ABSTRACT
Pseudomonas aeruginosa strains of non-cystic fibrosis (non-CF) origin do not produce significant amounts of extracellular alginate and are nonmucoid. In CF, such isolates can become mucoid through mutation of one of the genes (mucA, mucB, mucC, or mucD) that produce regulatory factors that sequester AlgU, required for increased expression of alginate genes. Mutation of the muc genes in the nonmucoid PAO1, PA14, PAKS-1, and Ps388 strains led to increased levels of extracellular alginate and an obvious mucoid phenotype, but only under iron-limiting growth conditions (≤5 µM), not under iron-replete conditions (≥10 µM). In contrast, >50% of P. aeruginosa isolates from chronic CF pulmonary infections expressed increased levels of alginate and mucoidy both under iron-limiting and iron-replete conditions (i.e., iron-constitutive phenotype). No single iron regulatory factor (e.g., Fur, PvdS) was associated with this loss of iron-regulated alginate expression and mucoidy in these CF isolates. However, the loss of only pyoverdine production, or its uptake, abrogated the ability of P. aeruginosa to produce a robust biofilm that represents the Psl-type of biofilm. In contrast, we show that mutation of the pyoverdine and pyochelin biosynthesis genes and the pyoverdine receptor (FpvA) lead to iron-constitutive expression of the key alginate biosynthesis gene, algD, and an explicitly mucoid phenotype in both iron-limiting and iron-replete conditions. These data indicate that alginate production and mucoidy, in contrast to other types of biofilms produced by P. aeruginosa, are substantially enhanced under iron limitation. These results also have compelling implications in relation to the use of iron chelators in the treatment of P. aeruginosa CF infections.
IMPORTANCE
Pseudomonas aeruginosa is a leading model for the investigation of biofilms. While data have been generated about the role of iron in alginate-independent (Psl/Pel) biofilm development, there is a paucity of data regarding the role of iron in alginate production and its associated mucoid phenotype. We demonstrate that biologically relevant levels of iron that exist in the airway mucus of cystic fibrosis (CF) patients have a substantial influence on production of alginate and the overt mucoid phenotype, pathognomonic of P. aeruginosa infections in CF. Mucoid mutants of non-CF P. aeruginosa isolates are mucoid only under iron limitation and do not express increased levels of alginate under iron-replete growth conditions. However, a significant number of long-term CF isolates lost their iron-regulated expression of increased alginate production and mucoidy and became iron constitutive for these properties. In contrast to the formation of Psl-type biofilms, increasing iron limitation ultimately leads to an iron-constitutive expression of alginate and mucoidy.
INTRODUCTION
Throughout the past decade, the ability of microbes to form complex communities called biofilms has become an exceedingly intense and worthy area of investigation in environmental and medical microbiology. Pseudomonas aeruginosa has become a leading paradigm based on its proclivity to form biofilms in diverse environments (e.g., pipelines, heart valves, bone), yet also in highly specialized situations, such as the bronchioles of cystic fibrosis (CF) patients (1–3). Exceptional research efforts during this time have revealed that P. aeruginosa is able to produce as many as three distinct exopolysaccharides, each of which is associated with specific types of biofilms and conditions under which they are formed. Overexpression of the alginate exopolysaccharide was first identified as being associated with P. aeruginosa mucoid isolates recovered from the lungs of chronically infected CF patients, but rarely from other types of infections (4–6). The Psl (polysaccharide synthesis locus) exopolysaccharide and its associated biofilm were uncovered by examining P. aeruginosa ∆algD mutants that were unable to produce alginate yet were still capable of forming biofilms on glass or plastic surfaces (6, 7). The third exopolysaccharide, Pel (named Pel for pellicle) was associated with P. aeruginosa biofilms at an air-liquid interface (8, 9). While it is not yet entirely clear which aspects of P. aeruginosa infection (e.g., colonization, survival) the Psl- and Pel-associated biofilms might contribute, particularly in CF patients, the long-term presence of highly mucoid strains, in the airways of CF patients, continues to provide a compelling rationale for further investigation into the environmental conditions and mechanisms that govern the production of alginate and the ensuing mucoid phenotype (1, 4, 10).
There is substantial knowledge relating to the biochemistry and molecular genetics of alginate biosynthesis, as well as the mucoid phenotype of P. aeruginosa. Additionally, during the past two decades, considerable insight has been garnered relating to how iron influences the expression of an array of P. aeruginosa virulence factors (exotoxins, proteases, and siderophores), basic metabolic processes (quorum sensing, intermediary metabolism, and resistance to redox stress) and even how this particular metal affects the formation of some types of P. aeruginosa biofilms (Psl and Pel) (11–17). In 2005, Banin et al. presented a comprehensive examination of the molecular and biochemical processes by which iron can affect biofilm formation by P. aeruginosa (11). However, the parental strain (PAO1) used in that study did not carry any muc mutations, and it did not produce significant levels of alginate. Moreover, the biofilms were grown on glass surfaces using a flowthrough system, thereby most likely representing a Psl type of biofilm. Nevertheless, it was demonstrated that strains with mutations in pyoverdine biosynthesis (∆pvdS and ∆pvdA) and the cognate receptor (∆fpvA) were all unable to form robust biofilms. In contrast, a strain carrying a missense mutation (Ala10Gly) in the ferric uptake regulator (Fur) that constitutively expresses Fur-repressed iron acquisition systems retained the ability to form structured biofilms both in the presence and absence of lactoferrin, an iron-binding protein that otherwise inhibits nonalginate biofilm formation by sequestering available iron. Taken together, sufficient levels of iron and intact P. aeruginosa iron acquisition systems (pyoverdine, pyochelin, and ferric dicitrate) were required for the formation of structured nonalginate biofilms.
The impact of iron and iron acquisition systems on alginate production has not been nearly as extensively examined. Boyce and Miller (18) presented the earliest investigation of the potential influence of iron on the mucoid phenotype. In cultures of mucoid CF isolates grown in the presence of iron, nonmucoid revertants accumulated to >80% of the population after 72 h (18). In a subsequent study, those observations were extended to demonstrate that the mucoid phenotype of a single CF isolate was more stable under iron-limited growth than under iron-replete growth (19). Subsequently, Terry et al. reported that ~2% of colonies isolated from strain PAO1 became mucoid after growing under iron limitation in a chemostat for at least 6 days (20). Since 1992, there have been no additional published reports that directly address how iron levels might differentially and specifically affect alginate production or its associated mucoid phenotype.
Herein, we examined whether iron has a regulatory effect on alginate production and mucoidy in non-CF and CF isolates. Our data illustrate that iron profoundly influences the production of alginate and the expression of the mucoid phenotype in strains with newly acquired muc mutations and that this regulation may be lost after prolonged survival in the CF airways. Further examination of the molecular mechanisms associated with the observed iron-regulated alginate production revealed that strains with mutations in iron acquisition systems lost the ability to regulate alginate production in response to iron. Taken together, this report describes the first in-depth assessment of the influence of iron on the expression of alginate, an important virulence factor. It also raises significant questions about the design and administration of therapeutics targeted at iron homeostasis to treat chronic P. aeruginosa infections, especially in the context of the CF pulmonary airways.
RESULTS
Iron influences alginate production and the mucoid phenotype of P. aeruginosa muc mutants.
