ABSTRACT
Burkholderia pseudomallei is the causative agent of melioidosis, an often fatal infectious disease for which there is no vaccine. B. pseudomallei is listed as a tier 1 select agent, and as current therapeutic options are limited due to its natural resistance to most antibiotics, the development of new antimicrobial therapies is imperative. To identify drug targets and better understand the complex B. pseudomallei genome, we sought a genome-wide approach to identify lethal gene targets. As B. pseudomallei has an unusually large genome spread over two chromosomes, an extensive screen was required to achieve a comprehensive analysis. Here we describe transposon-directed insertion site sequencing (TraDIS) of a library of over 106 transposon insertion mutants, which provides the level of genome saturation required to identify essential genes. Using this technique, we have identified a set of 505 genes that are predicted to be essential in B. pseudomallei K96243. To validate our screen, three genes predicted to be essential, pyrH, accA, and sodB, and a gene predicted to be nonessential, bpss0370, were independently investigated through the generation of conditional mutants. The conditional mutants confirmed the TraDIS predictions, showing that we have generated a list of genes predicted to be essential and demonstrating that this technique can be used to analyze complex genomes and thus be more widely applied.
IMPORTANCE
Burkholderia pseudomallei is a lethal human pathogen that is considered a potential bioterrorism threat and has limited treatment options due to an unusually high natural resistance to most antibiotics. We have identified a set of genes that are required for bacterial growth and thus are excellent candidates against which to develop potential novel antibiotics. To validate our approach, we constructed four mutants in which gene expression can be turned on and off conditionally to confirm that these genes are required for the bacteria to survive.
INTRODUCTION
Burkholderia pseudomallei is the causative agent of the human disease melioidosis, a severe disease that can manifest as a lethal acute infection or lay dormant as a chronic infection with the potential to reactivate decades later. Infection can occur through inhalation or ingestion of the bacteria or through skin abrasions. Dependent on the nature of the exposure, melioidosis can present as a localized skin ulcer or an ulceroglandular, intestinal, or acute pulmonary infection and can progress to systemic septicemia (1, 2). Due to the potential severity of melioidosis and its presumed ability to be spread by aerosols, B. pseudomallei is classified as a biosafety level 3 pathogen and has also been listed as a tier 1 select agent and potential bioterrorism threat by the U.S. Centers for Disease Control and Prevention. There is no licensed vaccine available to prevent melioidosis, and because B. pseudomallei demonstrates extraordinary resistance to many antibiotics, current therapeutic options are limited (3). Thus, the identification of novel drug targets is a research imperative.
Burkholderia pseudomallei has one of the largest and most complex genomes of any species of bacteria. The first strain to be fully sequenced, B. pseudomallei K96243, was found to contain approximately 6,332 predicted coding sequences within 7.25 Mb of DNA spread across two circular chromosomes (4, 5). This large genome encodes factors enabling the bacterium to persist in the environment as a soil saprophyte and also to act as a potent intracellular pathogen. The B. pseudomallei genome contains an unprecedented arsenal of potential virulence factors, including three type III secretion systems (T3SS), six type VI secretion systems (T6SS), multiple antibiotic resistance factors, and at least four polysaccharide gene clusters, including a capsular polysaccharide (4, 6, 7). In addition, the B. pseudomallei genome is highly plastic, demonstrating frequent acquisition of genomic islands by horizontal transfer (8). The size and recombinogenic nature of the genome mean that our understanding of the survival and pathogenesis of this important bacterium at the genetic level is still rudimentary.
The size and plasticity of the B. pseudomallei genome as well as the necessity to handle the pathogen under high-level containment conditions have made a comprehensive analysis of the genome difficult to achieve by traditional forward-genetics screening methods. Previous studies have used signature-tagged mutagenesis (STM) to identify novel virulence factors by screening pools of bacterial mutants (9, 10). However, these studies were limited by the technical constraints of STM screens, which allow pools of only 102 to 103 mutants to be analyzed. While these studies proved useful for identifying a limited number of virulence factors and even potential live-vaccine candidates (11), they were able to assay only a small portion of the genome and did not saturate the two chromosomes. More recently, technological advances have allowed transposon mutagenesis screens to be significantly scaled up by taking advantage of next-generation sequencing technology to efficiently identify transposon insertion sites. This facilitates the analysis of much larger pools of mutants using a technique known as transposon-directed insertion site sequencing (TraDIS) or a similar technique known as Tn-seq (12, 13).
Here we report the construction and sequencing of a large-scale transposon mutant library consisting of over 106 B. pseudomallei K96243 mutants and the analysis of this library by TraDIS. The ability to screen pools of this size has facilitated the characterization of a library with sufficient insertion density for the application of robust statistics to identify genes that are essential for the in vitro growth of B. pseudomallei K96243. This has enabled us to compile a comprehensive list of putative essential genes that can be exploited for select targets for antimicrobial development. The list includes known housekeeping genes, such as those encoding the ribosomal proteins and primary metabolic pathways, as well as core lipopolysaccharide biosynthesis genes and many genes encoding hypothetical proteins that have not previously been established as essential. Three genes selected from our list of genes predicted to be essential, pyrH, accA, and sodB, were independently confirmed to be essential through the generation of conditional mutants, validating our screening approach and providing a robust method to confirm gene essentiality. Furthermore, an additional gene, bpss0370, which is predicted to be essential through an in silico analysis but was shown to be nonessential through our TraDIS screen was confirmed as inessential for bacterial growth, suggesting that this method is more precise than a bioinformatic strategy. Our results provide new insight into the genome of B. pseudomallei K96243, present new targets for drug development, and demonstrate the potential of this technology to greatly expand our ability to characterize large and complex bacterial genomes on a comprehensive scale.
RESULTS
Construction and sequencing of a library of 1 million B. pseudomallei K96243 mutants.
To provide an appropriate saturation density for the identification of essential genes, a library of over 1 million bacterial mutants was constructed using a modified miniTn5 transposon (10). The transposon was delivered by direct mating with Escherichia coli 19851 pir+ carrying the plasmid pUTminiTn5Km2 (14). Approximately 250 individual pools of either 103 or 104 mutants were individually collected and combined to create a final library of approximately 106 mutants. To confirm that random single-insertion events were occurring, Southern blotting was performed using a probe directed against the kanamycin cassette within the transposon. Assays of randomly selected mutants showed that unique insertion events were occurring in every mutant tested, and analysis of pools of 5,000 mutants showed a range of insertions distributed across the genome (see Fig. S1 in the supplemental material).
