Abstract
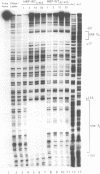
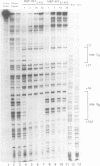
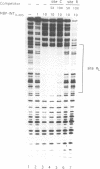
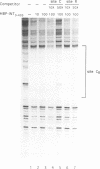
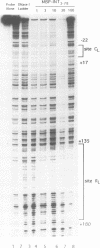
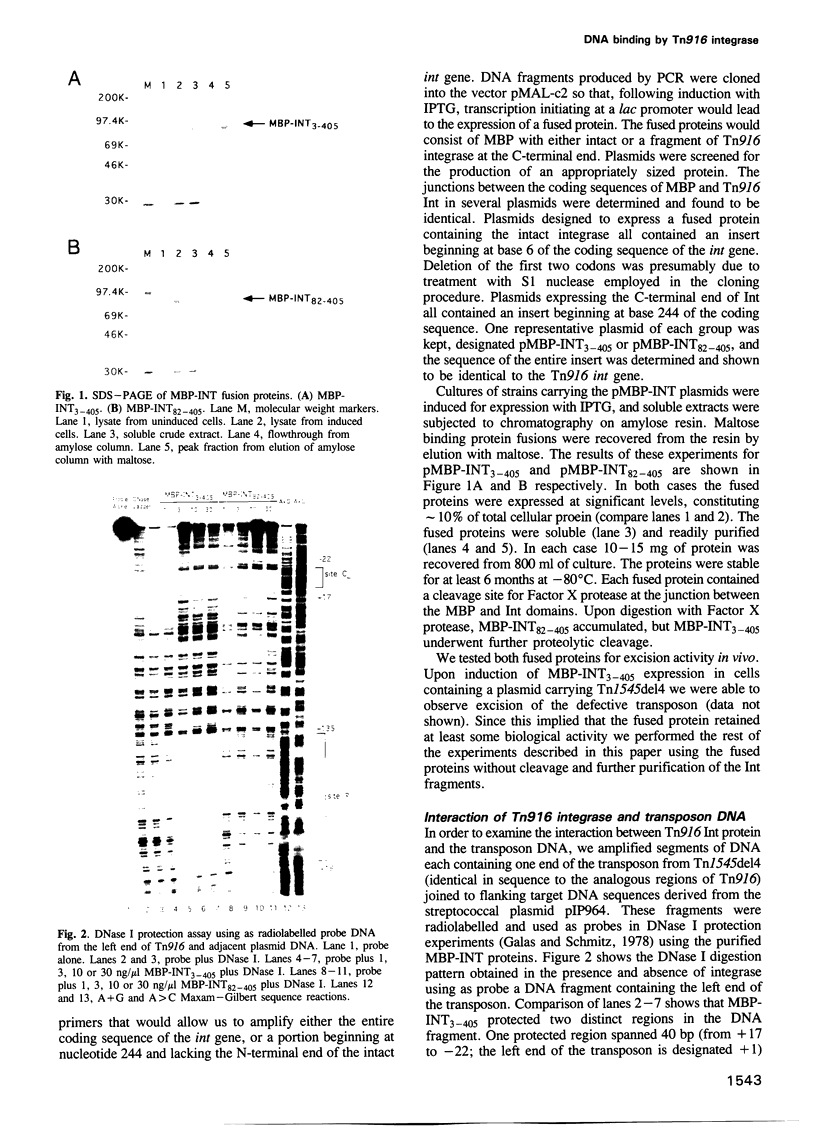
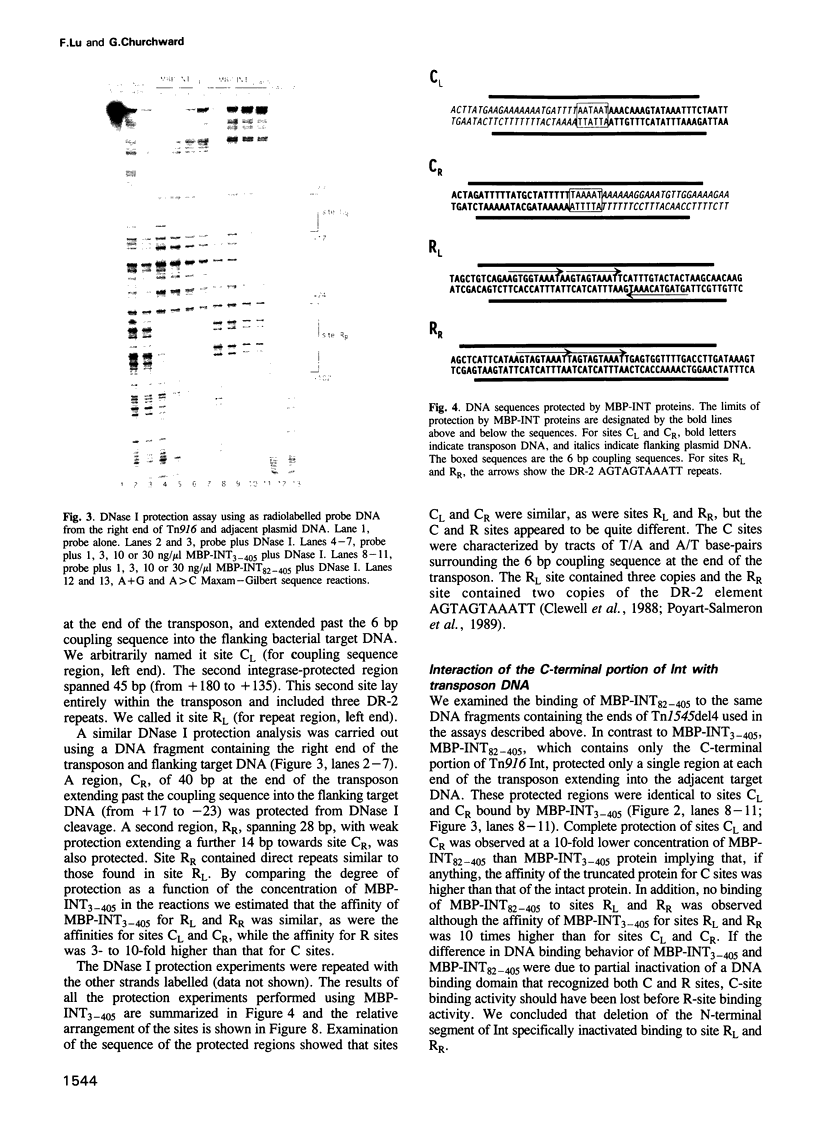
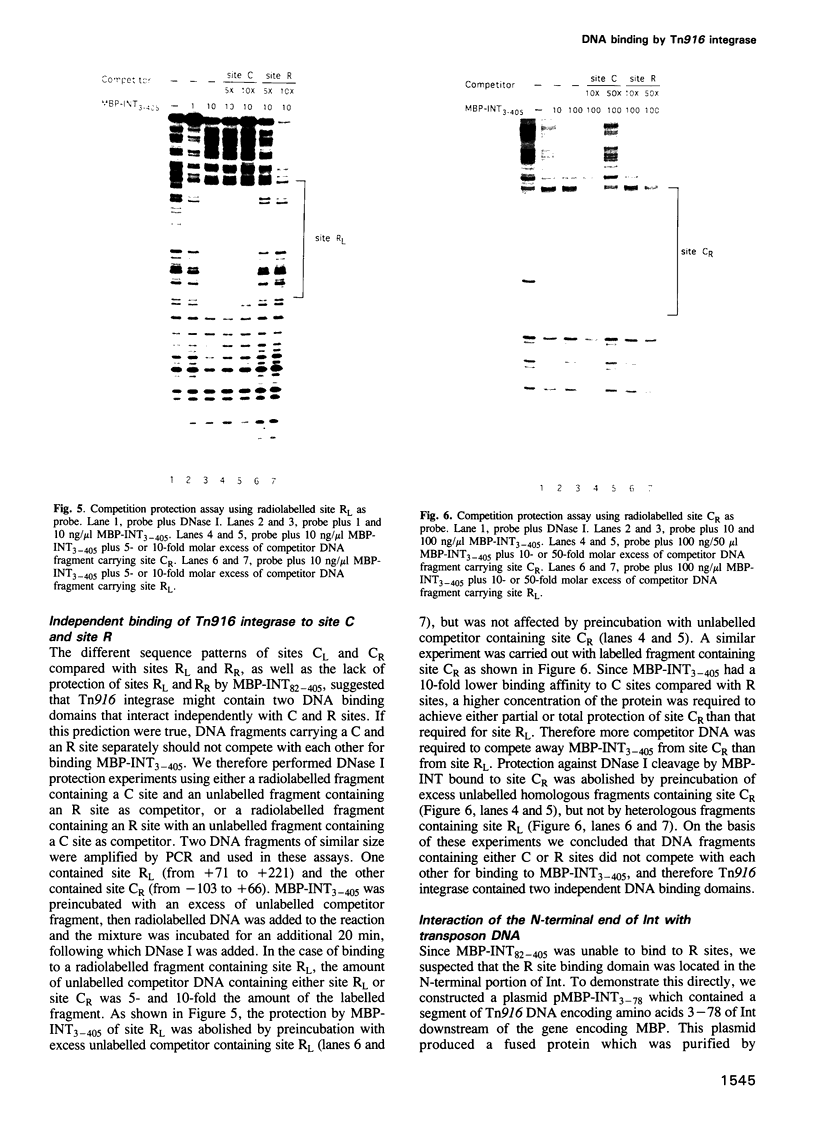
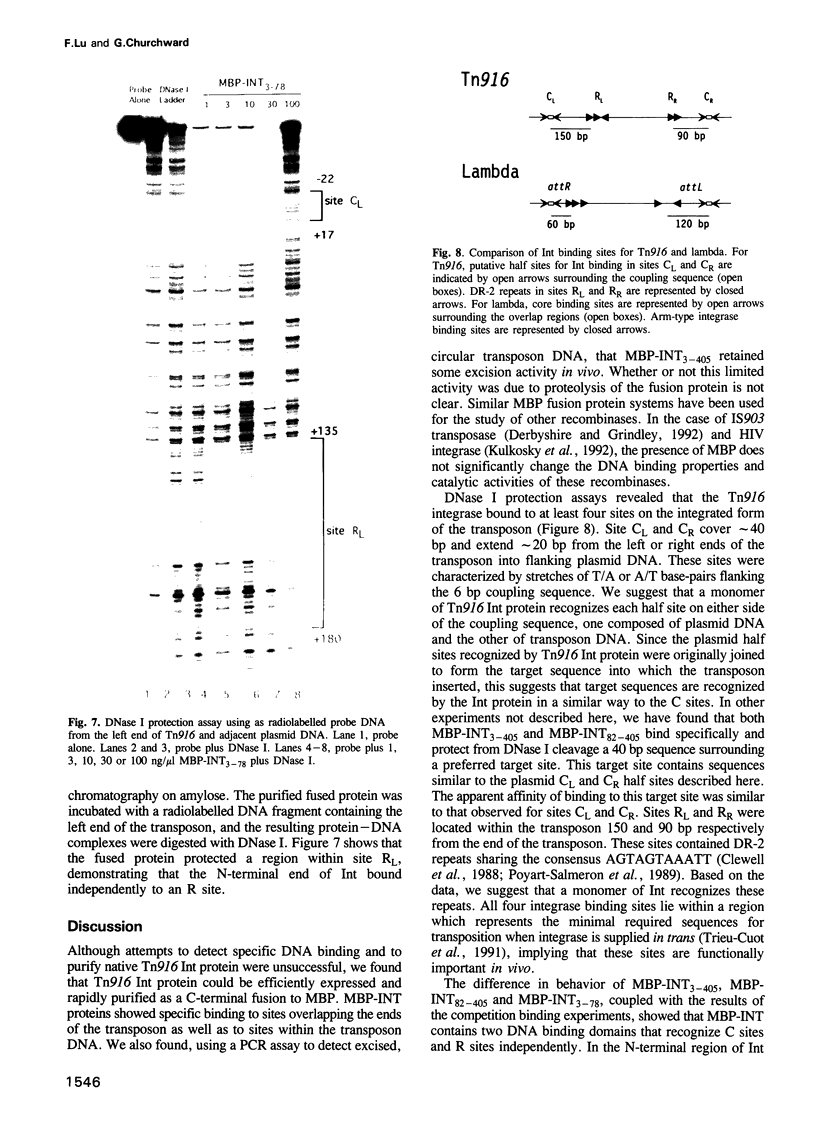
Transposition of the conjugative transposon Tn916 requires the activity of a protein, called Int, which is related to members of the integrase family of site-specific recombinases. This family includes phage lambda integrase as well as the Cre, FLP and XerC/XerD recombinases. Different proteins, consisting of fragments of Tn916 Int protein fused to the C-terminal end of maltose binding protein (MBP) were purified from Escherichia coli. DNase I protection experiments showed that MBP-INT proteins containing the C-terminal end of Int bound to the ends of the transposon and adjacent plasmid DNA. MBP-INT proteins containing the N-terminal end of Int bound to sequences within the transposon close to each end. Competition binding experiments showed that the sites recognized by the C- and N-terminal regions of Int did not compete with each other for binding to MBP-INT. We suggest that Tn916 and related conjugative transposons are unique among members of the integrase family of site-specific recombination systems because the presence of two DNA binding domains in the Int protein might allow Int to bridge recombining sites, and this bridging seems to be the sole mechanism ensuring that only correctly aligned molecules undergo recombination.