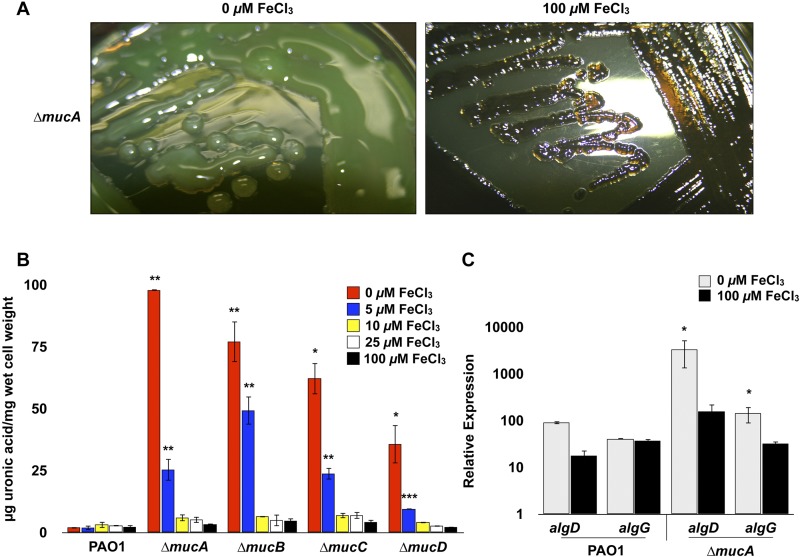
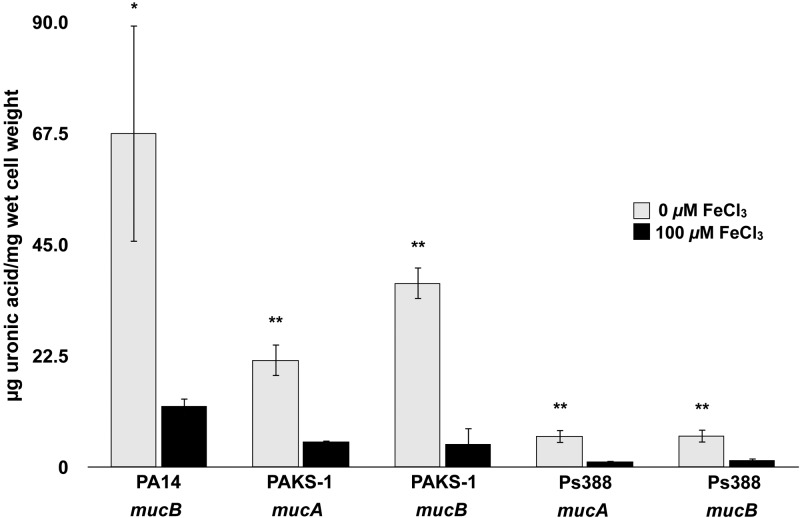
Human lungs, including those of CF patients, have historically been considered iron-limited environments. However, recent comprehensive studies indicate that there can be diverse microenvironments in the lung and that iron levels may show considerable variation and are not necessarily as limited by iron as previously thought. Reid et al. suggested that Fe3+ ions (13 to 134 µM) and ferritin (15 to 300 µg/liter) are major sources of iron in the lungs of CF patients and that patients with chronic obstructive pulmonary disease (COPD) commonly have much higher levels of elemental iron and ferritin (21–24). Additionally, recent findings show that Fe2+ ions are present in sputum samples from CF patients (25). In order to directly investigate the impact of iron on alginate biosynthesis, we constructed ∆mucA, ∆mucB, ∆mucC, or ∆mucD isogenic mutants of P. aeruginosa PAO1 or mucA and mucB insertion mutants of P. aeruginosa PA14, PAKS-1, and Ps388 and examined their production of extracellular alginate and mucoid phenotypes in the presence of variable iron levels (≤5 µM to 100 µM). These strains are all nonmucoid, non-CF-related isolates originating from widely differing types of infections and geographic regions. PAO1 is a wound isolate from Melbourne, Australia. PA14 is a burn isolate from Boston, MA. PAKS-1 is a urine isolate from Stockholm, Sweden. Ps388 is a blood isolate from Seattle, WA, USA. The strains were grown on various concentrations of iron reported to be present in the sputa of CF patients (<5 µM to >100 µM) and were evaluated for alginate production by measurement of extracellular uronic acid levels (see Materials and Methods). Unlike all of their parental strains, these strains carry single gene deletions in mucA, mucB, mucC, or mucD (PAO1) or insertion mutations in mucA or mucB (PA14, PAKS-1, or Ps388), are phenotypically mucoid, and produce substantially increased amounts of extracellular alginate when grown under iron limitation (≤5 µM) than when grown under more iron-replete conditions (≥10 µM) (Fig. 1A and B and 2). No alginate was detected from any of the parental strains grown under either iron-limited or iron-replete conditions (data not shown).
FIG 1.
Effect of iron on alginate production by muc mutants of P. aeruginosa PAO1. (A) P. aeruginosa PAO1 ∆mucA mutant was grown on DTSA with or without 100 µM FeCl3 and imaged after 48 h. (B) The PAO1 strain and ∆mucA, ∆mucB, ∆mucC, and ∆mucD mutants were grown on DTSA containing 5, 10, 25, or 100 µM FeCl3 for 48 h, and the levels of extracellular alginate were evaluated using a uronic acid assay (see Materials and Methods). Values that are significantly different from the value for PAO1 (wild type [WT]) by Student’s t test are indicated by asterisks as follows: ***, P < 0.0005; **, P < 0.005; *, P < 0.05. (C) Relative expression of algD and algG in wild-type PAO1 and PAO1 ∆mucA mutant was analyzed using qRT-PCR after 24 h of growth on DTSA with and without 100 µM FeCl3. Values that are significantly different from the value for PAO1 (WT) by Student’s t test are indicated by an asterisk.
FIG 2.
Effects of iron levels on alginate production in ∆mucA and ∆mucB mutants of P. aeruginosa PA14, PAKS-1, and Ps388. Alginate production was evaluated by measurement of extracellular uronic acid levels produced by PA14, PAKS-1, and Ps388 with insertion mutations in either mucA or mucB after 48 h of growth on DTSA with and without 100 µM FeCl3. Values that are significantly different from the value found with 100 µM FeCl3 by Student’s t test are indicated by asterisks as follows: **, P < 0.005; *, P < 0.05.
Moreover, when a mucB insertion mutation was introduced into a nonmucoid CF isolate, it clearly displayed a mucoid phenotype and regulated its alginate production in response to iron, as did the non-CF isolates (PAO1, PA14, PAKS-1, and Ps388) (see Table S2 in the supplemental material).
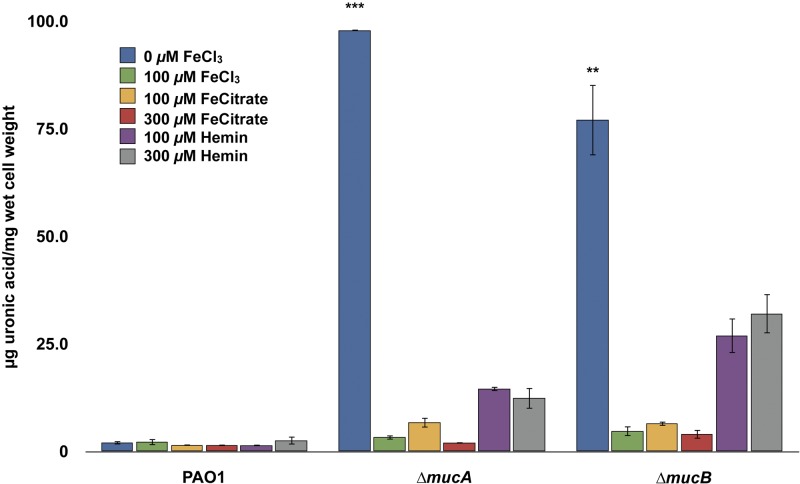
Iron is present in various forms in the CF lungs; therefore, we wanted to know whether iron sources other than iron salts (FeCl3) would result in the repression of alginate production. P. aeruginosa PAO1 and ∆mucA and ∆mucB mutants were cultured in the presence of alternative iron sources, such as ferric dicitrate or hemin. Significant decreases in extracellular alginate levels in these mutants were observed compared to mutants grown without the addition of these iron sources (Fig. 3). It should also be noted that corresponding increases in the concentrations of other metals (Zn2+, Cu2+, Ca2+, and Mg2+) that are similar to the concentrations used for iron (≤5 µM to 300 µM) failed to cause parallel decreases in alginate levels in any of the muc mutants examined (see Fig. S1 in the supplemental material; also data not shown).
FIG 3.
Influences of various iron sources on alginate production by P. aeruginosa PAO1 and ∆mucA and ∆mucB mutants. The strains were grown on DTSA supplemented with various sources and concentrations of iron-carrying compounds. Extracellular alginate production was measured by uronic acid assay after 48 h of growth. Values that are significantly different from the value for PAO1 (WT) by Student’s t test are indicated by asterisks as follows: ***, P < 0.0005; **, P < 0.005.