Precise insertion sites for each mutant were identified using TraDIS (12). Briefly, genomic DNA was isolated from two biological replicates representing separate cultures of the entire pool of 106 mutants and sequenced using a primer specific to the 5′ end of the transposon and reading directly into the surrounding genome sequence (15). The reads were filtered for the presence of the transposon sequence and then mapped to the B. pseudomallei genome, revealing coverage across the entire genome (Fig. 1A). Over 80% of the total reads in both biological replicates contained the transposon-specific sequence, and 70 to 90% of those reads were conclusively mapped to the bacterial genome, resulting in a total of over 33 million reads mapped. Sequence reads that matched more than one region were discarded if we were unable to assign the read to a unique location on the genome. From the library of 106 mutants, we identified approximately 240,000 unique transposon insertion sites. Approximately 170,000 of the insertion sites were mapped to chromosome 1, resulting in an average of 1 transposon insertion every 25 bp, while only approximately 70,000 mapped to chromosome 2, resulting in 1 insertion every 45 bp (12).
FIG 1 .
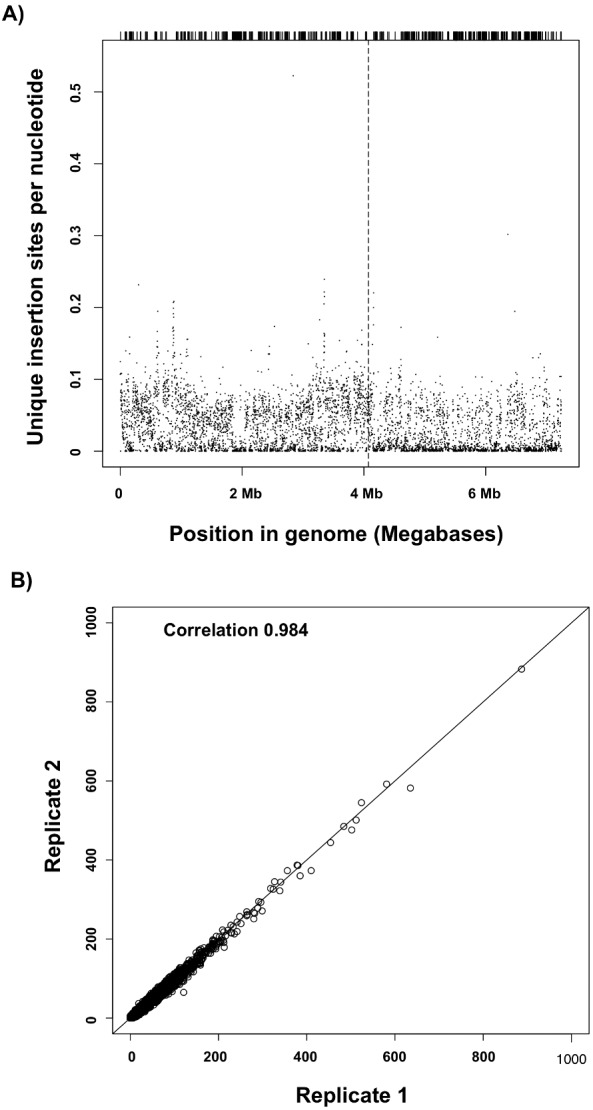
Distribution of transposon insertions along the B. pseudomallei K96243 genome. (A) The numbers of TraDIS reads mapped to each single nucleotide location are plotted along both chromosomes of the B. pseudomallei K96243 genome, demonstrating a representation of the entire genome. Chromosomes 1 and 2 are shown contiguously, with the dashed vertical line marking the boundary between chromosomes. The short lines along the top of the figure indicate the exact nucleotide location of each unique insertion site. (B) Gene insertion statistics for biological replicates overlaying the gene insertion indexes of the two biological replicates. A Spearman’s rho correlation of 0.984 was determined after we discarded two outliers, and a line of slope 1 through the origin is shown.
Identification of the essential genome of Burkholderia pseudomallei K96243.
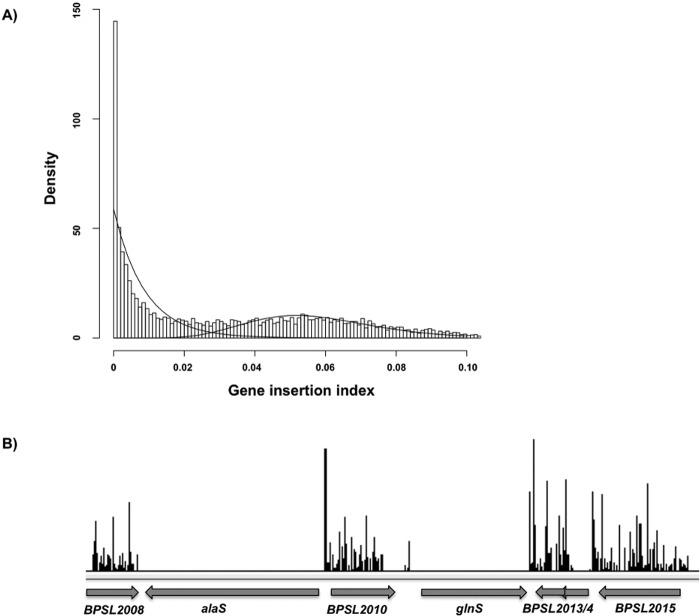
With the high transposon insertion density observed in our library, we predicted that genes with no or very few insertion sites are likely to be essential genes. To confirm this, we analyzed the number of insertion sites per gene after normalizing for gene length to create a gene insertion index. The gene insertion indexes determined over the two biological replicates were highly correlated (Spearman’s rho = 0.984) and concordant, validating the accuracy of our sequencing results (Fig. 1B). Performance of a density estimate of the frequency distribution of gene insertion indexes results in a bimodal curve in which the first sharp peak represents genes in which a transposon insertion would be lethal to the bacteria and the second elongated peak represents genes which can be mutated without affecting the viability of the bacterium (Fig. 2A). The gamma distributions of the density plot were used to estimate log2 likelihood ratios to distinguish essential from nonessential genes. With this method, we were able to predict which B. pseudomallei genes were essential for in vitro growth and survival based on a statistically significant lack of insertion sites. Two examples of genes which contain few or no insertion sites and are thus defined as essential are shown in Fig. 2B. As the frequency of transposon insertion was notably higher in chromosome 1 than in chromosome 2, we increased the stringency of our analysis for the second chromosome. This resulted in a list of 505 B. pseudomallei genes predicted to be essential (Table S1).