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Bringel F., Van Alstine G. L., Scott J. R. Conjugative transposition of Tn916: the transposon int gene is required only in the donor. J Bacteriol. 1992 Jun;174(12):4036–4041. doi: 10.1128/jb.174.12.4036-4041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman W., Thompson J. F., Vargas L., Landy A. Control of directionality in lambda site specific recombination. Science. 1985 Nov 22;230(4728):906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon M. G., Scott J. R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989 Dec 22;59(6):1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P., Carlier C. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol Gen Genet. 1986 Nov;205(2):291–297. doi: 10.1007/BF00430441. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992 Sep;11(9):3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982 Nov 18;300(5889):281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Wierzbicki A., Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986 Mar 11;14(5):2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992 Apr 10;256(5054):198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Moitoso de Vargas L., Nunes-Düby S. E., Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990 Nov 16;63(4):773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J Mol Biol. 1988 Nov 5;204(1):95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Moffatt B. A., Studier F. W. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987 Apr 24;49(2):221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Pargellis C. A., Hasan N. M., Bushman E. W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988 Sep 23;54(7):923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989 Nov;8(11):3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Parsons R. L., Evans B. R., Zheng L., Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. Possible role of Arg-308 in coupling substrate binding to catalysis. J Biol Chem. 1990 Mar 15;265(8):4527–4533. [PubMed] [Google Scholar]
- Parsons R. L., Prasad P. V., Harshey R. M., Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol Cell Biol. 1988 Aug;8(8):3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989 Aug;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Scott J. R., Kirchman P. A., Caparon M. G. An intermediate in transposition of the conjugative transposon Tn916. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4809–4813. doi: 10.1073/pnas.85.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R. Sex and the single circle: conjugative transposition. J Bacteriol. 1992 Oct;174(19):6005–6010. doi: 10.1128/jb.174.19.6005-6010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecoff J. F., Cox M. M. Directionality in FLP protein-promoted site-specific recombination is mediated by DNA-DNA pairing. J Biol Chem. 1986 Jun 5;261(16):7380–7386. [PubMed] [Google Scholar]