The data described above indicated that iron exhibits a selective regulatory effect on alginate biosynthesis. The regulation, biosynthesis, and export of the alginate exopolysaccharide are complex processes involving numerous adjacently located structural genes (PA3540 to PA3551) and an assortment of genes encoding regulatory factors (e.g., algR, algC, algB, and kinB) scattered throughout the P. aeruginosa genome (26–29). The biosynthesis and export of this exopolysaccharide are initiated at the algD (PA3540) promoter, which encodes the first enzyme (GDP-mannose 6-dehydrogenase) in the pathway leading to alginate biosynthesis. In addition, various proteases (e.g., AlgW and MucP) function in posttranscriptional regulation by liberating AlgU/T from the MucA/B/C/D sequestration complex at the membrane, thereby leading to the constitutive transcription of algD and the alginate biosynthetic operon (29). Quantitative reverse transcription-PCR (qRT-PCR) was used to validate the expression of algD and algG (encoding alginate-c5-mannuronan-epimerase in the alginate biosynthesis operon) in the PAO1 ∆mucA mutant grown under iron limitation and iron-replete conditions. Triplicate cultures of P. aeruginosa wild type PAO1 and the ∆mucA mutant were grown on DTSA solid medium with and without 100 µM FeCl3, and cells were harvested after 24 h. Relative expression analysis by qRT-PCR showed that the transcript levels of both algD and algG in the ∆mucA mutant were significantly higher than the levels in the wild-type PAO1 strain under iron limitation (Fig. 1C). As predicted from the uronic acid analysis shown in Fig. 1A and B, the expression of both algD and algG transcripts in the ∆mucA mutant decreased to the same levels as those for the wild type under iron-replete conditions (Fig. 1C). These data showed that iron levels of ≥10 µM result in a substantial reduction in the transcription of the alginate biosynthetic operon corresponding to the decrease in the amount of alginate measured by the uronic acid analysis described above.
Taken together, these data provide the first evidence that biologically significant levels of iron can have substantial effects on the ability of newly derived muc mutants from a very diverse group of P. aeruginosa strains to express high levels of extracellular alginate and manifest mucoid phenotypes.
Chemical analysis of P. aeruginosa ∆mucA and ∆mucB extracellular polymeric substance (EPS).
The data described above indicate that iron strongly influences the expression of the alginate exopolysaccharide. To ensure that the increased levels of uronic acids measured in our uronic acid analysis under iron-limiting conditions accurately mirror those of alginate [(1–4)-linked β-d-mannuronate and α-l-guluronate polymer], extracellular polysaccharides extracted from P. aeruginosa PAO1 ∆mucA and ∆mucB mutants were evaluated by size exclusion chromatography and glycosyl composition analysis with combined gas chromatography-mass spectrometry (GC-MS). Because it was possible that other biofilm-associated exopolysaccharides (Psl or Pel) could be produced by these strains, particularly from cells harvested from iron-replete cultures, exopolysaccharides were extracted from PAO1, the wild-type parent, and ∆mucA and ∆mucB mutants grown in both iron-limited and iron-replete conditions and evaluated (see Materials and Methods). The most abundant carbohydrate detected in the ∆mucA and ∆mucB mutants grown under iron limitations was mannuronic acid (Table 1). In contrast, the carbohydrates detected from samples from wild-type PAO1 were glucose and rhamnose, with a small amount of 3-deoxy-d-manno-octulosonate (KDO), which was not detected in the ∆mucA or ∆mucB samples. The samples from the ∆mucA and ∆mucB mutants grown under iron limitation contained significantly lower levels of glucose and rhamnose than those from the iron-limited parent, PAO1. The extracellular polysaccharide composition of Pel biofilms is mainly glucose, rhamnose, and mannose, whereas the extracellular polysaccharide composition of Psl biofilms is mainly galactose and mannose (6, 30). These results are consistent with previous findings that mannuronic acid is the predominant extracellular polysaccharide of alginate biofilms and mucoidy and confirmed the results of uronic acid analysis (6).
TABLE 1 .
EPS carbohydrate composition profiles for P. aeruginosa PAO1 and mutants
| Carbohydrate | Carbohydrate composition (µg/107 CFU ml−1) for the following strains grown under the indicated condition: |
|||||
|---|---|---|---|---|---|---|
| PAO1 |
∆mucA mutant |
∆mucB mutant |
||||
| 0 µM FeCl3 | 100 µM FeCl3 | 0 µM FeCl3 | 100 µM FeCl3 | 0 µM FeCl3 | 100 µM FeCl3 | |
| Mannuronic acid | 0 | 0 | 88.2 | 12.1 | 176 | 33.7 |
| Glucose | 45.8 | 25.6 | 18.6 | 13.6 | 34.56 | 1.4 |
| Rhamnose | 9.0 | 9.5 | 6.6 | 4.0 | 8.96 | 0.8 |
| Mannose | 0.7 | 0.4 | 1.8 | 0.5 | 3.84 | 0.4 |
| Ribose | 0.5 | 1.6 | 1.8 | 1.4 | 2.56 | 0.6 |
| Xylose | 0.5 | 0.1 | 1.2 | 0.2 | 3.84 | 0.4 |
| KDO | 1.0 | 3.6 | 0 | 0.7 | 0 | 0 |
| N-Acetylglucosamine | 1.6 | 1.6 | 0.6 | 0.9 | 0.64 | 0.1 |
Global transcriptional analysis of wild-type PAO1 and PAO1 ∆mucA mutant under iron limitation.
While there have been a considerable number of transcriptional studies of P. aeruginosa grown under different levels of iron, there have not been any comparable assessments of transcriptional responses to variable levels of this biologically consequential metal for isogenic mucoid variants and their nonmucoid parents (14, 15). Based on the observations described above, it was deemed worthwhile to examine the global transcriptional responses of P. aeruginosa PAO1 (wild type) compared to those of a ∆mucA mutant under iron-limited and iron-replete conditions using GeneChip microarray analysis (see Materials and Methods for growth conditions used for these microarray experiments). Table S1 in the supplemental material provides a complete list of the genes and operons of PAO1, the wild-type parent, and its isogenic ∆mucA mutant, which exhibited the most robust responses in iron-limited growth compared to iron-replete growth. As with most microarray-based approaches, a surfeit of data was generated. Nevertheless, there are several interesting outcomes based on these experiments that warrant specific comments.
There were a number of genes and operons with increased expression under iron limitation in the ∆mucA mutant that had significantly lower expression or were not expressed in the PAO1 parent under iron limitation (Table 2; see Table S1 in the supplemental material). Genes with increased expression in the ∆mucA mutant may provide insights into the basic metabolism of mucoid strains and the stresses related to increased production and secretion of extracellular alginate under iron limitation. The most striking and obvious examples in this regard are the increased transcriptional responses of genes involved in alginate biosynthesis and export (algD-algA) in the ∆mucA mutant under iron limitation, but not in the PAO1 parent (Table 2). These results and the fact that we did not observe corresponding increases in the expression of genes involved in the biosynthesis of the Psl (pslA-pslO; PA2231-PA2245) or the Pel (pelA-pelG; PA3058-PA3064) exopolysaccharides strongly reinforce the idea that iron limitation selectively influences the increased production of alginate, in contrast to either of the other biofilm-associated extracellular polysaccharides.
TABLE 2 .
Select genes with increased gene expression under iron limitation versus iron-replete conditions in P. aeruginosa PAO1 and ∆mucA mutant
| Locus tag | Gene | Change in gene expressiona |
Description/functionb | |
|---|---|---|---|---|
| PAO1 | ∆mucA mutant | |||
| PA0509 | nirN | NC | 3 | Probable c-type cytochrome |
| PA0510 | NC | 3 | Uroporphyrin III c-methyltransferase | |
| PA0511 | nirJ | NC | 4 | Heme d1 biosynthesis protein |
| PA0512 | NC | 3 | Conserved hypothetical protein | |
| PA0513 | NC | 3 | Probable transcriptional regulator | |
| PA0514 | nirL | NC | 4 | Heme d1 biosynthesis protein |
| PA0515 | NC | 4 | Probable transcriptional regulator | |
| PA0516 | nirF | NC | 4 | Heme d1 biosynthesis protein |
| PA0517 | nirC | NC | 5 | Probable c-type cytochrome precursor |
| PA0518 | nirM | NC | 6 | Cytochrome c-551 precursor |
| PA0519 | nirS | NC | 7 | Nitrite reductase precursor |
| PA0520 | nirQ | NC | 3 | Regulatory protein; central metabolism |
| PA0523 | norC | NC | 11 | Nitric oxide reductase subunit C |
| PA0524 | norB | 2 | 11 | Nitric oxide reductase subunit B |
| PA0525 | norD | 4 | 7 | Probable dinitrification protein |
| PA2147 | katE | NC | 13 | Heme-binding catalase |
| PA2185 | katN | 4 | 2 | Mn-containing catalase |
| PA3540 | algD | 2 | 23 | GDP-mannose 6-dehydrogenase |
| PA3541 | alg8 | NC | 7 | Alginate biosynthesis |
| PA3542 | alg44 | NC | 4 | Alginate biosynthesis |
| PA3543 | algK | NC | 60 | Scaffold protein |
| PA3544 | algE | NC | 6 | Outer membrane protein |
| PA3545 | algG | NC | 11 | Alginate-C5-mannuronan-epimerase |
| PA3546 | algX | NC | 4 | Alginate biosynthesis |
| PA3547 | algL | NC | 4 | Alginate lyase |
| PA3548 | algI | NC | 8 | Acetylase |
| PA3549 | algJ | D | 5 | Alginate O-acetyltransferase |
| PA3550 | algF | NC | 5 | Alginate O-acetyltransferase |
| PA3551 | algA | 2 | 11 | Phosphomannose isomerase |
| PA5097 | hutT | 3 | 6 | γ-Aminobutyrate permease |
| PA5098 | hutH | 2 | 14 | Histidine ammonia-lyase |
| PA5099 | NC | 47 | Nucleoside transporter family | |
| PA5100 | hutU | 5 | 23 | Urocanase |
Change in gene expression for P. pseudomonas PAO1 or the ∆mucA mutant grown in 0 µM FeCl3 versus 100 µM FeCl3. The numbers are the fold increase in gene expression. Abbreviations: NC, no change; D, decreased expression.