FIG 2 .
Defining the essential genes of B. pseudomallei K96243. (A) Density plot showing the frequency distribution of the gene insertion index. Shown is a clear bimodal distribution in which the leftmost peak represents genes in which a transposon insertion would be lethal to the bacteria, while the rightmost peak represents genes in which transposons were able to insert without causing lethality. Bars represent increments of 0.001. The lines shown indicate the gamma distributions used to estimate likelihood ratios and P values. (B) TraDIS reads are plotted along a small section of B. pseudomallei K96243 chromosome 1, demonstrating a lack of insertion sites in the putatively essential genes alaS and glnS. The height of each line along the y axis indicates the number of insertion sites at that location. The average numbers of reads from two biological replicates are shown
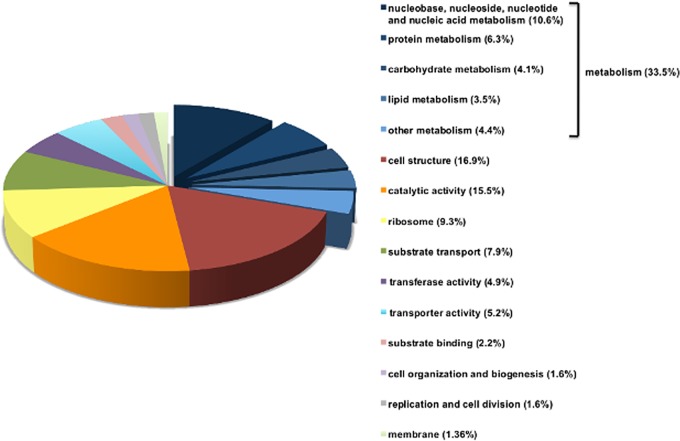
Included in our list of putative essential genes were a large number of genes which are essential in related bacterial species and genes which were previously predicted to be part of the core B. pseudomallei genome, as defined by conservation between multiple strains of B. pseudomallei (16). These include the ribosomal protein genes that are located in a large cluster of essential genes on chromosome 1 (bpsl3187 to bpsl3228), core components of the bacterium’s metabolic pathways, including a second large cluster containing the nuoA to nuoM genes, required for oxidative phosphorylation (bpsl1212 to bpsl1224), and numerous other genes predicted to be involved in amino acid and nucleotide biosynthesis. In addition, many of the genes involved in the biosynthesis of the lipopolysaccharide (LPS) inner core (bpsl2510, bpsl2665, bpsl0791) and peptidoglycan (bpsl3023 to bpsl3030) were also found to be essential, consistent with their roles in other bacteria. Of the 505 genes predicted to be essential, 319 were annotated with gene ontology (GO) terms which defined the predicted function of their gene product. The most highly represented functional categories are shown in Fig. 3. The remaining putative essential genes were categorized as encoding hypothetical proteins or conserved hypothetical proteins with no functional data available, presumably due to the high numbers of genes in Burkholderia pseudomallei that are as-yet uncharacterized.
FIG 3 .
Functional distribution of essential genes. Functional distribution of the 319 B. pseudomallei K96243 genes in our data set annotated with Gene Ontology (GO) terms. The largest subset of these genes was that associated with metabolic function (33.5%), including nucleic acid metabolism, protein and amino acid metabolism, and carbohydrate metabolism. The remainder of the known essential genes were distributed between essential functions, such as transcription, translation, and transport.
Confirmation of selected essential genes.
In order to confirm the utility of TraDIS for predicting essential genes, we chose four targets to validate individually. To conclusively determine whether these genes were required for growth in vitro, we utilized a method based on the construction of conditional lethal mutants. These were constructed by the insertion of a rhamnose-inducible promoter upstream of the predicted open reading frame (ORF). This results in the target gene being transcribed only in the presence of l-rhamnose, while transcription is abolished in the presence of glucose. Thus, if bacterial growth is demonstrated in media containing l-rhamnose but not in media containing glucose, an essential role for the target gene is established. Conditional lethal mutants were created in three B. pseudomallei genes selected from the TraDIS gene set and in a fourth that was predicted to be essential by other related methods but was not predicted to be essential in B. pseudomallei K962434 by TraDIS.
The first target gene chosen from the TraDIS gene set was pyrH (bpsl2157), which encodes uridylate kinase. Uridylate kinase catalyzes the reversible phosphorylation of UMP to UDP, thereby playing an essential role in the synthesis of pyrimidines (17). PyrH has been shown to be essential in 17 different bacterial species according to the database of essential genes (DEG), including E. coli, Pseudomonas aeruginosa, Mycobacterium tuberculosis, and Francisella tularensis (18). Our TraDIS analysis showed no insertions within the coding sequence of pyrH in any library mutants (Fig. 4A), suggesting that pyrH is also essential in B. pseudomallei. To confirm essentiality, the conditional pyrH mutant of B. pseudomallei was grown in minimal medium supplemented with either l-rhamnose (promoter on) or glucose (promoter off) for two passages at 37°C overnight. A second passage in glucose resulted in no visible growth (Fig. 4A), thereby confirming that pyrH is essential for B. pseudomallei viability.
FIG 4 .
