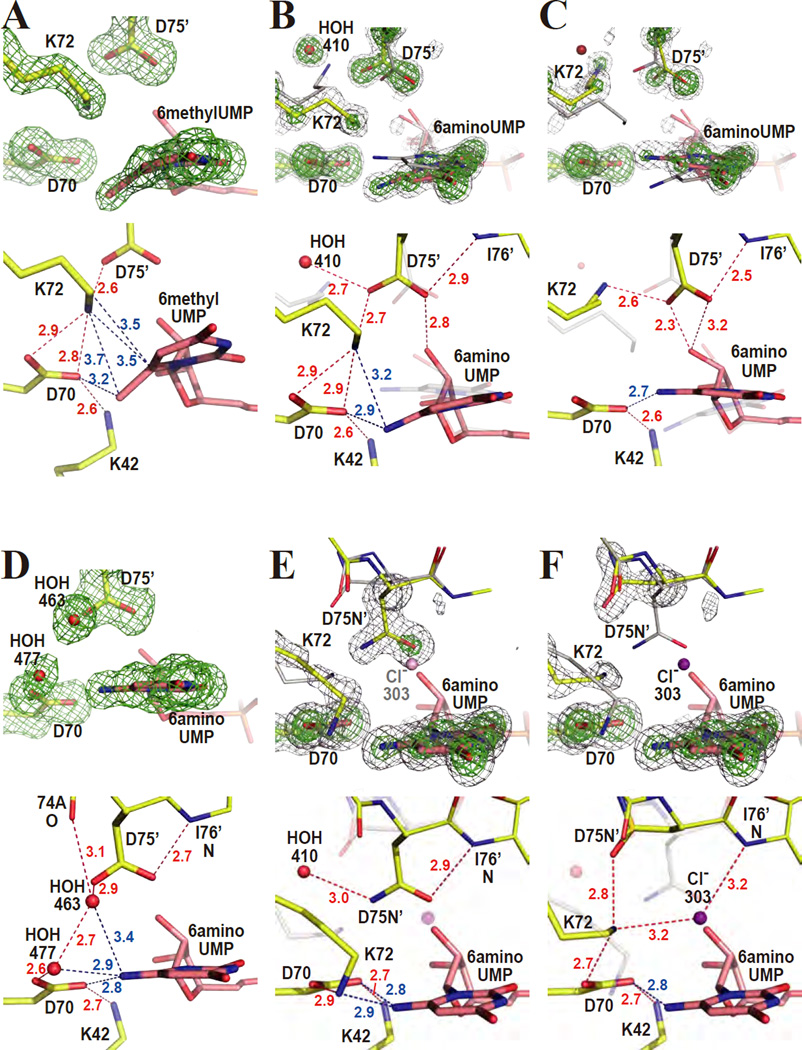
As seen in Figure 2A, the substituent of 6-methyl-UMP is significantly bent from the pyrimidine plane as we had observed earlier for the longer cyano moiety of 6-cyano-UMP in complex with WTMtODCase.15 The methyl C7 atom is ~16 degrees displaced from the C5-C6-N1 plane of the pyrimidine ring. In addition, the pyrimidine ring itself is also distorted. The dihedral angles of C4-C5-C6-N1 and C5-C6-N1-C2 refined to 8 and 18 degrees, respectively, whereas they are typically less than 5 degrees for a flat pyrimidine ring. The C7 atom is very close to Oδ2 of D70 (3.2 Å). K42, D70, K72 and D75’ engage in a strong electrostatic interaction, restricting the positions of their side chains. The distorted ligand structure is fully consistent with the corresponding weak Ki value (Ki = ~30 × KM), with the distortion consuming the binding energy.
Figure 2.
6-methyl-UMP and 6-amino-UMP complexes. The upper panels show omit electron density maps superimposed on the refined model. Lower panels describe the binding-modes of the various ligands in detail. Red and blue numbers give distances in Ångstroms. (A) WT-MtODCase with 6-methyl-UMP. The omit electron density map was phased without the atoms of the pyrimidine ring, D70, K72, and D75’ included and contoured at 3.5 σ. (B) Conformation A of WT-MtODCase with 6-amino-UMP. The omit electron density map was phased without the atoms of D70 and HOH410 as well as those of the conformation A of K72, D75’ and the pyrimidine ring included and was contoured at 5.5 σ (green) and 3.0 σ (gray). Yellow and gray sticks are conformers A (or the part without alternate conformation) and B, respectively. (C) Conformation B of WT-MtODCase with 6-amino-UMP. The omit electron density map was phased without the atoms of the residues of conformer B (and the part without alternate conformations) included, corresponding to panel B. The green and gray meshes are contoured at 5.5 σ and 3.0 σ, respectively. Yellow and gray sticks are conformers B and A, respectively. (D) K72A-MtODCase with 6-amino-UMP. The omit electron density map (3.0 σ) was phased without the atoms of the pyrimidine ring, D70, D75’, HOH463 and HOH477 included. (E) Conformation A of D75N-MtODCase with 6-amino-UMP. The omit electron density map corresponds to D70 and the pyrimidine ring as well as conformations A of K72 and D75N’. The green and gray meshes represent 8.0 σ and 3.0 σ, respectively. Yellow and gray sticks are conformers A (or the residues without alternate conformation) and B, respectively. (F) Conformation B of D75N-MtODCase with 6-amino-UMP. The omit electron density map was calculated without residues in conformer B (and the part without alternate conformations) included. The green and gray meshes are contoured at 8.0 σ and 3.0 σ, respectively. Yellow and gray sticks are conformers B and A, respectively. This panel is drawn in the same way as panel E, except that conformer B is emphasized (in color) instead of conformer A. The chloride anion, Cl303 (purple), is only observed in conformer B.
The structure of this complex shows that the ODCase active site obviously has the power to distort C6-substituents regardless of their size, even methyl groups, which - next to hydrogen - are one of the smallest substituents possible. The four-residues engaged in the electrostatic network assume positions almost identical to those seen in the complex of WT-MtODCase with BMP.17,43,44 The strong electron delocalization in effect for the O4-C4-C5-C6-O6‡ atoms in the flat BMP structure17,43,44 may play a considerable role in BMP’s resistance to bond-distortion by the catalytic center.
For 6-amino-UMP bound to WT-MtODCase, double conformers A and B are observed (Figs. 2B and 2C). Omit electron density maps phased from models without one of the two conformers clearly indicate their presence (Figs. 2B and 2C). The occupancies were assigned as 60% and 40% for conformers A and B, respectively, based on the relative electron density heights. No significant residual density is found at the 3.0 σ level (gray mesh in Figs. 2B and 2C), indicating that the two conformations explain the experimental electron density reasonably well. The pyrimidine ring of conformer A (pink in Fig. 2B) rotates counterclockwise ~20 degrees around the C2-N3 bond from the orientation of conformer B (gray in Fig. 2B). The amino group N7(A) of the ligand lies ~9 degrees outside the C5-C6-N1 plane of the pyrimidine ring (Fig. 2B). The pyrimidine plane is much less distorted than that of the 6-methyl-UMP complex. N7(A) is located 2.9 Å from Oδ2(A) of D70 and 3.2 Å from Nζ(A) of K72. In conformer A, the electrostatic network superimposes well onto the corresponding residues in the complexes of WT-MtODCase with both 6-methyl-UMP and BMP (Supporting Fig. S2A).