Descriptions were obtained from the Pseudomonas Genome Database (www.pseudomonas.com).
One additional salient outcome from these experiments is the considerably increased levels of expression of PA5097 (hutT), PA5098 (hutH), and PA5099 (Table 2). Transcription of these genes, which comprise an operon, was significantly increased in the PAO1 ∆mucA mutant under iron limitation, but to a substantially lesser extent, or not at all in PAO1 (wild type) under iron limitation. Increased expression of these genes in P. aeruginosa PAO1, which are involved in histidine utilization, leads to decreased cytotoxicity and the expression of at least one of the type III secreted cytotoxins (ExoS) (31). Taken together, such results could provide insights into why a substantial number of mucoid CF isolates display diminished type III cytotoxin-mediated cytotoxicity (32).
Do CF pulmonary isolates regulate alginate production in response to iron levels?
Mucoidy and alginate production by P. aeruginosa are commonly associated with chronic CF pulmonary infections. Therefore, it was of interest to ascertain whether variable levels of iron (≤5 µM to 100 µM) known to exist in the sputa of CF patients can influence the expression of alginate and mucoidy in alginate-producing isolates obtained directly from CF-associated infections. For this purpose, two groups of CF pulmonary isolates were examined. The first group, early CF isolates (Early-CFI) consisted of 8 isolates derived from individual children with CF who were <3 years of age (yoa) at the time of sputum collection (see Table S2 in the supplemental material). The second group, late CF isolates (Late-CFI), consisted of 17 isolates originating from older CF patients (≥10 yoa) who had been colonized with P. aeruginosa for at least 5 years (Table S3). While both the Early-CFI and Late-CFI groups contained isolates with mucoid phenotypes, there were some appreciable differences between these groups.
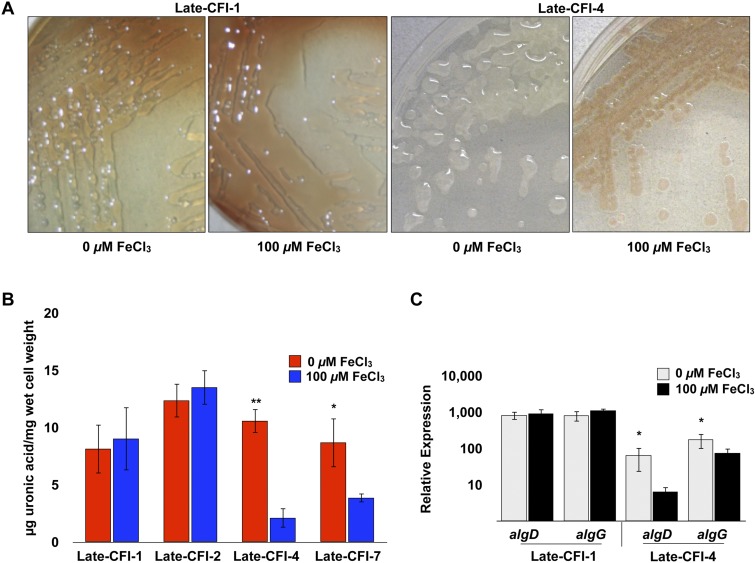
Only 50% (4 of 8) of the Early-CFI intrinsically produced alginate along with an obvious mucoid phenotype, while most (16/17 [94%]) of the Late-CFI produced significant levels of extracellular alginate, along with an obvious mucoid phenotype (see Table S2 and Table S3 in the supplemental material). Of the mucoid Early-CFI examined, 1 out of 4 showed constitutive expression of alginate levels and a mucoid phenotype when grown under iron-limited and iron-replete conditions, while 56% (9/16) of the mucoid Late-CFI were constitutive for alginate expression and mucoidy, even with the highest level of iron (100 µM) tested (Fig. 4A and B and Table S2 and Table S3). Figure 4 illustrates the differences between a select group of constitutively producing alginate isolates and those that regulate alginate production in response to iron levels as shown by uronic acid analysis of extracellular alginate and qRT-PCR analysis of algD and algG.
FIG 4.
Alginate production and mucoidy of late CF isolates (Late-CFI) in response to variable iron levels. (A) Late-CFI were grown on DSTA with and without 100 µM FeCl3 and imaged at 48 h. (B) The level of extracellular alginate produced by various Late-CFI grown on DSTA with and without 100 µM FeCl3 was measured by uronic acid assay at 48 h. Values that are significantly different from the value for late CF isolate 4 (Late-CFI-4) or Late-CFI-7 with 100 µM FeCl3 by Student’s t test are indicated by asterisks as follows: **, P < 0.005; *, P < 0.05. (C) Relative expression of algD and algG of Late-CFI-1 and Late-CFI-4 was analyzed using qRT-PCR after 24 h of growth on DSTA with and without 100 µM FeCl3. Values that are significantly different (P < 0.05) from the value for Late-CFI-4 with 100 µM FeCl3 by Student’s t test are indicated by an asterisk.
These results demonstrate the robust variations that occur in the expression of a highly relevant virulence determinant (alginate production and mucoidy) as the consequence of the fluctuating iron levels that are known to occur during P. aeruginosa CF pulmonary infections. Although the number of CF isolates examined in this study is relatively small, taken together, our results do support the idea that over time (years to decades), an initially vigorous regulatory influence of iron on the expression of alginate and mucoidy can be lost, resulting in alginate production that is indifferent to the variable levels of iron known to exist in the lungs of CF patients.
How does iron initially control the increased production of extracellular alginate and mucoidy and how can this regulation be lost?
Based on the high percentage of Late-CFI that had lost iron-regulated alginate production, it was of interest to examine whether these Late-CFI were capable of acquiring iron. Chrome Azurol S (CAS) agar plate assay is a technique used to study siderophore production in bacteria (12). Using this technique, reduced siderophore production was observed in several CFI that exhibit constitutive alginate production (see Table S2 and Table S3 in the supplemental material). For example, late CF isolate 1 (Late-CFI-1), Late-CFI-2, Late-CFI-12, and Late-CFI-16, which all exhibit iron-constitutive alginate production produce very low levels of siderophores (pyoverdine and pyochelin) on both iron-limited and iron-replete CAS agar plates (Table S3). However, no compelling correlation could be discerned between this and the loss of iron-regulated alginate production. There are a variety of ways that a constitutive mucoid phenotype, most often associated with Late-CFI, can occur (see Discussion below).
In the Banin et al. study, the impact of iron and iron limitation on the formation of Psl/Pel-type biofilms was examined in an assortment of mutants altered in their ability to (i) express key iron-related regulatory factors (Fur and PvdS), (ii) produce enzymes involved in siderophore biosynthesis (PvdA), or (iii) express receptors for specific siderophores (FpvA) (11). Based on those data, it was clear that interfering with such iron regulatory or iron acquisition systems resulted in a significantly diminished ability of wild-type PAO1 to produce robust Psl-type biofilms under the conditions used in that study.