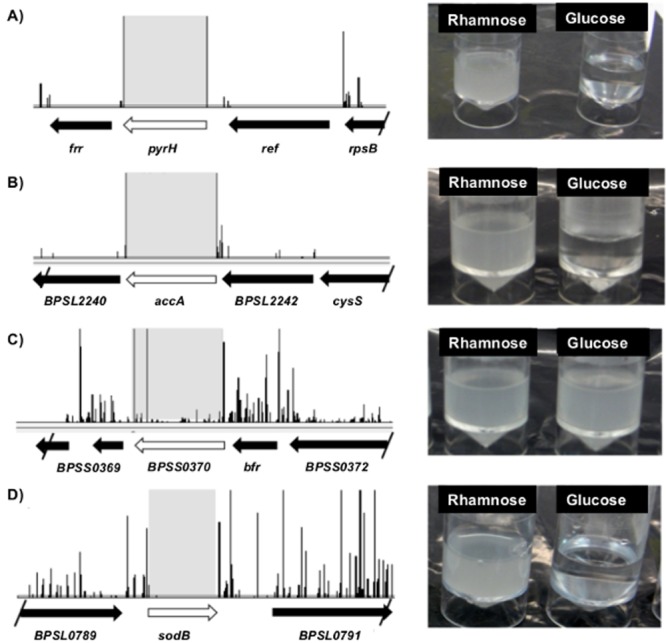
Confirmation of selected essential genes. (A to D) Distribution of hits along the B. pseudomallei genome in the regions surrounding the genes pyrH (A), accA (B), bpss0370 (C), and sodB (D). The height of the y axis represents the number of unique insertion sites located at each position. To the right of each plot is shown overnight broth cultures of each conditional mutant supplemented with either l-rhamnose (promoter on) or glucose (promoter off), demonstrating a lack of growth when essential genes are not induced.
The second target selected was accA (bspl2241), which encodes the acetyl coenzyme A (acetyl-CoA) carboxylase A enzyme. AccA is a subunit of an essential enzyme complex in the biosynthesis of fatty acids by catalyzing the carboxylation of acetyl-CoA to form malonyl-CoA (19). AccA has been shown to be essential in 12 different bacterial species according to the DEG, including E. coli, P. aeruginosa, M. tuberculosis, and F. tularensis. Our TraDIS analysis showed no insertions within the coding sequence of accA in one biological replicate and one hit in the second biological replicate and was predicted to be an essential gene (Fig. 4B). As for pyrH, the conditional accA mutant of B. pseudomallei failed to grow after repeated passage in the presence of glucose, demonstrating that this gene is essential (Fig. 4B).
The third target gene analyzed was bpss0370, encoding a glutamate racemase enzyme. These enzymes convert l-glutamate to d-glutamate, an essential component of bacterial peptidoglycan (20). According to the DEG, glutamate racemases are essential in 12 different bacterial species, including E. coli, P. aeruginosa, and F. tularensis. Surprisingly, our TraDIS analysis showed 34 unique transposon insertion sites within the coding sequence of bpss0370 (Fig. 4C). This suggested that the glutamate racemase enzyme of B. pseudomallei is not essential for growth under the conditions of this study. Indeed, the conditional bpss0370 mutant was able to grow in the presence of glucose (Fig. 4C).
Finally, we assessed the essentiality of sodB (bpsl0880), encoding an iron- and manganese-cofactored superoxide dismutase (Sod). Sod enzymes catalyze the degradation of toxic superoxide radicals to hydrogen peroxide and oxygen and thereby play an essential role in the resistance to host cell killing and in intracellular survival in many intracellular pathogens (21). SodB is not essential in E. coli, and the DEG contains only three hits for this enzyme in Francisella novicida, Acinetobacter baylyi, and P. aeruginosa. TraDIS screening showed only one transposon insertion site within the sodB sequence, compared to 68 unique insertion sites in the surrounding intergenic regions (Fig. 4D). Thus, our windowed algorithm determined that sodB is predicted to be essential in B. pseudomallei. Using the conditional lethal sodB mutant of B. pseudomallei, we were able to confirm that the mutant was unable to grow in the presence of glucose, and thus sodB is an essential gene (Fig. 4D). This demonstrated that in four out of four cases tested, the data from our conditional mutants confirmed the TraDIS prediction, confirming the validity of applying this technique to identify putative essential genes.
DISCUSSION
In order to survive in multiple environments and cause clinically diverse forms of disease, B. pseudomallei has evolved a large and highly plastic genome consisting of two chromosomes. In addition, the highly virulent nature of this pathogen and the fact that it is a listed select agent and thus a potential bioterrorism threat makes developing new vaccines and therapies against B. pseudomallei a high priority. Previous studies using transposon insertion mutants have had some success at identifying B. pseudomallei virulence factors and even candidates for live attenuated vaccines (9, 10). However, due to the technology at the time, none of these studies were capable of being performed at sufficient scales to achieve genome saturation and thus were of limited use for identifying potential essential genes.
TraDIS technology represents a breakthrough in the methods available to study bacterial pathogens. Previously, TraDIS has successfully been applied to identify the essential genes of Salmonella enterica serovar Typhi (12) and to extend an in vivo STM study of Escherichia coli O157 (15). In addition, a similar approach known as Tn-seq has been used to investigate the genomes of a growing list of pathogens, including P. aeruginosa, Vibrio cholerae, and Streptococcus pneumoniae (22–24). More recently, a Tn-seq analysis of smaller pools of approximately 200,000 mutants has been applied to the related nonpathogenic species Burkholderia thailandensis (25). The work described here expands on these previous studies by demonstrating that TraDIS can be applied to one of the largest and most complex bacterial genomes in a highly virulent biosafety level 3 pathogen. The identification of essential genes is of particular importance for B. pseudomallei because it is naturally extremely resistant to many commonly used antibiotics and because essential gene products may be excellent targets for novel antimicrobial drugs. A large and comprehensive list of essential targets provides a significant advantage because it allows us both to pick and choose targets, such as enzymes, that may be more easily inhibited with a lethal effect on the bacterium and to design drugs that can target more than one essential gene and thus minimize the development of resistance. In addition, this list of putative essential genes provides an invaluable resource for the B. pseudomallei research community, providing information on every single gene in the B. pseudomallei K96243 genome and predicting whether mutating a gene of interest would be viable.
We found that the modified miniTn5 transposon used to create our TraDIS library randomly inserted into the genome, consistent with the results of previous studies. However, we noticed a higher density of insertions in chromosome 1 than in the smaller chromosome 2. This means that there is potentially a higher rate of false discovery in the second chromosome than in the first chromosome due to an increased chance of genes being unrepresented in the transposon library by random chance. A lower frequency of transposon insertion is particularly evident in the regions surrounding the three type III secretion systems (T3SS) and type VI secretion systems (T6SS). However, the insertional bias has largely been overcome by analyzing a large number of mutants, and we have compensated for the differences in insertion density by increasing the stringency of our analysis of data originating from the second chromosome.