As stated above, the pyrimidine ring of conformer B (pink in Fig. 2C) is rotated clockwise ~20 degrees from its position in conformer A (gray in Fig. 2C). It refines as a flat ring with only minimal deviations. This also includes N7, although no angle and planarity restraints were applied on N7 during the refinement (Fig. 2C). The long side chain of K72(B) (yellow in Fig. 2C) has moved from the position that it adopts in conformer A (transparent gray in Fig. 2C), and has lost the electrostatic interaction with D70, leading to the disruption of the K42-D70-K72-D75’ network. Nζ(B) of K72 conserves its binding to D75’ but is now located at the position previously occupied by HOH410(A). From these results, it seems that N7 of 6-amino-UMP and Nζ of K72 compete for the position closer to D70. Because of the two conformations of the ligand-binding site and several bond formations/disruptions around the ligand, it is difficult to deduce a direct relationship between structure and Ki value for this compound.
Such a competition between the amino groups of K72 and the ligand is supported further by the crystal structure of the K72A mutant complexed with 6-amino-UMP. It shows that with the side chain of K72 removed, the position of the pyrimidine ring, with N7 of 6-amino-UMP in plane, corresponds to that of conformer B of the WT-MtODCase complex (Fig. 2D). Two water molecules, HOH463 and HOH477, replace the amino group of K72 as the electrostatic link between D70 and D75’ (Fig. 2D).
The potential role of K72 in bending the bond between C6 of the pyrimidine ring and various substituents, here methyl and amino groups, is also reflected in the way 6-amino-UMP binds to D75N-MtODCase. This mutation weakens the originally strong interaction between K72 and D75’ to the point that K72 is free to move away from D75N’ and its location in WT-MtODCase conformation A (Fig. 2B), a position it also assumes in the ligand-free enzyme. In the crystal structure of D75N-MtODCase complexed with 6-amino-UMP (Figs. 2E and 2F), residues K72 and D75N refine as double conformers, together with V73 and A74. Both K72(A) and K72(B) move significantly when compared to conformation A of the same complex in WT-MtODCase (Fig. 3A). N7 of 6-amino-UMP is located in between the A and B conformations of the wild-type complex (Fig. 3A). In this case, both amino groups, the 6-substituent of the nucleotide and the primary amine of K72, are not successful in claiming the original K72 position. K72(B) moves with D75N’(B), whereas D75N’(A) remains at the position it assumes in the wild-type complex. The pyrimidine ring is flat. Note that in conformer B of this complex, a chloride anion, Cl303(B), is found close to the position of D75N’ in conformation A (Figs. 2E and 2F). A corresponding chloride ion appears in other D75N-MtODCase structures.16
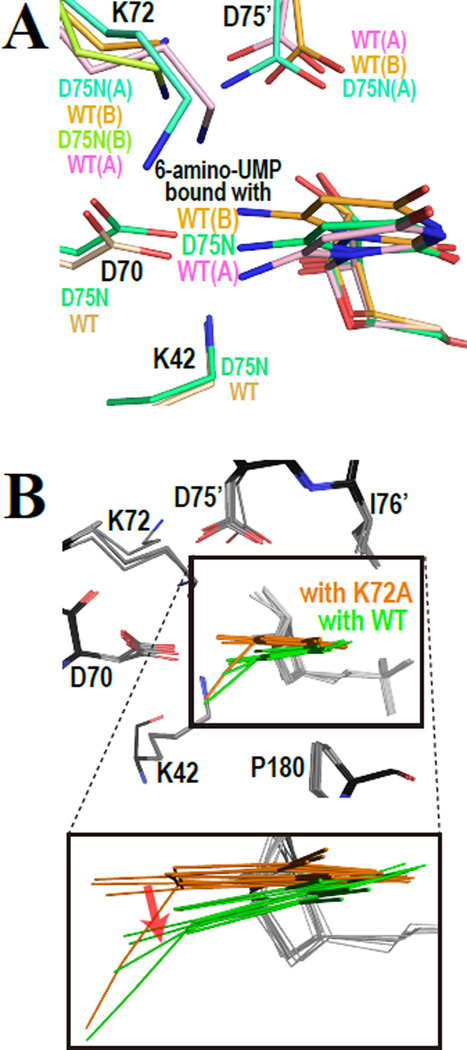
Figure 3.
Superimpositions of the active sites of various enzyme-ligand complexes. (A) 6-amino-UMP in complex with WT-MtODCase and D75N-MtODCase. (B) Comparison of various ligands as bound to the active site in complexes of WTMtODCase and K72A-MtODCase. The BMP, UMP, 6-cyano-UMP, 6-methyl-UMP, and 6-amino-UMP complexes with WT-MtODCase and K72A-MtODCase are superimposed. The models colored in orange and green are the ligands with K72A-MtODCase and WT-MtODCase, respectively. In the lower panel, which shows an enlargement of the superimposed pyrimidine rings, the red arrow indicates the effect on ligand binding positions caused by K72. Note that the conformation B of 6-amino-UMP in complex with WTMtODCase is included in orange, since K72(B) in this complex is flipped from the typical K72 position in other complexes and can not influence the ligand positions.
The effects of the competition between K72 and the ligand can be seen when comparing wild-type and K72A mutant structures complexed with various ligands. As seen in Figure 3B, the pyrimidine rings of the ligands complexed to K72A-MtODCase move towards where K72 is located in wild-type MtODCase. Unless positive charge on the ligand replaces its amino headgroup, K72 in the wild-type enzyme is strongly held in the four-residue electrostatic network and pushes down onto the C6-substituent (red arrow in Fig. 3B). If the resonance energy of the pyrimidine ring is not sufficiently strong to counteract, this interaction can lead to the distortion of the bond from C6 to the substituent.
OMP methyl and ethyl esters
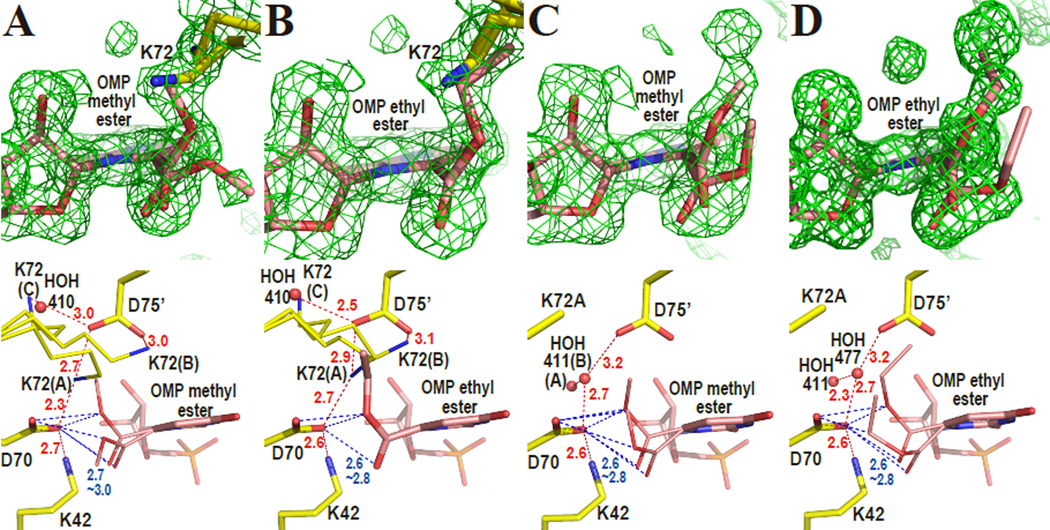
OMP esters were chosen to test ligands quite similar to the substrate OMP but not reactive due to ester formation. Figure 4A shows the omit electron density map around the active site superimposed on the atomic model of WT-MtODCase with OMP-methyl-ester. Although the electron density is not consistent with a single orientation of the substituent, it is clear that the carboxyl group is located very close to D70 (2.7–3.0 Å) and the carboxyl plane of the ester (the C6, C7, O71 and O72 plane) is rotated from the pyrimidine plane (Fig. 4A). Two major conformations have been fitted. Depending on the rotation of the ester group, they deviate by 35~60 degrees from the plane of the carboxylate in WT-MtODCase. A partial overlap of the electron density corresponding to K72 with the density representing the ester group leads to further difficulty in their interpretation. Similar results were obtained when WT-MtODCase formed a complex with OMP-ethyl-ester (Fig. 4B), which also includes significant rotation (~55 degrees) of the ester group and its overlap with parts of the side chain of K72.