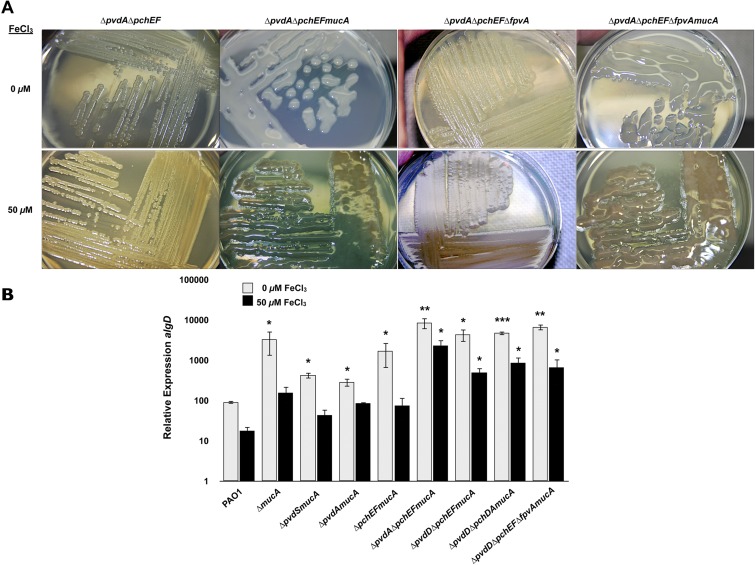
In the present study, a similar approach was used. However, mucoid strains were created by introducing either mucA or mucB mutations into a variety of single, double, and triple mutants of the PAO1 strain carrying deletions in genes encoding (i) an iron regulatory factor (pvdS), (ii) assorted pyoverdine (pvdA and pvdD) and pyochelin (pchEF and pchDA) siderophore biosynthesis enzymes, or (iii) the pyoverdine receptor (fpvA). The single, double, and triple iron acquisition mutants of PAO1, which also carry mucA or mucB insertion mutations, were grown on dialyzed tryptic soy agar (DTSA) with or without 50 µM FeCl3 for 24 h or 48 h and then examined for the mucoid phenotype and the relative expression of algD by qRT-PCR (Fig. 5A and B). None of the single, double, or even triple iron acquisition mutations caused significant decreases in the expression of the algD transcript under iron limitation (Fig. 5A and B). Notably, however, the following mutants, ΔpvdA ΔpchEF mucA mutant, ΔpvdD ΔpchEF mucA mutant, ΔpvdD ΔpchDA mucA mutant, and ΔpvdD ΔpchEF ΔfpvA mucA mutant produced alginate and were mucoid under iron-replete conditions at the same levels observed under iron limitation (Fig. 5A and data not shown). Further, the relative expression of algD remained unchanged in these mutants regardless of the iron concentration (Fig. 5B). Interestingly, in stark contrast to previous data (22) regarding the requirement of PvdS for the formation of structured Psl/Pel-type biofilms, PvdS was not required for the production of alginate and was not involved in iron-regulated alginate production (Fig. 5B). Taken together, the present data suggest that strains with mutations in multiple iron acquisition systems (pyoverdine and pyochelin), which likely result in the reduced acquisition of iron, lose the ability to regulate alginate in response to iron.
FIG 5.
Investigation of alginate production in iron acquisition mutants of P. aeruginosa PAO1. (A) P. aeruginosa PAO1 and several isogenic iron acquisition mutants were grown on DTSA with and without 50 µM FeCl3 and imaged after 48 h. (B) Relative expression of algD in PAO1 (wild type) and several isogenic iron acquisition mutants were analyzed using qRT-PCR after 24 h of growth on DTSA with and without 50 µM FeCl3. Values that are significantly different from the value for PAO1 (WT) by Student’s t test are indicated by asterisks as follows: ***, P < 0.0005; **, P < 0.005; *, P < 0.05.
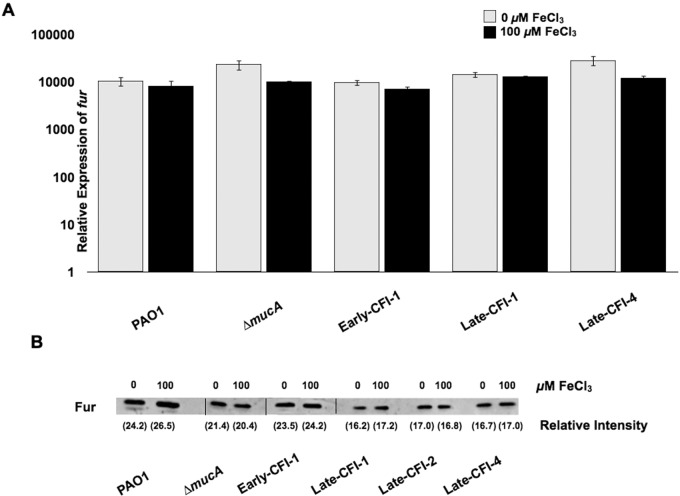
Due to the decreased ability of the majority of Late-CFI to regulate alginate production in response to iron and because mutations in both siderophore iron acquisition systems resulted in constitutive alginate expression regardless of iron levels, it is possible that either mutations in Fur or a change in the expression of Fur contributes to constitutive alginate expression in some of the Late-CFI. Fur is the master regulator of iron homeostasis, and a mutation in Fur would result in the derepression of a large number of regulatory and structural genes, including those that might be involved in alginate production (33). While Fur is essential in P. aeruginosa, strains with point mutations that partially affect Fur function or reduce Fur levels show a constitutive phenotype with regard to the expression of a plethora of iron-regulated factors, including two siderophores (pyoverdine and pyochelin) and exotoxin A (12). It is possible that CF isolates, which exhibit a constitutive phenotype in terms of an increased expression of alginate and mucoidy under both iron-limited and iron-replete conditions, could have acquired point mutations that either affect Fur function or reduce its levels (12). The involvement of Fur was examined in two ways. First, the sequence of the fur gene, including its regulatory region, was examined in both Early-CFI and Late-CFI, including those showing an iron-regulated phenotype and an iron-constitutive phenotype with regard to the production of alginate. No base changes in the structural and regulatory region of fur were detected in any of these isolates (data not shown). Second, the levels of Fur protein and fur transcript, as detected by Western blotting and qRT-PCR were examined in CFI that exhibit both an iron-regulated phenotype and an iron-constitutive phenotype with regard to alginate production (Fig. 6A and B). There were no differences in the levels of Fur protein or fur transcript produced under iron-limited or iron-replete conditions that could be associated with either an iron-regulated or iron-constitutive phenotype in terms of increased alginate production or mucoidy. Fur in P. aeruginosa is not autoregulated. Consequently, we did not expect to find differences between iron-limiting and iron-replete growth conditions in the amount of protein or transcript if mutations were not present. However, while we did not find Fur mutations or expression deviations, the indirect involvement of Fur cannot be ruled out. It is still possible that Fur represses an activator of alginate biosynthesis or indirectly controls a repressor involved in the iron-regulated alginate phenotype.
FIG 6.
Investigation of the role of Fur in alginate production. (A) P. aeruginosa PAO1, ∆mucA mutant, Early-CFI-1, Late-CFI-1, and Late-CFI-4 were grown on DTSA with and without 100 µM FeCl3. The relative expression of fur was measured by qRT-PCR after 24 h of growth. (B) Strain PAO1, ∆mucA mutant, Early-CFI-1, Late-CFI-1, Late-CFI-2, and Late-CFI-4 were grown on DTSA with and without 100 µM FeCl3. Fur protein levels (shown as relative intensities within parentheses below the blot) were analyzed by Western blotting and quantified as described in Materials and Methods.
DISCUSSION
Iron, probably more so than any other metal, has been shown to have profound effects on the expression of bacterial virulence determinants, host-pathogen interactions, and the ensuing outcomes of bacterial infectious diseases. While the levels of free iron in mammalian hosts are ordinarily exceedingly limited, most prokaryotes have at their disposal multiple strategies (siderophores, heme uptake, utilization of host iron-binding proteins) that can overcome the paucity of free iron they might encounter upon colonization of a eukaryotic host (34). Yet, an abnormal increase in the levels of free iron in sera or tissues of mammalian hosts can lead to rapidly fulminating infections (5, 21, 22, 24, 35). During an infectious disease, the level of free iron can vary from one that is extremely restrictive to one where free iron or a readily available source of iron may become more accessible as a consequence of infection-related pathology (hemorrhage, inflammation, proteolysis of host proteins) (21, 22, 24, 36). For instance, the levels of free iron in the airway mucus of CF patients during acute exacerbations are known to fluctuate between very low, growth-limiting levels (<1 µM) to more than 100 times these concentrations (>100 µM) (22–24). Notably, however, such low levels of free iron are those that are typically responsible for an increase in the in vitro expression of iron-regulated genes encoding virulence determinants (e.g., exotoxins, proteases, and siderophores), while higher levels most often suppress their expression, favor increased growth rates, and in certain cases, favor the eventual formation of particular types of biofilms (11, 15, 33, 37).