We also saw a slight bias in the frequency of transposon insertions based on the GC content of the DNA sequence. MiniTn5 transposons are thought to have relatively unbiased insertion sites compared to those of other transposon systems due to their flexibility in target site recognition (26). However, an insertion site bias has previously been observed in regions with a GC skew, such as in the origin and terminus of replication (27). The B. pseudomallei genome is highly GC rich compared to other bacteria but also contains a number of genomic islands that have enriched AT contents (4), in which we found higher concentrations of transposon insertions. In addition, the capsular polysaccharide synthesis I (CPS I) locus of B. pseudomallei K96243 located in the region of bpsl2787 to bpsl2810, which has an overall lower GC content than the rest of the genome, was highly represented by transposon insertion mutants, with many of the genes in this locus containing over 100 unique insertion sites. This may explain why mutations in this region were preferentially identified in previous STM screens (9, 10). In regions with a high transposon density, it is more likely that essential genes contain nonattenuating insertions, as the frequency of insertion makes it more likely that any regions of the gene that are not required for essential gene function contain transposon insertions by chance alone. However, the windowed algorithm used in this study improves our ability to differentiate these genes from nonessential genes, though as with any high-throughput screen, genes must be further assessed on an individual basis.
Previous large-scale genomic sequencing studies comparing different strains of B. pseudomallei have identified a core genome that is conserved between all strains (5, 16). The core genome was predicted to consist of 4,619 open reading frames (ORFs), of which 2,590 were assigned to functional categories based on homology. Genes from a number of functional categories were highly enriched within the core genome, including genes for amino acid transport and metabolism (377 genes), inorganic ion transport and metabolism (199 genes), nucleotide transport and metabolism (78 genes), protein translation (158 genes), and virulence components (321 genes) (16). As expected, we found that all of these functional gene categories, with the exception of the virulence component category, are strongly represented within our data set (Fig. 3). Our finding that virulence factor genes are enriched in the core genome but not in the essential-gene list is consistent with the likelihood that these genes provide an advantage to the pathogen in a mammalian host but are not required for bacterial growth or survival in vitro.
We found that of the 394 essential genes previously identified in B. thailandensis (25), we were able to identify orthologues of 224 within B. pseudomallei K96243. The similarity of the results from these two studies is interesting, as the two species are thought to have diverged approximately 47 million years ago and both have highly plastic genomes, resulting in many divergent loci. Notably, the metabolic pathways of B. pseudomallei and B. thailandensis are known to be divergent, as the ability of B. thailandensis but not B. pseudomallei to utilize arabinose and xylose as carbon sources is well characterized, which would explain the larger number of essential genes predicted in B. pseudomallei (28).
The robustness of our study is supported by the finding that all four of our conditional essential mutants confirmed our TraDIS predictions. In each example that we explored, we found that our TraDIS predictions were more accurate at predicting essential B. pseudomallei genes than both an in silico prediction of essential B. pseudomallei K96243 genes (29) and the previous Tn-seq experiment performed with the related nonpathogenic species B. thailandensis E264 (3). For example, our B. pseudomallei K96243 TraDIS study predicted both pyrH and accA to be essential, and this was confirmed with our conditional mutants. In contrast, only pyrH was predicted to be essential by the in silico analysis of B. pseudomallei K96243 and by B. thailandensis TraDIS. Similarly, we found sodB to be an essential gene, whereas it was not identified in the in silico set of essential genes, and the B. thailandensis homologue (bth_I0744) was not predicted to be essential by Tn-seq. Since sodB often plays a role in resistance to oxidative stress and macrophage killing, it is not necessarily predicted to have an essential role in in vitro growth (21). However, in other bacteria, including F. tularensis and L. pneumophila, this gene is required for aerobic growth in vitro (30, 31). Clearly, our data set includes essential genes that have not been described in previous studies reporting smaller gene lists.
As a final proof of principle, we examined the putative murI homologue bpss0370. Glutamate racemases encoded by murI play a role in peptidoglycan synthesis and are essential in many other bacteria (20). Unsurprisingly, this gene was predicted to be essential in the in silico analysis of B. pseudomallei K96243 (29). However, we conclusively demonstrated by TraDIS and through our subsequent studies with the conditional mutant that murI is not essential in B. pseudomallei K96243. This suggests that B. pseudomallei either is able to obtain d-glutamate through a different pathway or has an altered peptidoglycan synthesis pathway compared to that of other bacteria. Confirming our TraDIS predictions for the four genes described in this work demonstrates the importance of providing biological evidence for essentiality by showing how much more accurate TraDIS is at predicting essential genes than a bioinformatic analysis. In addition, our TraDIS data highlight some of the unique features of the B. pseudomallei K96243 genome compared to the genomes of other species of bacteria and demonstrate the utility of performing this analysis directly in the pathogenic species.
In conclusion, the application of TraDIS to the study of B. pseudomallei K96243 has been successful at identifying putative essential genes and has provided a wealth of new targets for future antimicrobial development. Further, we have validated our technique through the generation of conditional mutants and demonstrated the robustness of this technique. The library constructed in this study provides an important tool for investigating the genome of this important human pathogen, and we welcome future collaborations for large-scale in vitro and in vivo screening that could potentially define the role of every B. pseudomallei K96243 gene in many aspects of pathogenicity and environmental survival.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. pseudomallei strain K96243, a clinical isolate from Thailand, was used for the construction of the TraDIS library (4). All experiments using the TraDIS library were performed in Luria-Bertani (LB) broth or agar at 37°C. Conditional-mutant experiments were performed in a modified M9 medium as described below. Escherichia coli 19851 (pir+) was used for direct conjugation. When necessary, plates and cultures were supplemented with antibiotics at the following concentrations: 100 µg/ml for phleomycin (Zeocin; Life Technologies), 400 µg/ml for kanamycin, and 100 µg/ml for ampicillin.
Transposon and plasmids.
The transposon used in these studies was a modified miniTn5Km2 transposon as described in the work of Cuccui et al. (10). The transposon was carried on the plasmid pUT, which can be maintained only in pir+ strains due to an R6K origin of replication and is thus a suicide plasmid in B. pseudomallei.