Figure 4.
Methyl and ethyl ester complexes of ODCase. The upper panels show omit electron density maps superposed on the refined models. The omit electron density maps (2.0 σ) were phased without the atoms of the ligand and K72 included in the calculations. The lower panels display the binding modes of the ligands in detail. Red and blue numbers give distances in Ångstroms. (A) WT-MtODCase with OMP-methyl-ester. (B) WT-MtODCase with OMP-ethyl-ester. (C) K72A-MtODCase with OMP-methyl-ester. (D) K72A-MtODCase with OMP-ethyl-ester.
Since some conformations of K72 contribute to the same electron density as does the ester part of the ligand, the crystal structures of the K72A mutant with the two esters were also determined (Figs. 4C and 4D). Despite the absence of an interfering side chain, the ester groups in both methyl- and ethyl-esters are still rotated 40~70 degrees. These two structures indicate that these rotations are not caused by K72 alone. As had been seen for HOH463 and HOH477 in K72A MtODCase complexed with 6-amino-UMP (Fig. 2D), the remaining members of the electrostatic network stay connected via a link from K42 to D75’ through water residues HOH411(B) and HOH477 in the OMP-methyl-ester and OMP-ethyl-ester complexes, respectively. Superpositions of these ester complexes of K72A-MtODCase with those of Wt-MtODCase display the same shifts of the pyrimidine location that had been seen in other complexes (Fig. 3B and Supporting Fig. S2B).
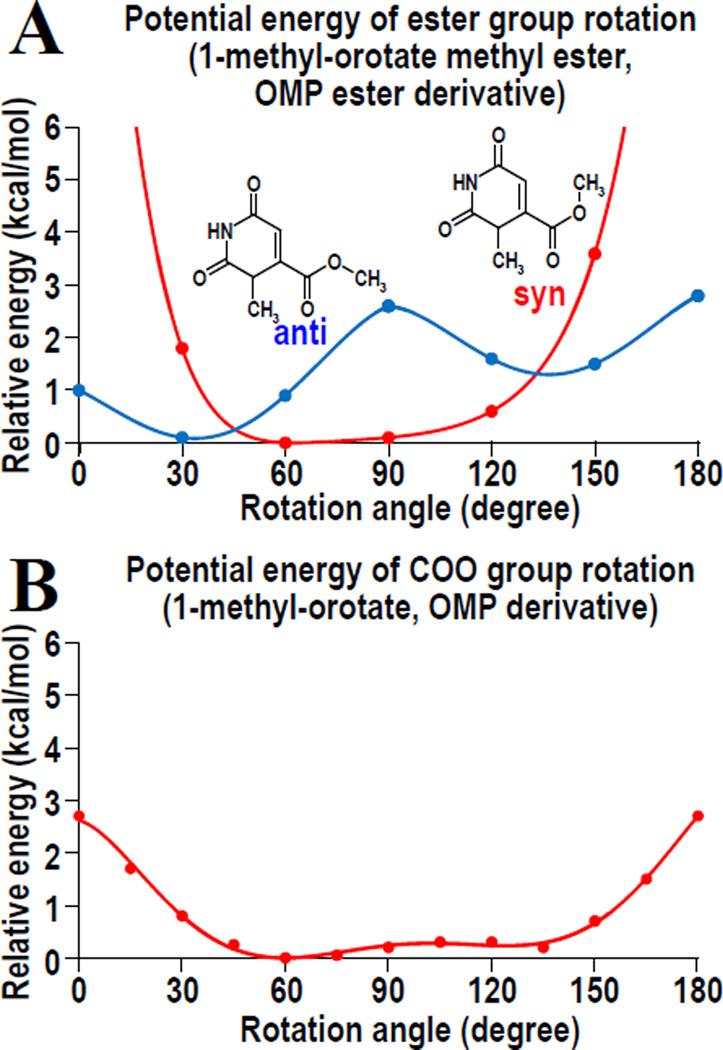
Although it is very difficult to refine precise coordinates for the overlapping parts and multiple conformations in the four structures of ester ligands, the rotation of the ester groups is clearly observed. To obtain an estimate of the energies involved in such a rotation, we calculated the potential energies for a rotating ester group with the methyl group in the syn and anti conformation. Figure 5A shows the potential energy profile of the ester group rotation in 1-methyl-orotate methyl ester. The panel indicates that the ester group is more stable in an out-of-plane rotational conformation than in a nonrotated structure. In addition, the anti-conformation of the methyl group is more stable than its syn-conformation. In the crystal structure of the complex, however, energetically unfavorable interactions between D70 and the anti-conformations of the methyl (or ethyl) groups of the esters forces them into the syn-conformation. The corresponding energy profile for 1-methyl-orotate, a model compound for OMP (Fig. 5B) shows that the carboxylate group of OMP, too, is more stable in its out-of-rotation conformation. The Cambridge Structural Database of small molecule structures also includes a large number of carboxylate groups that are attached onto aromatic rings and rotated out of the ring plane. Together, all these findings suggest that an out-of-plane rotation of the carboxylate group is not a rare event. It is therefore most probable that OMP esters mimic the OMP molecule approaching the active site of ODCase, with the carboxylate group of OMP also rotated away from the in-plane orientation.
Figure 5.
Potential energy profiles of the rotation around the C6-C7 bond of OMP analogs. (A) Profile of 1-methyl-orotate methyl ester, a model compound for OMP methyl ester. Blue and red lines indicate the profiles for anti and syn conformations of the methyl group, respectively, whose chemical formulae are also shown. (B) Profile of the carboxylate group rotation in 1-methyl-orotate.
Simulating the structure of the enzyme-substrate, transition-state, and intermediate complexes
Our computational findings also suggest that ODCase has the power to distort the C6 substituent of ligands bound to its active site. Although the distortion is not very obvious in the present ester complexes due to overlapping electron densities, the bonds linking the C6 atoms and the respective substituents of 6-methyl-UMP, 6-cyano-UMP and 6-acetyl-UMP are clearly distorted in their ODCase complexes (Fig 2A and refs.15,20). In addition, our WT-UMP complex structure, recently determined at atomic resolution (1.03 Å), indicates that the pyrimidine ring of UMP itself is slightly distorted, too.45 These structures imply that ODCase can utilize substrate distortion to achieve the enormous acceleration of the reaction it catalyzes. Although various groups have undertaken computational simulations of ODCase catalysis, detailed analyses of the distortion effects have not been performed thus far. Both transition state stabilization and ground state destabilization have been suggested as the major contributing factors to ODCase catalysis.46,47 Warshel et al. proposed that ODCase utilizes transition state stabilization based on their binding energy analyses of the ligand and the enzyme.5,47 In contrast, calculations by Gao et al. indicated that the protein part of the enzyme-substrate complex is distorted compared to the transition state structure, and thus they proposed a ground state destabilization mechanism.46 More recently, Hu et al. presented a mechanistic proposal that has ODCase exerting its catalytic function through direct decarboxylation to form an anionic intermediate supported by K72 stabilization of the vinyl anion intermediate of the almost decarboxylated transition state structure.48 They also noted that the water molecule located ~2.7 Å from the O6 atom of the BMP molecule in the ODCase-BMP complex17,43,44 must be excluded from the calculation, since no corresponding water molecules are found in all other complexes. However, all reported simulations did not consider potentially distorted substrate structures and did not analyze the interaction between the ligand and each of the protein’s residues. We have now performed ab initio QM/MM calculations to evaluate the contribution of substrate distortion to catalysis and to determine the interaction energy between each residue and each of the ES – enzyme-substrate-complex/TS – transition state/INT – intermediate stages of the substrate.