- Senghas E., Jones J. M., Yamamoto M., Gawron-Burke C., Clewell D. B. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol. 1988 Jan;170(1):245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Catalysis by site-specific recombinases. Trends Genet. 1992 Dec;8(12):432–439. [PubMed] [Google Scholar]
- Storrs M. J., Poyart-Salmeron C., Trieu-Cuot P., Courvalin P. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase. J Bacteriol. 1991 Jul;173(14):4347–4352. doi: 10.1128/jb.173.14.4347-4352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Waechter-Brulla D., Gumport R. I., Gardner J. F., Moitoso de Vargas L., Landy A. Mutations in an integration host factor-binding site: effect on lambda site-specific recombination and regulatory implications. J Bacteriol. 1986 Dec;168(3):1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. An integrative vector exploiting the transposition properties of Tn1545 for insertional mutagenesis and cloning of genes from gram-positive bacteria. Gene. 1991 Sep 30;106(1):21–27. doi: 10.1016/0378-1119(91)90561-o. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot P., Poyart-Salmeron C., Carlier C., Courvalin P. Sequence requirements for target activity in site-specific recombination mediated by the Int protein of transposon Tn 1545. Mol Microbiol. 1993 Apr;8(1):179–185. doi: 10.1111/j.1365-2958.1993.tb01214.x. [DOI] [PubMed] [Google Scholar]