Library construction.
The miniTn5Km2 transposon was delivered into B. pseudomallei K96243 by direct conjugation with an E. coli strain carrying the plasmid. The conjugations were performed overnight on LB plates at 24°C. The colonies were then scraped and resuspended in phosphate-buffered saline (PBS) and plated onto antibiotic plates supplemented with kanamycin and phleomycin to select for B. pseudomallei containing transposon insertions. Conditions were optimized to produce roughly 100 to 300 colonies per plate to allow selection and prevent clonal expansion of mutants. Colonies were then collected and frozen directly from plates in 25% glycerol (PBS). Pools of 103, 104, and 5 × 104 organisms were collected and frozen to create a total of 106 mutants. The TraDIS library will be made available as a resource to the community (contact the corresponding author).
Genomic DNA extraction.
Ten milliliters of each overnight, shaken culture was spun down at 4,000 rpm in a benchtop centrifuge and resuspended in 10 ml of lysis buffer (100 µg/ml proteinase K, 10 ml NaCl, 20 ml Tris-HCl, pH 8, 1 mM EDTA, 0.5% SDS). Three milliliters of sodium perchlorate was added to the solution, and the solution was incubated for 1 h at room temperature. Genomic DNA was isolated using a phenol-chloroform-isoamyl alcohol (25:24:1) extraction, precipitated with ethanol, and spooled into deionized water.
Southern blots.
Southern blotting was used to test individual colonies and to confirm individual random-insertion events. Briefly, genomic DNA was extracted from individual colonies, and 3 µg was digested using the SacI restriction enzyme (NEB). The resulting fragments were run on a 1% agarose gel overnight at 25 V and 500 mA. The DNA was denatured for 30 min in 1.5 M NaCl, 0.5 M NaOH, and the gel was then neutralized for 30 min in 0.5 M Tris-Cl, pH 7.2, 1 M NaCl. The samples were transferred overnight to a Hybond N membrane (Amersham) via capillary action in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The DNA was then cross-linked to the membrane with UV light using a UV Stratalinker 1800 (Stratagene). A DNA probe against the kanamycin cassette in the transposon was generated by PCR using the primers KanF (5′ CGACTGAATCCGGTGAGAAT 3′) and KanR (5′ CCGCGATTAAATTCCAACAT 3′). The probe was labeled and hybridized to the membrane using an AlkPhos direct labeling and detection kit (GE Healthcare) as per the manufacturer’s instructions.
Illumina sequencing.
For the sequencing of the TraDIS libraries, approximately 2 µg of genomic DNA from each sample was fragmented to ~200 bp by ultrasonication using a Covaris instrument. The fragmented DNA was end repaired and A-tailed using the NEBNext DNA library preparation reagent kit for Illumina sequencing (NEB). Annealed adapters, Ind_Ad_T (ACACTCTTTCCCTACACGACGCTCTTCCGATC*T, where * indicates phosphorothioate) and Ind_Ad_B (pGATCGGAAGAGCGGTTCAGCAGGAATGCCGAGACCGATCTC) were ligated to the samples. PCR was performed using primers PE_PCR_V3.3 (CAAGCAGAAGACGGCATACGAGATCGGTACACTCTTTCCCTACACGACGCTCTTCCGATC) and MnTn5_P5_3pr_3 (AATGATACGGCGACCACCGAGATCTACACCTAGGCtGCGGCtGCACTTGTG), which include flow cell binding sites. The PCR program used was 2 min at 94°C; 22 cycles of 30 s at 94°C, 20 s at 65°C, and 30 s at 72°C; and 10 min at 72°C. Sequences were then size selected to between 200 and 400 bp in a 2% agarose gel made up with 1× Tris-borate-EDTA (TBE) buffer, with purification with a Qiagen gel extraction kit. The final concentrations of the samples were checked both with a Bioanalyzer and by quantitative PCR (qPCR). Preparation products were sequenced on an Illumina Hi-Seq 2000 platform as 36-bp single-end reads. The concentrations of the samples were established using qPCR with the primers Syb_FP5 (ATGATACGGCGACCACCGAG) and Syb_RP7 (CAAGCAGAAGACGGCATACGAG). They were then size selected to between 300 and 500 bp in a 2% agarose gel made up with 1× TBE buffer, with purification with a Qiagen gel extraction kit. The final concentrations of the samples were checked both with a Bioanalyzer and by qPCR. Preparation products were sequenced on an Illumina Hi-Seq 2000 sequencer as 100 bp single-end reads.
Generation of conditional mutants.
Conditional lethal mutants of B. pseudomallei were created using the pSC200 plasmid system (32). The plasmid encodes a multiple-cloning site downstream of the rhamnose-inducible promoter (PrhaB), its activators (rhaR and rhaS), a dhfr cassette mediating resistance to trimethoprim, a mob gene for conjugation, and an oriR6K gene, which forces the plasmid to become integrated into the chromosome in the absence of pir genes. K96243 genomic DNA fragments spanning the first 250 to 260 bp of the coding sequence of each target gene were amplified by PCR using the Failsafe PCR system (Epicenter) and B. pseudomallei K96243 DNA as a template. The fragments were cloned into pSC200 via its NdeI/XbaI restrictions sites, and resulting plasmids were maintained in E. coli DH5α λpir. The pSC200 derivatives were introduced into B. pseudomallei strain K96243 by triparental mating using an E. coli helper strain carrying plasmid pRK2013 (33). Chromosomal integrants were selected for on LB agar plates containing 100 µg/ml trimethoprim and 0.5% (wt/vol) l-rhamnose. All mutants were confirmed by PCR using either the primers pSC1300 (TAACGGTTGTGGACAACAAGCCAGGG) and pSC_5183_fw (CTCCTGATGTCGTCAACACGG), which bind within the plasmid sequence, or the following primers, which are located 500 bp into the flanking regions within the chromosome of each gene: pyrH-wt-rv (5′ ACCTTGCCCTCCTCGAGCTG 3′), pyrH-up-fw (5′ GATCGAGCAGATGCTCAAGG 3′), accA-wt-rv (5′ GGCGCGGCATCCCGAAATTG 3′), murI-wt-rv (5′ GCGTCGCCTGGGTGGCGAG 3′), sodB-wt-rv (5′ GCGCGTTGCGGTAATCGATG 3′), and sodB-up-fw (5′ GATGCACGTGGGGCAGCTCG 3′). All conditional mutants were also confirmed by sequencing. Fragments (700 bp) spanning the rhaB promoter region and 500 bp into the downstream target gene were PCR amplified using primer pSC_5183_fw and the wt-rv primer for each target gene. PCR products were sequenced using primer pSC_5183_fw.