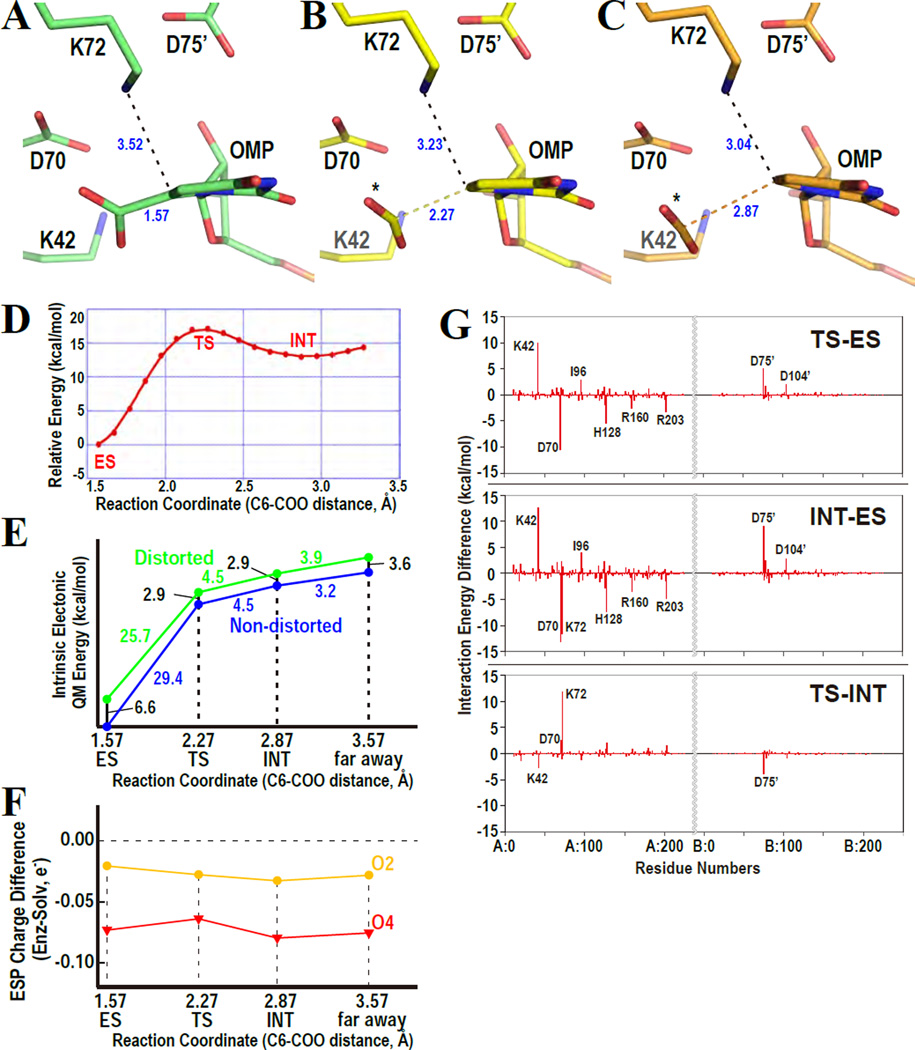
The calculations were performed based on a direct decarboxylation pathway, which forms a vinyl anion intermediate and is experimentally supported,38,40,41 and at the QM/MM MP2/6–31(+)G**//RHF/6–31(+)G**/ AMBER level. The water molecule located close to the O6 atom of BMP was not included in the calculations. All ten QM/MM optimized structures of the ES complex clearly demonstrate that the C6–C7 bond is bent from the pyrimidine plane. A typical structure of the ES complex (OMP with WT-MtODCase) is shown in Figure 6A. The C6–C7 bond is distorted by 7.5–14 degrees from the C5-C6-N1 plane, and the planarity of the pyrimidine ring is apparently disrupted. The K42-D70-K72-D75’ network is intact (distances between interacting side chain atoms <3.2 Å) in nine of ten simulated structures, which shows the four residues are networked in almost all states generated by the dynamic motion of the enzyme. These computational ES complex structures indicate that the bent C6 substituent from the pyrimidine plane is a "stable" structure in the ODCase environment, which is consistent with various experimental structures, including the structures presented in this report (Fig. 2 and refs.15,19,20)
Figure 6.
Simulations of the ODCase reaction. Representative structures of enzyme-substrate complex (ES, A), transition state (TS, B), and intermediate state (INT, C) are drawn. The three states are defined in panel D. The asterisks represent the released carboxylate. (D) Direct decarboxylation profile forming vinyl anion intermediate along the reaction coordinate determined by ab initio QM/MM calculations (QM/MM MP2/6–31(+)G**//RHF/6–31(+)G**//AMBER (parm.96)). The x- and y- axes indicate the C6-COO bond distance (Å) and the relative energy of the reaction (kcal/mol), respectively. TS and INT are defined in the peak and the metastable part along the decarboxylation path, respectively. (E) Potential energy profiles of the electronic energy (QM energy) of the distorted (green, inside enzyme) and non-distorted (blue, in aqueous solution) orotate portion of the OMP substrate. The distorted energy penalty is implicitly incorporated into the potential energy curves, and these intrinsic energy profiles are mapped onto the reduced one-dimensional reaction coordinate as shown in panel D. The x- and y-axes indicate the reaction coordinate of the C6-COO distance and the relative energy of the decarboxylation reaction, respectively. Energy differences between the two profiles (numbered in black) represent the additional energy cost to deform the ligand structure relative to the reference, which is the relaxed structure in the aqueous solution. (F) Electrostatic potential (ESP) charge difference between enzyme-bound ligand and the ligand in solution. The x- axis indicates the reaction coordinate of the C6-COO distance. The y-axis indicates the ESP charges in enzyme subtracted by those in solution. The charges of all atoms in the pyrimidine ring are represented in the supporting Fig. S3. (G) Interaction energy differences between two selected states along the decarboxylation coordinate determined by FMO2-RHF/6–31G* calculations. Upper, middle and lower panels indicate energy difference between TS and ES, between INT and ES, and between TS and INT, respectively. The x-axis corresponds to the amino acid residue serial number, and the y-axis designates the interaction energy differences between the two selected states.
The direct decarboxylation pathway38–41 was followed using QM/MM geometry optimizations, and located TS and INT onto the reaction energy profile (Fig. 6D). This energy profile fits well with the experimentally determined kinetic values of MtODCase. ΔΔG‡ (difference of the transition state free energies between enzyme-catalyzed and non-enzymatic reactions) of MtODCase calculated from the [(kcat/KM)/knon] value is ca. 29–30 kcal/mol,7,14,18,34,43,49,50 whereas that of yeast ODCase is ~31 kcal/mol.7,51 The ΔΔG‡ value puts the energy required to go from the ES complex of MtODCase to its TS at 17–18 kcal/mol,14,18,34,43,49,50 which is in excellent agreement with the present computational value of ~17 kcal/mol (Fig. 6D). In this typical QM/MM result, the C6–C7 bond distances of TS and INT are 2.27 Å and 2.87 Å, respectively. These two structures are shown in Figures 6B and 6C. The C6–C7 bonds are apparently bent from the C5-C6-N1 planes and the pyrimidine rings are distorted in both TS and INT structures as seen before in the ES complex.