As previously discussed, Banin et al. demonstrated that the robust formation of a wild-type P. aeruginosa nonalginate biofilm grown on glass slides requires either the biosynthesis of pyoverdine or the expression of its cognate receptor (FpvA) (11). The diminution of biofilm formation by these mutants could be overcome by providing alternative sources of iron (1 µM ferric dicitrate or 1.5 µM desferrioxamine). Furthermore, in those studies, nonalginate biofilm formation was significantly inhibited by exposing the bacteria to the iron-binding protein lactoferrin (20 µg/ml). Overall, the results from that study provided cogent support for the view that the normal ability to form a robust nonalginate biofilm by P. aeruginosa can be affected merely by disruption of only one of its iron acquisition systems (pyoverdine biosynthesis or uptake) or by exposing it to an exogenous competitor for iron (lactoferrin). The bottom line is that it is probably the ability of P. aeruginosa to acquire sufficient levels of cytoplasmic iron that is crucial to the formation of robust nonalginate biofilms. It should be noted, however, that in the Banin et al. studies, the actual production of a biofilm-associated exopolysaccharide (Psl or Pel) was never directly examined. However, it could be argued that that the striking mucoid presentation on a solid agar surface as shown in the figures from the present study (Fig. 1, 4, and 5) could at least in some manner, be akin to alginate-related biofilms as they exist in mucus-plugged CF airways.
In the present study, we investigated whether biologically relevant levels of iron (≤5 µM to ≥100 µM) have an influence on the production of alginate and the mucoid phenotype by P. aeruginosa. Since wild-type P. aeruginosa does not normally produce alginate, we constructed muc mutants from a variety of non-CF strains from diverse geographic regions and determined whether these levels of iron in any way affected alginate production and the ensuing mucoid phenotype. Similarly, a nonmucoid isolate from an early CF patient (Early-CFI) was converted to the mucoid phenotype by introducing an insertion mutation into mucB. All of these newly constructed muc mutants produced increased levels of alginate under iron-limiting (≤5 µM) growth and significantly reduced levels of alginate under iron-replete (≥10 µM) growth. In contrast, in a group of CF isolates that intrinsically produce alginate by acquisition of mutations within the CF lung, a significant proportion (56%) had lost iron-regulated control over alginate production and are mucoid irrespective of the levels of iron to which they were exposed.
These observations posed additional salient questions about iron, the expression of alginate, and the overtly mucoid phenotype. (i) How does iron initially exhibit its tight control over alginate production? (ii) How can this regulatory influence be nullified during the course of chronic pulmonary disease? To address these questions, we considered whether alteration of one of more known iron regulatory factors (Fur or PvdS) or iron acquisition systems (pyoverdine or pyochelin) could specifically affect the iron-regulated expression of alginate production and mucoidy, as they did in relation to the production of nonalginate biofilms (22). Our results demonstrated in a variety of ways that abrogation of multiple acquisition systems (pyoverdine or pyochelin) failed to diminish the expression of alginate under iron limitation. Interestingly, strains with mutations in two or more iron acquisition systems exhibited enhanced production of extracellular alginate under iron-limiting and iron-replete conditions, similar to what was observed with more than half (56%) of the mucoid Late-CFI examined.
While at this time it is not clear whether our observations with the PAO1 muc mutants carrying multiple mutations in iron acquisition (e.g., ∆pvdD ∆pchEF ∆fpvA mucA mutant) directly relate to the iron-constitutive phenotypes of the Late-CFI, several of the Late-CFI did exhibit reduced levels of siderophores (Late-CFI-1, -2, -12, and -16 [see Table S3 in the supplemental material]). Importantly, a significant number of P. aeruginosa isolates from CF patients have been shown to be unable to produce detectable levels of pyoverdine (38). On the other hand, not all of the Late-CFI with an iron-constitutive alginate-expressing phenotype (Late-CFI-3, -4, -5, -11, and -13 [Table S3]) exhibited decreased siderophore production in the CAS assay. Perhaps, as suggested in the review by Poole and McKay entitled “Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome” (39) or in a more recent report by Konings et al. (25), there are multiple ways for this pathogen to strategically acquire iron. At this point, it has not yet been discerned whether alginate expression is controlled by specific iron-responsive regulatory factors or whether generally low cytoplasmic iron levels generate further stresses leading to increased alginate expression, even in an iron-replete environment. Nevertheless, we demonstrated that none of the examined iron-constitutive alginate-producing CFI carried mutations that could alter the function or level of Fur, the master regulator of iron homeostasis. Likewise, a possible role for the Fur-regulated sigma factor PvdS was eliminated. These observations, however, do not as of yet exclude the possibility that mutation of an unidentified Fur-regulated gene that controls the expression of one or more alginate biosynthesis genes (e.g., algD and algR) occurs. A mutation in a Fur-regulated gene could in some cases be the cause of the iron-constitutive mucoid phenotype seen in the majority of the Late-CFI. Along these lines, it is of interest that recently two genes of Pseudomonas syringae associated with alginate biosynthesis (algJ and algF) were identified as being Fur regulated (40). In P. aeruginosa, AlgJ and AlgF seem to be involved in the acetylation of alginate (41), rather than in its biosynthesis. These genes should be further examined. While unlikely, it is also possible that some of the Late-CFI in this study exhibit an iron-constitutive phenotype because they have acquired mutations in other alginate regulatory genes (e.g., KinB and type IV pili) instead of the commonly acquired muc mutations most often seen in CF isolates (42, 43).
Finally, iron homeostasis, particularly in the context of biofilm formation, has become an attractive target for therapeutic interventions in non-CF and CF P. aeruginosa infection models (44–46). In some cases (i.e., gallium nitrate), they are the focus of current ongoing clinical trials (see study NCT01093521 at ClinicalTrials.gov). Gallium (Ga3+) is a FDA-approved ferric (Fe3+) iron analog and while it is not an iron chelator, it is thought to interfere with how iron is processed by P. aeruginosa (45, 47). Even though Ga3+ can kill P. aeruginosa in vitro, including mucoid strains and those growing in biofilms, there is a significant population that shows resistance to this agent. There are some properties of Ga3+ that, very likely, would not allow it to affect all the systems that P. aeruginosa possesses in its armamentarium for scavenging iron and maintaining iron homeostasis. For example, unlike iron, Ga3+ cannot be reduced under physiological conditions, and therefore, it cannot substitute for ferrous (Fe2+) iron, which P. aeruginosa can acquire through its ferrous (Feo) uptake system (15). What is more, gallium cannot interfere with heme uptake by P. aeruginosa, because iron is in the ferrous form (i.e., Fe2+) in heme.
Another approach that targets iron homeostasis to interfere with biofilm formation involves the use of FDA-approved iron chelators deferoxamine (DFO) and deferasirox (DSX) (Exjade), along with tobramycin, a primary antibiotic used to treat CF lung infections (48, 49). Investigators have examined P. aeruginosa biofilm formation on human CF airway epithelial cells and reported that tobramycin and DFO together do not reduce biofilm formation to a greater extent than tobramycin alone. However, tobramycin and DSX together significantly decreased the ability of P. aeruginosa PAO1 to form biofilms on human CF airway epithelial cells compared with tobramycin alone. These investigators used wild-type PAO1, which does not produce alginate, as their model organism. It is also worth mentioning that P. aeruginosa has a receptor for DFO, and it is not yet known whether PAO1 has a DSX receptor.
Consequently, studies that have investigated the use of therapeutics targeting iron homeostasis and biofilm formation used only P. aeruginosa strains that, while capable of biofilm formation, do not express high levels of extracellular alginate or exhibit a notable mucoid phenotype. Further, thus far, iron-targeted therapeutics have frequently focused on the ability of this pathogen to acquire iron. It would be predicted, based on the observations presented in this report that reduction of iron acquisition by this opportunist would actually lead to increased alginate production and the maintenance of the mucoid phenotype. Such a strategy, in reality, is potentially detrimental in some cases, especially in the context of CF lung infections where alginate-producing strains dominate the population.
MATERIALS AND METHODS
Strains, mutant construction, and culture conditions.
P. aeruginosa PAO1, PA14, PAKS-1, and Ps388 originated from various types of non-CF infections and have been continuously maintained at −80°C. Jane Burns, Seattle Children’s Hospital and Department of Pediatrics, University of Washington School of Medicine, provided the Early CF isolates (Early-CFI). Benjamin Staudinger, Pulmonary and Critical Care Medicine, University of Washington School of Medicine, provided the Late CF isolates (Late-CFI). P. aeruginosa PAO1 ∆mucA, ∆mucB, ∆mucC, and ∆mucD mutants were constructed as previously described (11–17). The mucA and mucB mutations of PA14, PAKS-1, and Ps388 strains and the mucB mutation of the nonmucoid Early-CFI were constructed by insertional mutagenesis. Briefly, internal fragments from mucA or mucB genes were amplified by PCR and ligated into pCR 2.1-TOPO (Invitrogen). The fragments were excised from the cloning vector and ligated into pSUP203, which is transferred to P. aeruginosa by conjugation, but cannot autonomously replicate in this organism (50). This approach was also used to create mucA mutations in previously constructed PAO1 ∆pvdS, ∆pvdA, ∆pchEF, ∆pvdA ∆pchEF, ∆pvdD ∆pchED, ∆pvdD ∆pchDA, and ∆pvdD ∆pchEF ∆fpvA mutants.