Lethality screening.
In order to assess whether a target gene is essential for viability, 5 ml of M9 minimal medium (1× M9 salts, 20 mM succinic acid, 2 mM MgSO4, and 0.1 mM CaCl2) supplemented with 100 µg/ml trimethoprim and either 0.5% (wt/vol) l-rhamnose or 0.5% (wt/vol) glucose was inoculated with one colony of a conditional mutant from a plate, and the cultures were incubated with aeration at 37°C for 24 h. Ten-microliter samples of these cultures were transferred to plates with fresh M9 medium containing either antibiotic, rhamnose, or glucose, and the cultures were incubated for an additional 24 h at 37°C. Growth was assessed by measuring the optical densities of the cultures at 595 nm.
Bioinformatic and statistical analysis.
Raw reads that passed Trimmomatic quality control filters and contained the transposon were mapped in the B. pseudomallei K96243 reference genome (version 6) using Bowtie (version 1.0.0) (34), allowing for zero mismatches and excluding non-uniquely mapped reads. An in-house pipeline based on the SAMtools (http://samtools.sourceforge.net) and BCFtools toolkits were applied to the alignment files to determine insertion sites and coverage. Raw data files showing the number of unique transposon insertion sites identified in each gene are available for download at http://lshtm.name/Tradis. An insertion index was calculated for each gene by dividing its number of unique insertion sites by its length. Insertion ratios across genes were observed to fit a bimodal distribution corresponding to essential and nonessential sets. In particular, each of the modes was modeled using a gamma distribution or an exponential distribution to fit genes with no observed insertion sites. This framework allowed the calculation of log2 likelihood ratios and corresponding P values for each gene, with their ranking determined by inferred essentiality. All statistical analysis was performed using the R software. Gene ontology classification was performed using CateGOrizer (http://www.animalgenome.org/tools/catego/) with a GOSlim modified for use with prokaryotes (35).
Nucleotide sequence accession number.
The fastq files containing the raw sequencing data have been uploaded to the European Nucleotide Archive (http://www.ebi.ac.uk) under study identification number PRJEB5123.
SUPPLEMENTAL MATERIAL
Southern blots of individual B. pseudomallei K96243 transposon mutants indicating unique insertion sites. Southern blots of 10 individual B. pseudomallei K96243 transposon insertion mutants in which we used a probe designed against the kanamycin cassette located within the transposon, indicating that the transposon has inserted into unique positions in each mutant. Download
Promoter regions of the conditional essential mutants. The sequences of the promoter regions of the accA, bpss0370, pyrH, and sodB mutants are shown aligned with the original sequence of the plasmid pSC200, from which they were derived. The ATG start sequence of each gene is highlighted in blue within the NdeI restriction enzyme site that was used for cloning. All promoter region sequences are shown to be identical. Download
Predicted essential genes of B. pseudomallei K96243.
ACKNOWLEDGMENTS
This work was partially funded by the Defence Science and Technology Laboratories (DSTL).
We thank Helen S. Atkins, Richard J. Saint, Tim Atkins, Gemma C. Langridge, Sabine E. Eckert, and Sam Willcocks for their discussions and contributions to this study.
Footnotes
Citation Moule MG, Hemsley CM, Seet Q, Guerra-Assunção JA, Lim J, Sarkar-Tyson M, Clark TG, Tan PBO, Titball RW, Cuccui J, Wren BW. 2014. Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5(1):e00926-13. doi:10.1128/mBio.00926-13.
REFERENCES
- 1. Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N. Engl. J. Med. 367:1035–1044. 10.1056/NEJMra1204699 [DOI] [PubMed] [Google Scholar]
- 2. Dance DA. 1991. Melioidosis: the tip of the iceberg? Clin. Microbiol. Rev. 4:52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thibault FM, Hernandez E, Vidal DR, Girardet M, Cavallo JD. 2004. Antibiotic susceptibility of 65 isolates of Burkholderia pseudomallei and Burkholderia mallei to 35 antimicrobial agents. J. Antimicrob. Chemother. 54:1134–1138. 10.1093/jac/dkh471 [DOI] [PubMed] [Google Scholar]
- 4. Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245. 10.1073/pnas.0403302101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nandi T, Ong C, Singh AP, Boddey J, Atkins T, Sarkar-Tyson M, Essex-Lopresti AE, Chua HH, Pearson T, Kreisberg JF, Nilsson C, Ariyaratne P, Ronning C, Losada L, Ruan Y, Sung WK, Woods D, Titball RW, Beacham I, Peak I, Keim P, Nierman WC, Tan P. 2010. A genomic survey of positive selection in Burkholderia pseudomallei provides insights into the evolution of accidental virulence. PLoS Pathog. 6:e1000845. 10.1371/journal.ppat.1000845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarkar-Tyson M, Thwaite JE, Harding SV, Smither SJ, Oyston PC, Atkins TP, Titball RW. 2007. Polysaccharides and virulence of Burkholderia pseudomallei. J. Med. Microbiol. 56:1005–1010. 10.1099/jmm.0.47043-0 [DOI] [PubMed] [Google Scholar]
- 7. Shalom G, Shaw JG, Thomas MS. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699. 10.1099/mic.0.2007/006585-0 [DOI] [PubMed] [Google Scholar]
- 8. Tumapa S, Holden MT, Vesaratchavest M, Wuthiekanun V, Limmathurotsakul D, Chierakul W, Feil EJ, Currie BJ, Day NP, Nierman WC, Peacock SJ. 2008. Burkholderia pseudomallei genome plasticity associated with genomic island variation. BMC Genomics 9:190. 10.1186/1471-2164-9-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atkins T, Prior R, Mack K, Russell P, Nelson M, Prior J, Ellis J, Oyston PC, Dougan G, Titball RW. 2002. Characterisation of an acapsular mutant of Burkholderia pseudomallei identified by signature tagged mutagenesis. J. Med. Microbiol. 51:539–547 [DOI] [PubMed] [Google Scholar]