The next issue is how substrate distortion can contribute to ODCase catalysis (Fig. 6E). We estimated the energy penalty to distort the OMP ligand inside the active site by computational experiments (gradually releasing the conformational as well as electrostatic strain originating from the protein environment). The intrinsic electronic energies (=QM energy of the orotate part of OMP) were calculated for the distorted and the relaxed conformations inside the enzyme and in an aqueous environment, respectively, and comparison of the calculated energies revealed the energetic penalty for the distortion. The C6-C7 bond-lengths of the relaxed conformations corresponding to TS, INT and “far away” (mimicking the completely dissociated form) were restrained to 2.27 Å, 2.87 Å and 3.57 Å, respectively, in order to compare equivalent states in the reaction coordinates. The intrinsic energy difference between the distorted and nondistorted structures corresponding to ES is 6.6 kcal/mol, whereas the corresponding number for TS is 2.9 kcal/mol (Fig 6E). These estimates indicate that the ligand structure in ES is more distorted than that in TS, and the corresponding intrinsic energy difference is 3.7 kcal/mol. This energy difference is considered to be the contribution of substrate distortion in ODCase catalysis. Since the ΔΔG‡ value of MtODCase is 29–30 kcal/mol, both when deduced from experimental data and when computed in our present simulation, the contribution of the substrate distortion in ODCase catalysis is estimated to be 10~15%. Incidentally, these numbers agree well with those found by Ruben et al. in their experimental study of ketosteroid isomerase, which found strong evidence for binding energy-lowering interactions between substrate analogs and residues in the enzyme’s active site.52
Then, we determined partial atomic charges derived from the molecular electrostatic potential (ESP) of each atom of OMP in order to estimate the charge distribution during the reaction both when catalyzed by the enzyme and in solution The simulated ESP charge is consistent with the mutational analyses performed thus far14,49. The O4 and O2 atoms are kept negative throughout the reaction (Supporting Fig. S3), and the negative charges are stronger in the enzyme complex than in solvent (Fig. 6F). This fact suggests that the enzyme preferentially accommodates negative charges at the O4 and O2 positions. Also, the charge difference of O2 is approximately one-third of that of O4 throughout the reaction (Fig. 6F). This result is consistent with the finding that the disruption of the interaction between the main chain amide of S127 and O4 more severely affects the kinetic parameters than the disruption of the interaction between Q185 and O2.14,49 It also places some carbene character on C6 due to significant delocalization of the negative charge from C6 to O4.
Finally, we estimated the molecular interaction energies between each residue and the ligand (Fig. 6G) by performing all-electron QM calculations based on QM/MM optimized geometries. In the calculations, D70, D75’ and OMP carboxylates are deprotonated whereas K42 and K72 are protonated. This assumption is consistent with the pKa values of individual residues calculated by the program PROPKA36,37 (Supporting Table S2). An experimental pKa analysis using the catalytically still active D70C and D75C mutants also supports this assumption by identifying the charge state of the cysteine side chains as thiolate ions.13 The computational results clearly show that K42 stabilizes the ES complex more than the TS and INT structures, D70 destabilizes the ES complex in comparison to the other two states and K72 stabilizes the INT structure relative to the ES and TS complexes. Smaller but still significant peaks are seen for several other residues but are regarded as artifacts due to the following reasons. The peaks for I96 may be due to the specific computational method, which imposes a constraint on C-C bond distance along the reaction coordinate as it follows the decarboxylation pathway. Just before bond-breakage, the carboxylate groups in TS and INT were located at a position where they would form considerably unfavorable steric interactions with I96. H128, R163, R203 and D104’ are not included in the QM region and may well be shielded more strongly if they were. The interaction between D75’ and the ligand is expected to be much weaker than that between K72 and D75’, since K72 is sandwiched between D75’ and the substrate’s carboxylate. The role for D75’ is thought to be to fix K72 at the appropriate position for the reaction, as seen in the structure of D75N-MtODCase with 6-amino-UMP (Figs. 2E and 2F). Only K42, D70 and K72 are located at positions in which they can directly interact with the carboxylate group, and thus display large energetic effects (Fig. 6G).
Integrating the results into a complete reaction scheme
Our present crystallographic results and computational simulations together with a careful analysis of the existing literature guide the following detailed discussion of the catalytic reaction mechanism of ODCase. All residues are numbered according to the MtODCase sequence. In solution, the pyrimidine ring of OMP is predicted to mainly (>90%) adopt the syn-conformation, whereas UMP strongly prefers the anti-conformation.53 We therefore assume that OMP approaches ODCase in the syn-conformation. There, the carboxylate group of OMP faces the characteristic K42-D70-K72-D75’ electrostatic network, which is conserved in all known ODCases.6,8–10,19,21,54 A medium resolution (2.65 Å) crystallographic analysis of Plasmodium falciparum ODCase (PfODCase) captured the binding of OMP, showing that the two carboxyl moieties of OMP and D70 are located very close to each other.55 Hydrogen bonds between the side chain of S127 and N3 of OMP as well as between Q185 and O2 stabilize the syn-conformation. Changing S127 to an alanine resulted in a 50:50 mixture of syn- and anti-conformation of 6-Aza-UMP,14,16 further supporting this assignment.