Chelex-treated and dialyzed tryptic soy broth (DTSB) or agar (DTSA) supplemented with 1% glycerol and 50 mM glutamate was used as an iron-limited base medium and iron-replete conditions were created by the addition of various concentrations of FeCl3, ferric dicitrate, or bovine hemin (Sigma). Brain heart infusion (BHI) agar or broth was used for the general maintenance of cultures.
Uronic acid assay.
P. aeruginosa cultures were grown at 37°C on DTSA with or without supplementation. All biological material was removed from the surface of each agar plate with a sterile petri dish cell scraper and suspended in 0.85% NaCl. The suspension was spun at 10,000 × g to pellet cells, and the supernatant fraction was removed. The levels of uronic acids were measured using a carbazole assay previously described by Knutson and Jeanes (51), with slight modifications. Alginate was quantified using a standard curve made from Macrocystis pyrifera alginate (Sigma) and reported as micrograms of uronic acid per milligram of cell weight (wet weight).
Glycosyl composition analysis of extracellular polysaccharides.
P. aeruginosa PAO1 (wild type) and ∆mucA and ∆mucB mutants were grown at 37°C for 24 h on DTSA with and without 100 µM FeCl3 covered with 12,000- to 14,000-molecular-weight-cutoff (MWCO) membrane (Life Science Products, Inc.). Total exopolysaccharides were isolated as previously described with slight modifications (52). Glycosyl composition analysis was performed at the Complex Carbohydrate Research Center, University of Georgia, under the supervision of Parastoo Azadi. This analysis was performed by combined gas chromatography-mass spectrometry (GC-MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methylglycosides produced from the sample by acidic methanolysis. The samples were placed into test tubes, and 20 µg of inositol was added to each test tube. Methylglycosides were then prepared from the dry samples by methanolysis in 1 M HCl in methanol at 80°C for 18 h, followed by reacetylation (N-acetylation) with pyridine and acetic anhydride in methanol. The samples were then per-O-trimethylsilylated by treatment with Tri-Sil (Pierce) at 80°C for 0.5 h. GC-MS analysis of the TMS methylglycosides was performed on an Agilent 6890 N GC interfaced to a 5975B MSD, using an Agilent DB-1 fused silica capillary column (30 m BY 0.25 mm inner diameter [ID]).
qRT-PCR and microarray analysis.
Total RNA was isolated using Qiagen RNeasy minicolumns and DNase treated with RNase-free DNase I (NEB). ImPromII reverse transcription system (Promega) was used for cDNA synthesis. Each cDNA reaction mixture contained 500 ng of RNA. Quantitative reverse transcription-PCR (qRT-PCR) experiments were carried out in the Roche LightCycler 480 system using LightCycler 480 probes master and gene-specific primers and probes. Relative transcript levels were determined by the comparative standard curve method. All samples were normalized to the constitutively produced omlA transcript (14).
Western blot analysis.
Western blotting was performed on whole-cell extracts of bacteria grown for 24 h at 37°C on DTSA medium with either 0 µM or 100 µM FeCl3. A total of 3 × 109 cells (optical density at 590 nm [OD590] of 1.0) were harvested from plates, washed with 0.85% NaCl, and then pelleted by centrifugation. Cell pellets were suspended in 500 µl of phosphate-buffered saline (PBS) and sonicated three times for 30 s each time at an output of 0.5 Watts. Western blotting was performed as previously described (12). The intensity of the bands was determined using Image Lab software (Bio-Rad) and then normalized to the OmlA protein.
SUPPLEMENTAL MATERIAL
Influence of assorted metals on alginate production. P. aeruginosa PAO1 (wild type) and the PAO1 ∆mucD mutant were grown on DTSA supplemented with 100 µM ZnCl2, CuCl2, CaCl2, MgCl2, or FeCl3. Extracellular alginate was measured by uronic acid assay after 48 h of growth. Download
Global gene expression increases under iron limitation versus iron-replete conditions in P. aeruginosa PAO1 and ∆mucA mutant. Duplicate cultures of P. aeruginosa wild-type PAO1 and ∆mucA and ∆mucD mutants were grown on DTSA solid medium with and without 100 µM FeCl3, and cells were harvested after 24 h. The global gene expression profiles were examined using GeneChip microarray analysis (15), and the results were compared and averaged.
Analysis of mucoid phenotypes of Pseudomonas aeruginosa strains from young CF patients at Seattle Children’s Hospital. Detailed descriptions of eight early CF isolates from individual children with CF are given in the table. The isolates were obtained from Jane Burns at Seattle Children’s Hospital. This table contains phenotypic analysis of isolates grown on BHI and DTSA with and without 100 µM FeCl3 and zone size (millimeters) of halos observed on CAS agar plates. Additionally, this table provides data relating to zone size (in millimeters) of several PAO1 muc mutants and PAO1 siderophore mutants that were used as controls for this study.
Late CF clinical isolates. This table contains descriptions of the phenotypes of 17 late CF isolates originating from older CF patients who had been colonized with P. aeruginosa for at least 5 years. These isolates were obtained from Benjamin Staudinger at the University of Washington. Strains were grown on BHI and DTSA with and without 100 µM FeCl3 and analyzed for alginate production and mucoidy. Zone size (in millimeters) of halos were noted after growth on CAS agar plates.
Supplemental Materials and Methods Download
ACKNOWLEDGMENTS
This work was supported by the NIH National Institute of Allergy and Infectious Diseases grant R37-AI15940-33 (M.L.V.) and by a National Institute of Allergy and Infectious Diseases predoctoral fellowship grant T32-AI0520660 (J.R.W.).
We thank Pradeep Singh and Benjamin Staudinger at the University of Washington and Jane Burns at Seattle Children’s Hospital for providing CF isolates.
Footnotes
Citation Wiens JR, Vasil A, Schurr MJ, Vasil ML. 2014. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio 5(1):e01010-13. doi:10.1128/mBio.01010-13.
REFERENCES
- 1. Boyd A, Chakrabarty AM. 1995. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J. Ind. Microbiol. 15:162–168. 10.1007/BF01569821 [DOI] [PubMed] [Google Scholar]
- 2. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- 3. Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21:595–599. 10.1016/j.pupt.2007.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194–222. 10.1128/CMR.15.2.194-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murray TS, Egan M, Kazmierczak BI. 2007. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr. Opin. Pediatr. 19:83–88. 10.1097/MOP.0b013e3280123a5d [DOI] [PubMed] [Google Scholar]
- 6. Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U. S. A. 100:7907–7912. 10.1073/pnas.1231792100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466–4475. 10.1128/JB.186.14.4466-4475.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J, Molin S. 2011. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ. Microbiol. 13:1705–1717. 10.1111/j.1462-2920.2011.02503.x [DOI] [PubMed] [Google Scholar]
- 9. Li Z, Chen JH, Hao Y, Nair SK. 2012. Structures of the PelD cyclic diguanylate effector involved in pellicle formation in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 287:30191–30204. 10.1074/jbc.M112.378273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banin E, Vasil ML, Greenberg EP. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 102:11076–11081. 10.1073/pnas.0504266102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barton HA, Johnson Z, Cox CD, Vasil AI, Vasil ML. 1996. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol. Microbiol. 21:1001–1017. 10.1046/j.1365-2958.1996.381426.x [DOI] [PubMed] [Google Scholar]
- 13. Ochsner UA, Johnson Z, Lamont IL, Cunliffe HE, Vasil ML. 1996. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 21:1019–1028. 10.1046/j.1365-2958.1996.481425.x [DOI] [PubMed] [Google Scholar]
- 14. Ochsner UA, Vasil ML. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 93:4409–4414. 10.1073/pnas.93.9.4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45:1277–1287. 10.1046/j.1365-2958.2002.03084.x [DOI] [PubMed] [Google Scholar]
- 16. Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792–9797. 10.1073/pnas.0403423101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilderman PJ, Vasil AI, Johnson Z, Wilson MJ, Cunliffe HE, Lamont IL, Vasil ML. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385–5394. 10.1128/IAI.69.9.5385-5394.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boyce JR, Miller RV. 1980. Effects of cations on stability of cystic fibrosis associated mucoid Pseudomonas. Lancet 2:268–269 [DOI] [PubMed] [Google Scholar]