- 10. Cuccui J, Easton A, Chu KK, Bancroft GJ, Oyston PC, Titball RW, Wren BW. 2007. Development of signature-tagged mutagenesis in Burkholderia pseudomallei to identify genes important in survival and pathogenesis. Infect. Immun. 75:1186–1195. 10.1128/IAI.01240-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haque A, Chu K, Easton A, Stevens MP, Galyov EE, Atkins T, Titball R, Bancroft GJ. 2006. A live experimental vaccine against Burkholderia pseudomallei elicits CD4+ T cell-mediated immunity, priming T cells specific for 2 type III secretion system proteins. J. Infect. Dis. 194:1241–1248. 10.1086/508217 [DOI] [PubMed] [Google Scholar]
- 12. Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19:2308–2316. 10.1101/gr.097097.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6:767–772. 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karlyshev AV, Oyston PC, Williams K, Clark GC, Titball RW, Winzeler EA, Wren BW. 2001. Application of high-density array-based signature-tagged mutagenesis to discover novel Yersinia virulence-associated genes. Infect. Immun. 69:7810–7819. 10.1128/IAI.69.12.7810-7819.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eckert SE, Dziva F, Chaudhuri RR, Langridge GC, Turner DJ, Pickard DJ, Maskell DJ, Thomson NR, Stevens MP. 2011. Retrospective application of transposon-directed insertion site sequencing to a library of signature-tagged mini-Tn5Km2 mutants of Escherichia coli O157:H7 screened in cattle. J. Bacteriol. 193:1771–1776. 10.1128/JB.01292-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sim SH, Yu Y, Lin CH, Karuturi RK, Wuthiekanun V, Tuanyok A, Chua HH, Ong C, Paramalingam SS, Tan G, Tang L, Lau G, Ooi EE, Woods D, Feil E, Peacock SJ, Tan P. 2008. The core and accessory genomes of Burkholderia pseudomallei: implications for human melioidosis. PLoS Pathog. 4:e1000178. 10.1371/journal.ppat.1000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davidson JN, Chen KC, Jamison RS, Musmanno LA, Kern CB. 1993. The evolutionary history of the first three enzymes in pyrimidine biosynthesis. Bioessays 15:157–164. 10.1002/bies.950150303 [DOI] [PubMed] [Google Scholar]
- 18. Zhang R, Lin Y. 2009. DEG 5:0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 37:D455–D458. 10.1093/nar/gkn858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freiberg C, Brunner NA, Schiffer G, Lampe T, Pohlmann J, Brands M, Raabe M, Häbich D, Ziegelbauer K. 2004. Identification and characterization of the first class of potent bacterial acetyl-CoA carboxylase inhibitors with antibacterial activity. J. Biol. Chem. 279:26066–26073. 10.1074/jbc.M402989200 [DOI] [PubMed] [Google Scholar]
- 20. Doublet P, van Heijenoort J, Bohin JP, Mengin-Lecreulx D. 1993. The murI gene of Escherichia coli is an essential gene that encodes a glutamate racemase activity. J. Bacteriol. 175:2970–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lynch M, Kuramitsu H. 2000. Expression and role of superoxide dismutases (SOD) in pathogenic bacteria. Microbes Infect. 2:1245–1255. 10.1016/S1286-4579(00)01278-8 [DOI] [PubMed] [Google Scholar]
- 22. van Opijnen T, Camilli A. 2010. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr. Protoc. Microbiol. Chapter 1:Unit 1E.3. 10.1002/9780471729259.mc01e03s19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-00310. 10.1128/mBio.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. 2013. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 110:2623–2628. 10.1073/pnas.1222783110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baugh L, Gallagher LA, Patrapuvich R, Clifton MC, Gardberg AS, Edwards TE, Armour B, Begley DW, Dieterich SH, Dranow DM, Abendroth J, Fairman JW, Fox D, III, Staker BL, Phan I, Gillespie A, Choi R, Nakazawa-Hewitt S, Nguyen MT, Napuli A, Barrett L, Buchko GW, Stacy R, Myler PJ, Stewart LJ, Manoil C, Van Voorhis WC. 2013. Combining functional and structural genomics to sample the essential Burkholderia structome. PLoS One 8:e53851. 10.1371/journal.pone.0053851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921–4930. 10.1128/JB.186.15.4921-4930.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, Hancock RE. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15:583–589. 10.1101/gr.3513905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu Y, Kim HS, Chua HH, Lin CH, Sim SH, Lin D, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. 2006. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol. 6:46. 10.1186/1471-2180-6-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chong CE, Lim BS, Nathan S, Mohamed R. 2006. In silico analysis of Burkholderia pseudomallei genome sequence for potential drug targets. In Silico Biol. 6:341–346 [PubMed] [Google Scholar]
- 30. Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, Sellati TJ. 2006. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188:6443–6448. 10.1128/JB.00266-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sadosky AB, Wilson JW, Steinman HM, Shuman HA. 1994. The iron superoxide dismutase of Legionella pneumophila is essential for viability. J. Bacteriol. 176:3790–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ortega XP, Cardona ST, Brown AR, Loutet SA, Flannagan RS, Campopiano DJ, Govan JR, Valvano MA. 2007. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J. Bacteriol. 189:3639–3644. 10.1128/JB.00153-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu ZL, Bao J, Reecy JM. 2008. CateGOrizer. A Web-based program to batch analyze gene ontology classification categories. Online J. Bioinform. 9:108–112 http://www.animalgenome.org/tools/catego/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern blots of individual B. pseudomallei K96243 transposon mutants indicating unique insertion sites. Southern blots of 10 individual B. pseudomallei K96243 transposon insertion mutants in which we used a probe designed against the kanamycin cassette located within the transposon, indicating that the transposon has inserted into unique positions in each mutant. Download
Promoter regions of the conditional essential mutants. The sequences of the promoter regions of the accA, bpss0370, pyrH, and sodB mutants are shown aligned with the original sequence of the plasmid pSC200, from which they were derived. The ATG start sequence of each gene is highlighted in blue within the NdeI restriction enzyme site that was used for cloning. All promoter region sequences are shown to be identical. Download
Predicted essential genes of B. pseudomallei K96243.