We also envision the carboxylate group of OMP rotated away from the plane of the pyrimidine ring, especially as our ab initio QM calculation confirmed the rotated conformation as the most stable one (Fig. 5B). Similar results had been obtained by DFT (density functional theory) calculations with B3LYP/6–31+G* and B3LYP/6–311+G* sets.48 In addition in all four of the OMP-ester complexes investigated, the carboxylate ester group is rotated away from the plane of the pyrimidine ring, further supporting the assumption that the carboxylate group of OMP binds to ODCase in the stable rotated conformation. The two juxtaposed carboxylate groups of OMP and D70 are approximately perpendicular to each other as shown in Figure 4. Given the very short distance (~3.6 Å) between Cγ of D70 and the carbon center of the ester carboxylate (C7), a relatively strong electrostatic repulsion, consistent with PROPKA assignments (Supporting Table S2), between the two carboxylates has to be expected. However, K42, one of the two proximal lysines, bridges the two carboxylates moderating the repulsive force. In order to achieve this effect, K42 moves from its position in the non-liganded enzyme; this shift and its interaction with the OMP carboxylate were captured both in our simulation (Fig. 6A) and in the crystal structure of the PfODCase-OMP complex.55 Consequently, mutation of this lysine resulted in a 1,000-fold increase of Km and a 100-fold decrease of kcat.12
As the two carboxylates of OMP and D70 approach each other, the bond environment of OMP C7 should become highly distorted. There has been a longstanding controversy about the potential contribution of such a substrate distortion to ODCase catalysis. The major argument against such an effect was seen in the relatively weak binding of the product UMP, which - having lost its carboxylate group - was expected to bind more tightly than the substrate OMP.5,47,56 However, by combining an atomic-resolution crystal structure of the UMP complex with surface plasmon resonance measurements, we recently established that the weak binding of UMP is caused by steric interference with K72; the binding constant of UMP to the K72A mutant was determined as 4 × 10−9 M, second only to the best known ODCase inhibitor BMP (barbituric acid monophosphate). These findings clearly invalidate the original argument.45 The disruption of the resonance between the rotated carboxylate group and the pyrimidine ring might ease the distortion. The C7 atom moves out of the pyrimidine ring plane, the planarity of which is also disrupted. The molecular structures of the present WT-MtODCase with 6-methyl-UMP as well as those of previously published complexes with 6-cyano-UMP15 clearly display the significant distortion at the C6 substituent of the pyrimidine ring, as do the crystal structures of the D312N mutant of human ODCase with OMP, and the native human ODCase with 6-aceto-UMP or various other OMP analogues.19,20 We now show that this extends to the smallest substituents, the methyl and amino groups. In addition, all of these structures display an apparent disruption of the pyrimidine ring planarity unless strong planarity restraint parameters are applied during the refinement. Although no such distortions are obvious in the present ester complexes, this may simply be due to the lower quality of the electron density maps (multiple conformations and overlap), which require just such tight restraints. Our 10 ab initio computational analyses resulted in 7.5–14 degrees deviations of the OMP carboxylate from the pyrimidine plane in the enzyme-substrate complex; in addition, the pyrimidine plane distortion was also reproduced (Fig. 6A). The energy necessary for such a break in the planarity of an aromatic ring could well be provided by the numerous hydrogen bonds between enzyme residues and the ligands, especially those to the phosphate group of OMP (Supporting Fig. S1).43,49,57–59 Various structures indicate that ligand binding leads to the closure of a loop (residues 182–190), which is disordered in the ligand-free enzyme.6,10,19,54,55,60–62 When locked down, this loop forms five interactions, directly or through water molecules, with the phosphate part of the ligand. 1-(β-D-erythrofuranosyl)orotic acid (EO), an OMP derivative missing O5 of the ribose ring and the phosphate group (Supporting Fig. S4), is decarboxylated by ODCase but the corresponding kcat/Km value is approximately 5 × 108 times smaller than that of OMP.63 The EO reaction could be accelerated 8 × 104–fold by adding HPO32− to the solution. Orotidine (Supporting Fig. S4) is decarboxylated by yeast ODCase with a ~1011–fold lower kcat/Km value than OMP.64 Mutations of the phosphate binding arginine (R203) result in 103–104–fold lower kcat/Km values.65 These drastic effects on catalytic efficiency confirm the importance of the phosphate part of OMP for the catalytic reaction. It is quite reasonable to assume that a part of the binding energy, especially the contribution from the phosphate group, is used to distort the pyrimidine ring.
The distorted carboxylate group then dissociates from the pyrimidine ring and a vinyl anion intermediate at C6 is formed.38–41 The ODCase reaction performed in H2O/D2O (1:1 ratio) buffer results in the same H/D ratio at the C6 hydrogen of UMP.39,40 This isotope analysis rules out any catalytic mechanism with a protonation step as the rate-determining step and supports the direct decarboxylation mechanism through the vinyl anion intermediate. The faster decarboxylation of 5-fluoro-OMP compared to OMP is consistent with the existence of such an anion intermediate.34,41,66 The pKa values of the UMP C6 proton are estimated as 29–34 (ref.67–69) and 29 (ref.70) in water and in DMSO, respectively, whereas the pKa in yeast ODCase is estimated as ≤ 22 (ref.38). The remarkable shift in pKa seen when substrate is transferred from solvent to the enzyme’s active site also points to a strong stabilization of the C6 anion intermediate. The C6-C7 bond distortion decreases the energy barrier for the decarboxylation by ~3.7 kcal/mol (Fig. 6E), which is 10~15% of the total ΔΔG‡ value. Mutation of the proximal D70 significantly decreases the kcat value, but hardly affects the stabilization of the reaction intermediate.34 Our interaction residue energy profile shows that K72 is involved in the stabilization of the developing negative charge in the intermediate (Fig. 6F). It is therefore not surprising that its mutation results in the loss of enzymatic activity (≤10−5 × WT).12,65 Sharing of the negative charge at C6 with the O4 atom of the ligand through delocalization, imparting partial carbene character, may also contribute to the stabilization of the intermediate.14 The O4 atom interacts with the main chain amide of S127. Our simulation showed that ODCase preferentially accommodates negative charge at the O4 atom (Fig. 6F). Mutation of S127 to proline, which disrupts the interaction with the O4 atom, results in a significant decrease of the kcat/KM values (~4 × 10−7 of WT)14. The strong inhibition of ODCase by BMP (Ki = ~9 × 10−12 M)71,72 might also relate to the negative charge at the O4 atom. This all points to a much larger contribution to transition state stabilization, and thereby the tight binding of the transition state analog BMP, by the charge accumulating at O4 than by the charge at O6.
Protonation by the ammonium group of K72 is considered the last step of the catalytic reaction, leading to the formation of the final product UMP. The low affinity of UMP to ODCase (Ki = ~10−4)50,65,73,74 will support product release, readying ODCase for the next cycle of catalysis, which encompasses aspects of both substrate distortion and transition state stabilization.
ODCase is not the only enzyme for which distortion in the conformation of its substrate molecule has been proposed in computational studies or measured in kinetic and structural experiments. Theoretical studies75–77 using the crystal structures of TS-analogue complexes78,79 as initial models predict that chorismate mutase holds its substrate chorismate in an energetically unfavored chair-like conformation. Computations on glutamate mutase suggest that the C4'-C5'-Co bond-angle of adocobalamin (vitamin B12) is distorted, facilitating the dissociation of the bond between C5' and Co producing the catalytically important radical at C5'.80 Measurement of the Binding Isotope Effect in orotate phosphoribosyltransferase also indicated that OMP binds to the enzyme distorted around the bond between its ribose and aromatic base components.81 Other recently published examples of enzymes for which substrate distortion is invoked in the catalytic mechanism include aspartate aminotransferase,82 α-1,3-galactosyltransferase,83 family II pyrophosphatase,84 and the already mentioned ketosteroid isomerase.52 Substrate distortion was already part of the first enzyme mechanism formulated based on the knowledge of the actual structure of the active site, the ‘Phillips’ mechanism of hen egg white lysozyme85 and it is still considered relevant in the recent, most elegant revision of the general mechanism of retaining β-glycosidases.86,87
Conclusion
Our work elucidated the contribution of the substrate distortion to ODCase catalysis in structural and computational detail. Crystal structures showed that ODCase can distort the bond between the aromatic ring of a ligand and its C6 substituent, regardless of the latter’s charge or size. Ab initio calculations revealed that the distortion contributes 3.7 kcal/mol to the catalysis, which corresponds to 10–15% of ΔΔG‡ of the enzyme-catalyzed reaction. Our proposed mechanism is consistent with the numerous experimental results obtained thus far. Various factors have been identified to contribute to the enormous acceleration of the decarboxylation rate by ODCase. They include substrate distortion, which is described here, stabilization of negative charge at C6 by K7239,40,68 and at O4 by S12714, a hydrophobic environment to destabilize the carboxylate part of OMP and to accommodate the product carbon dioxide14, hydrophobic core formation to close the mobile loop43, the large binding energy of the phosphate part63–65, and probably others not yet considered.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to the staff members at the beamlines of the BioCARS sector of the Advanced Photon Source, at SPring-8, and at the Photon Factory for their help with data collection, to Ms. Wing M. Lau, Ms. Wanda Gillon and Mr. Kousuke Mouri for sample preparation and to Dr. Jian-Rong Su for helpful discussions. This work was supported in part by Grant-in-Aid for Scientific Research (C) (24570130 to MF and 23550035 to TI), Grants-in-Aid for Young Scientists (B) (20770081 and 2277012 to MF) from Japan Society for the Promotion of Science (JSPS) and Uehara Memorial Foundation (to MF) as well as the Canadian Institutes for Health Research (to LPK and EFP) and the Canada Research Chairs Program to EFP. Use of the beamlines at the Photon Factory and SPring-8 was approved by the Photon Factory Advisory Committee (Proposal 2006G164) and by the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal 2006B1212), respectively. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02-06CH11357. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Institute of General Medical Science grant P41GM103543 (formerly National Center for Research Resources P41RR007707).