- 19. Boyce JR, Miller RV. 1982. Selection of nonmucoid derivatives of mucoid Pseudomonas aeruginosa is strongly influenced by the level of iron in the culture medium. Infect. Immun. 37:695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Terry JM, Piña SE, Mattingly SJ. 1992. Role of energy metabolism in conversion of nonmucoid Pseudomonas aeruginosa to the mucoid phenotype. Infect. Immun. 60:1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reid DW, Anderson GJ, Lamont IL. 2009. Role of lung iron in determining the bacterial and host struggle in cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 297:L795–L802. 10.1152/ajplung.00132.2009 [DOI] [PubMed] [Google Scholar]
- 22. Reid DW, Carroll V, O’May C, Champion A, Kirov SM. 2007. Increased airway iron as a potential factor in the persistence of Pseudomonas aeruginosa infection in cystic fibrosis. Eur. Respir. J. 30:286–292. 10.1183/09031936.00154006 [DOI] [PubMed] [Google Scholar]
- 23. Reid DW, Lam QT, Schneider H, Walters EH. 2004. Airway iron and iron-regulatory cytokines in cystic fibrosis. Eur. Respir. J. 24:286–291. 10.1183/09031936.04.00104803 [DOI] [PubMed] [Google Scholar]
- 24. Reid DW, Withers NJ, Francis L, Wilson JW, Kotsimbos TC. 2002. Iron deficiency in cystic fibrosis: relationship to lung disease severity and chronic Pseudomonas aeruginosa infection. Chest 121:48–54. 10.1378/chest.121.1.48 [DOI] [PubMed] [Google Scholar]
- 25. Konings AF, Martin LW, Sharples KJ, Roddam LF, Latham R, Reid DW, Lamont IL. 2013. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect. Immun. 81:2697–2704. 10.1128/IAI.00418-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deretic V, Gill JF, Chakrabarty AM. 1987. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J. Bacteriol. 169:351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2:167. 10.3389/fmicb.2011.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hershberger CD, Ye RW, Parsek MR, Xie ZD, Chakrabarty AM. 1995. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative sigma factor (sigma E). Proc. Natl. Acad. Sci. U. S. A. 92:7941–7945. 10.1073/pnas.92.17.7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiu D, Eisinger VM, Rowen DW, Yu HD. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8107–8112. 10.1073/pnas.0702660104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak DJ. 2007. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 189:8353–8356. 10.1128/JB.00620-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rietsch A, Wolfgang MC, Mekalanos JJ. 2004. Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect. Immun. 72:1383–1390. 10.1128/IAI.72.3.1383-1390.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jain M, Bar-Meir M, McColley S, Cullina J, Potter E, Powers C, Prickett M, Seshadri R, Jovanovic B, Petrocheilou A, King JD, Hauser AR. 2008. Evolution of Pseudomonas aeruginosa type III secretion in cystic fibrosis: a paradigm of chronic infection. Transl. Res. 152:257–264. 10.1016/j.trsl.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vasil ML. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20:587–601 [DOI] [PubMed] [Google Scholar]
- 34. Nairz M, Schroll A, Sonnweber T, Weiss G. 2010. The struggle for iron—a metal at the host-pathogen interface. Cell. Microbiol. 12:1691–1702. 10.1111/j.1462-5822.2010.01529.x [DOI] [PubMed] [Google Scholar]
- 35. Fischer R, Simmerlein R, Huber RM, Schiffl H, Lang SM. 2007. Lung disease severity, chronic inflammation, iron deficiency, and erythropoietin response in adults with cystic fibrosis. Pediatr. Pulmonol. 42:1193–1197. 10.1002/ppul.20717 [DOI] [PubMed] [Google Scholar]
- 36. Ward RJ, Crichton RR, Taylor DL, Della Corte L, Srai SK, Dexter DT. 2011. Iron and the immune system. J. Neural Transm. 118:315–328. 10.1007/s00702-010-0479-3 [DOI] [PubMed] [Google Scholar]
- 37. Vasil ML, Ochsner UA. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399–413. 10.1046/j.1365-2958.1999.01586.x [DOI] [PubMed] [Google Scholar]
- 38. De Vos D, De Chial M, Cochez C, Jansen S, Tümmler B, Meyer JM, Cornelis P. 2001. Study of pyoverdine type and production by Pseudomonas aeruginosa isolated from cystic fibrosis patients: prevalence of type II pyoverdine isolates and accumulation of pyoverdine-negative mutations. Arch. Microbiol. 175:384–388. 10.1007/s002030100278 [DOI] [PubMed] [Google Scholar]
- 39. Poole K, McKay GA. 2003. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8:d661–d686. 10.2741/1051 [DOI] [PubMed] [Google Scholar]
- 40. Butcher BG, Bronstein PA, Myers CR, Stodghill PV, Bolton JJ, Markel EJ, Filiatrault MJ, Swingle B, Gaballa A, Helmann JD, Schneider DJ, Cartinhour SW. 2011. Characterization of the Fur regulon in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 193:4598–4611. 10.1128/JB.00340-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franklin MJ, Ohman DE. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000–3007. 10.1128/JB.184.11.3000-3007.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Damron FH, Qiu D, Yu HD. 2009. The Pseudomonas aeruginosa sensor kinase KinB negatively controls alginate production through AlgW-dependent MucA proteolysis. J. Bacteriol. 191:2285–2295. 10.1128/JB.01490-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ryan Withers T, Heath Damron F, Yin Y, Yu HD. 2013. Truncation of type IV pilin induces mucoidy in Pseudomonas aeruginosa strain PAO579. MicrobiologyOpen 2:459–470. 10.1002/mbo3.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Imperi F, Massai F, Facchini M, Frangipani E, Visaggio D, Leoni L, Bragonzi A, Visca P. 2013. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 110:7458–7463. 10.1073/pnas.1222706110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banin E, Lozinski A, Brady KM, Berenshtein E, Butterfield PW, Moshe M, Chevion M, Greenberg EP, Banin E. 2008. The potential of desferrioxamine-gallium as an anti-Pseudomonas therapeutic agent. Proc. Natl. Acad. Sci. U. S. A. 105:16761–16766. 10.1073/pnas.0808608105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith DJ, Lamont IL, Anderson GJ, Reid DW. 2013. Targeting iron uptake to control Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir. J. 42:1723–1736 [DOI] [PubMed] [Google Scholar]
- 47. Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J. Clin. Invest. 117:877–888. 10.1172/JCI30783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anderson GG, Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect. Immun. 76:1423–1433. 10.1128/IAI.01373-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moreau-Marquis S, O’Toole GA, Stanton BA. 2009. Tobramycin and FDA-approved iron chelators eliminate Pseudomonas aeruginosa biofilms on cystic fibrosis cells. Am. J. Respir. Cell Mol. Biol. 41:305–313. 10.1165/rcmb.2008-0299OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Knutson CA, Jeanes A. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal. Biochem. 24:470–481. 10.1016/0003-2697(68)90154-1 [DOI] [PubMed] [Google Scholar]
- 52. Read RR, Costerton JW. 1987. Purification and characterization of adhesive exopolysaccharides from Pseudomonas putida and Pseudomonas fluorescens. Can. J. Microbiol. 33:1080–1090. 10.1139/m87-189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Influence of assorted metals on alginate production. P. aeruginosa PAO1 (wild type) and the PAO1 ∆mucD mutant were grown on DTSA supplemented with 100 µM ZnCl2, CuCl2, CaCl2, MgCl2, or FeCl3. Extracellular alginate was measured by uronic acid assay after 48 h of growth. Download
Global gene expression increases under iron limitation versus iron-replete conditions in P. aeruginosa PAO1 and ∆mucA mutant. Duplicate cultures of P. aeruginosa wild-type PAO1 and ∆mucA and ∆mucD mutants were grown on DTSA solid medium with and without 100 µM FeCl3, and cells were harvested after 24 h. The global gene expression profiles were examined using GeneChip microarray analysis (15), and the results were compared and averaged.
Analysis of mucoid phenotypes of Pseudomonas aeruginosa strains from young CF patients at Seattle Children’s Hospital. Detailed descriptions of eight early CF isolates from individual children with CF are given in the table. The isolates were obtained from Jane Burns at Seattle Children’s Hospital. This table contains phenotypic analysis of isolates grown on BHI and DTSA with and without 100 µM FeCl3 and zone size (millimeters) of halos observed on CAS agar plates. Additionally, this table provides data relating to zone size (in millimeters) of several PAO1 muc mutants and PAO1 siderophore mutants that were used as controls for this study.
Late CF clinical isolates. This table contains descriptions of the phenotypes of 17 late CF isolates originating from older CF patients who had been colonized with P. aeruginosa for at least 5 years. These isolates were obtained from Benjamin Staudinger at the University of Washington. Strains were grown on BHI and DTSA with and without 100 µM FeCl3 and analyzed for alginate production and mucoidy. Zone size (in millimeters) of halos were noted after growth on CAS agar plates.
Supplemental Materials and Methods Download