Footnotes
‘ indicates that the residue belongs to the second subunit.
Throughout this paper, O6 of BMP refers to the oxygen atom connected to the C6 atom as shown in Fig. 1D. In the PDB deposition, the same atom is named O1 according to the data bank’s nomenclature.
ASSOCIATED CONTENT
Supporting Information:
Experimental details, crystallographic data and refinement statistics, estimated pKa values of the residues around the reaction center, and additional figures describing the summary of mutagenesis works thus far, the superimpositions, ESP charge difference and chemical structures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Pauling L. Nature. 1948;161:707. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 2.Amyes TL, Richard JP. Biochemistry. 2013 [Google Scholar]
- 3.Warshel A, Florian J. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5950. doi: 10.1073/pnas.95.11.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan BP, Wolfenden R. J. Am. Chem. Soc. 2004;126:14698. doi: 10.1021/ja0450049. [DOI] [PubMed] [Google Scholar]
- 5.Warshel A, Strajbl M, Villa J, Florian J. Biochemistry. 2000;39:14728. doi: 10.1021/bi000987h. [DOI] [PubMed] [Google Scholar]
- 6.Wu N, Mo Y, Gao J, Pai EF. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2017. doi: 10.1073/pnas.050417797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radzicka A, Wolfenden R. Science. 1995;267:90. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 8.Appleby TC, Kinsland C, Begley TP, Ealick SE. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2005. doi: 10.1073/pnas.259441296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris P, Navarro Poulsen JC, Jensen KF, Larsen S. Biochemistry. 2000;39:4217. doi: 10.1021/bi992952r. [DOI] [PubMed] [Google Scholar]
- 10.Miller BG, Hassell AM, Wolfenden R, Milburn MV, Short SA. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2011. doi: 10.1073/pnas.030409797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller BG, Butterfoss GL, Short SA, Wolfenden R. Biochemistry. 2001;40:6227. doi: 10.1021/bi0028993. [DOI] [PubMed] [Google Scholar]
- 12.Miller BG, Snider MJ, Wolfenden R, Short SA. J. Biol. Chem. 2001;276:15174. doi: 10.1074/jbc.M011429200. [DOI] [PubMed] [Google Scholar]
- 13.Yuan J, Cardenas AM, Gilbert HF, Palzkill T. Protein Sci. 2011;20:1891. doi: 10.1002/pro.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iiams V, Desai BJ, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. Biochemistry. 2011;50:8497. doi: 10.1021/bi2012355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujihashi M, Wei L, Kotra LP, Pai EF. J. Mol. Biol. 2009;387:1199. doi: 10.1016/j.jmb.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu N, Gillon W, Pai EF. Biochemistry. 2002;41:4002. doi: 10.1021/bi015758p. [DOI] [PubMed] [Google Scholar]
- 17.Fujihashi M, Bello AM, Poduch E, Wei L, Annedi SC, Pai EF, Kotra LP. J. Am. Chem. Soc. 2005;127:15048. doi: 10.1021/ja054865u. [DOI] [PubMed] [Google Scholar]
- 18.Poduch E, Bello AM, Tang S, Fujihashi M, Pai EF, Kotra LP. J. Med. Chem. 2006;49:4937. doi: 10.1021/jm060202r. [DOI] [PubMed] [Google Scholar]
- 19.Wittmann JG, Heinrich D, Gasow K, Frey A, Diederichsen U, Rudolph MG. Structure. 2008;16:82. doi: 10.1016/j.str.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Heinrich D, Diederichsen U, Rudolph MG. Chemistry. 2009;15:6619. doi: 10.1002/chem.200900397. [DOI] [PubMed] [Google Scholar]
- 21.Bello AM, Poduch E, Liu Y, Wei L, Crandall I, Wang X, Dyanand C, Kain KC, Pai EF, Kotra LP. J. Med. Chem. 2008;51:439. doi: 10.1021/jm7010673. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H, Hayakawa H, Miyasaka T. Tetrahedron. 1982;38:2635. [Google Scholar]
- 23.Sowa T, Ouchi S. Bull. Chem. Soc. Jpn. 1975;48:2084. [Google Scholar]
- 24.Ueda T, Yamamoto M, Yamane A, Imazawa M, Inoue H. J. Carb.-Nucleos.-Nucl. 1978;5:261. [Google Scholar]
- 25.Wu N, Christendat D, Dharamsi A, Pai EF. Acta Crystallogr. Sect. D. 2000;56:912. doi: 10.1107/s090744490000576x. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. In: Macromolecular Crystallography, Part A. Carter CW Jr, Sweet RM, editors. Vol. 276. Academic Press; 1997. p. 307. [Google Scholar]
- 27.Collaborative Computional Project, N. Acta Crystallogr. Sect. D. 1994;50:760. [Google Scholar]
- 28.Emsley P, Cowtan K. Acta Crystallogr. Sect. D. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Acta Crystallogr. Sect. D. 1999;55:247. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 30.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins. 2003;50:437. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 31.Ishida T. J. Chem. Phys. 2008;129:125105. doi: 10.1063/1.2977458. [DOI] [PubMed] [Google Scholar]
- 32.Ishida T. J. Am. Chem. Soc. 2010;132:7104. doi: 10.1021/ja100744h. [DOI] [PubMed] [Google Scholar]
- 33.Ishida T, Kato S. J. Am. Chem. Soc. 2003;125:12035. doi: 10.1021/ja021369m. [DOI] [PubMed] [Google Scholar]
- 34.Chan KK, Wood BM, Fedorov AA, Fedorov EV, Imker HJ, Amyes TL, Richard JP, Almo SC, Gerlt JA. Biochemistry. 2009;48:5518. doi: 10.1021/bi900623r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smiley JA, Paneth P, O'Leary MH, Bell JB, Jones ME. Biochemistry. 1991;30:6216. doi: 10.1021/bi00239a020. [DOI] [PubMed] [Google Scholar]
- 36.Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH. J. Chem. Theory Comput. 2011;7:525. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 37.Sondergaard CR, Olsson MHM, Rostkowski M, Jensen JH. J. Chem. Theory Comput. 2011;7:2284. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 38.Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. J. Am. Chem. Soc. 2008;130:1574. doi: 10.1021/ja710384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toth K, Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. J. Am. Chem. Soc. 2007;129:12946. doi: 10.1021/ja076222f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toth K, Amyes TL, Wood BM, Chan K, Gerlt JA, Richard JP. J. Am. Chem. Soc. 2010;132:7018. doi: 10.1021/ja102408k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Vleet JL, Reinhardt LA, Miller BG, Sievers A, Cleland WW. Biochemistry. 2008;47:798. doi: 10.1021/bi701664n. [DOI] [PubMed] [Google Scholar]
- 42.Fedorov D, Kitaura K. The Fragment Molecular Orbital Method: Practical Applications to Large Molecular Systems. Boca Raton: CRC Press; 2009. [Google Scholar]
- 43.Wood BM, Amyes TL, Fedorov AA, Fedorov EV, Shabila A, Almo SC, Richard JP, Gerlt JA. Biochemistry. 2010;49:3514. doi: 10.1021/bi100443a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu N, Pai EF. J. Biol. Chem. 2002;277:28080. doi: 10.1074/jbc.M202362200. [DOI] [PubMed] [Google Scholar]
- 45.Fujihashi M, Mito K, Pai EF, Miki K. J. Biol. Chem. 2013;288:9011. doi: 10.1074/jbc.M112.427252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao J. Curr. Opin. Struct. Biol. 2003;13:184. doi: 10.1016/s0959-440x(03)00041-1. [DOI] [PubMed] [Google Scholar]
- 47.Kamerlin SC, Chu ZT, Warshel A. J. Org. Chem. 2010;75:6391. doi: 10.1021/jo100651s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu H, Boone A, Yang W. J. Am. Chem. Soc. 2008;130:14493. doi: 10.1021/ja801202j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desai BJ, Wood BM, Fedorov AA, Fedorov EV, Goryanova B, Amyes TL, Richard JP, Almo SC, Gerlt JA. Biochemistry. 2012;51:8665. doi: 10.1021/bi301188k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poduch E, Wei L, Pai EF, Kotra LP. J. Med. Chem. 2008;51:432. doi: 10.1021/jm700968x. [DOI] [PubMed] [Google Scholar]
- 51.Bell JB, Jones ME. J. Biol. Chem. 1991;266:12662. [PubMed] [Google Scholar]
- 52.Ruben EA, Schwans JP, Sonnett M, Natarajan A, Gonzalez A, Tsai Y, Herschlag D. Biochemistry. 2013;52:1074. doi: 10.1021/bi301348x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips LM, Lee JK. J. Org. Chem. 2005;70:1211. doi: 10.1021/jo040279s. [DOI] [PubMed] [Google Scholar]
- 54.Vedadi M, Lew J, Artz J, Amani M, Zhao Y, Dong A, Wasney GA, Gao M, Hills T, Brokx S, Qiu W, Sharma S, Diassiti A, Alam Z, Melone M, Mulichak A, Wernimont A, Bray J, Loppnau P, Plotnikova O, Newberry K, Sundararajan E, Houston S, Walker J, Tempel W, Bochkarev A, Kozieradzki I, Edwards A, Arrowsmith C, Roos D, Kain K, Hui R. Mol. Biochem. Parasitol. 2007;151:100. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 55.Tokuoka K, Kusakari Y, Krungkrai SR, Matsumura H, Kai Y, Krungkrai J, Horii T, Inoue T. J. Biochem. 2008;143:69. doi: 10.1093/jb/mvm193. [DOI] [PubMed] [Google Scholar]
- 56.Warshel A, Florian J, Strajbl M, Villa J. Chem Bio Chem. 2001;2:109. doi: 10.1002/1439-7633(20010202)2:2<109::AID-CBIC109>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 57.Amyes TL, Ming SA, Goldman LM, Wood BM, Desai B, Gerlt JA, Richard JP. Biochemistry. 2012;51:4630. doi: 10.1021/bi300585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goryanova B, Amyes TL, Gerlt JA, Richard JP. J. Am. Chem. Soc. 2011;133:6545. doi: 10.1021/ja201734z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barnett SA, Amyes TL, Wood BM, Gerlt JA, Richard JP. Biochemistry. 2010;49:824. doi: 10.1021/bi902174q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harris P, Poulsen JC, Jensen KF, Larsen S. J. Mol. Biol. 2002;318:1019. doi: 10.1016/S0022-2836(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 61.Toth K, Amyes TL, Wood BM, Chan KK, Gerlt JA, Richard JP. Biochemistry. 2009;48:8006. doi: 10.1021/bi901064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood BM, Chan KK, Amyes TL, Richard JP, Gerlt JA. Biochemistry. 2009;48:5510. doi: 10.1021/bi9006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amyes TL, Richard JP, Tait JJ. J. Am. Chem. Soc. 2005;127:15708. doi: 10.1021/ja055493s. [DOI] [PubMed] [Google Scholar]
- 64.Sievers A, Wolfenden R. Bioorg. Chem. 2005;33:45. doi: 10.1016/j.bioorg.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Miller BG, Snider MJ, Short SA, Wolfenden R. Biochemistry. 2000;39:8113. doi: 10.1021/bi000818x. [DOI] [PubMed] [Google Scholar]
- 66.Shostak K, Jones ME. Biochemistry. 1992;31:12155. doi: 10.1021/bi00163a026. [DOI] [PubMed] [Google Scholar]
- 67.Sievers A, Wolfenden R. J. Am. Chem. Soc. 2002;124:13986. doi: 10.1021/ja021073g. [DOI] [PubMed] [Google Scholar]
- 68.Tsang WY, Wood BM, Wong FM, Wu W, Gerlt JA, Amyes TL, Richard JP. J. Am. Chem. Soc. 2012;134:14580. doi: 10.1021/ja3058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong FM, Capule CC, Wu W. Org. Lett. 2006;8:6019. doi: 10.1021/ol0624981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeoh FY, Cuasito RR, Capule CC, Wong FM, Wu W. Bioorg. Chem. 2007;35:338. doi: 10.1016/j.bioorg.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Levine HL, Brody RS, Westheimer FH. Biochemistry. 1980;19:4993. doi: 10.1021/bi00563a010. [DOI] [PubMed] [Google Scholar]
- 72.Miller BG, Traut TW, Wolfenden R. Bioorg. Chem. 1998;26:283. [Google Scholar]
- 73.Krungkrai SR, DelFraino BJ, Smiley JA, Prapunwattana P, Mitamura T, Horii T, Krungkrai J. Biochemistry. 2005;44:1643. doi: 10.1021/bi048439h. [DOI] [PubMed] [Google Scholar]
- 74.Smiley JA, Saleh L. Bioorg. Chem. 1999;27:297. [Google Scholar]
- 75.Bruice TC. Acc. Chem. Res. 2002;35:139. doi: 10.1021/ar0001665. [DOI] [PubMed] [Google Scholar]
- 76.Giraldo J, Roche D, Rovira X, Serra J. FEBS Lett. 2006;580:2170. doi: 10.1016/j.febslet.2006.03.060. [DOI] [PubMed] [Google Scholar]
- 77.Hur S, Bruice TC. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12015. doi: 10.1073/pnas.1534873100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chook YM, Ke H, Lipscomb WN. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8600. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee AY, Karplus PA, Ganem B, Clardy J. J. Am. Chem. Soc. 1995;117:3627. [Google Scholar]
- 80.Jensen KP, Ryde U. J. Am. Chem. Soc. 2005;127:9117. doi: 10.1021/ja050744i. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y, Schramm VL. Biochemistry. 2011 doi: 10.1021/bi200638x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hayashi H, Mizuguchi H, Miyahara I, Islam MM, Ikushiro H, Nakajima Y, Hirotsu K, Kagamiyama H. Biochim. Biophys. Acta. 2003;1647:103. doi: 10.1016/s1570-9639(03)00068-2. [DOI] [PubMed] [Google Scholar]
- 83.Jamaluddin H, Tumbale P, Withers SG, Acharya KR, Brew K. J. Mol. Biol. 2007;369:1270. doi: 10.1016/j.jmb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 84.Fabrichniy IP, Lehtio L, Tammenkoski M, Zyryanov AB, Oksanen E, Baykov AA, Lahti R, Goldman A. J. Biol. Chem. 2007;282:1422. doi: 10.1074/jbc.M513161200. [DOI] [PubMed] [Google Scholar]
- 85.Phillips DC. Proc. Natl. Acad. Sci. U. S. A. 1967;57:483. [Google Scholar]
- 86.Vocadlo DJ, Davies GJ, Laine R, Withers SG. Nature. 2001;412:835. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- 87.Zechel DL, Withers SG. Acc. Chem. Res. 2000;33:11. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.