Abstract
The production of pluripotent stem cells (iPSCs) for therapeutic applications will require practical methods to achieve tight temporal and quantitative control of reprogramming factor (RF) expression, while avoiding the mutagenic potential of gene transfer. Toward this end, we have developed cell-permeable RF proteins (CP-RFs) incorporating newly developed macromolecule transduction domains (MTDs). Treatment of human dermal fibroblasts (HDFs) with combinations of cell-permeable OCT4, SOX2, KLF4, CMYC and either NANOG or LIN28 proteins induced the outgrowth of stem cell-like colonies (iSCs). iSC colonies generated with CP-RFs resembled embryonic stem cells with regard to morphology, biomarker expression, and extended capacity for self-renewal, but failed to expand as iPSC or ES cell lines. Partial reprogramming appears to be a common response to protein-based delivery of programming factors into somatic cells.
Terminally differentiated somatic cells can be reprogrammed to become induced pluripotent stem cells (iPSCs) by enforced expression of reprogramming factors (RFs) that promote self-renewal and render cells pluripotent with regard to cellular differentiation1,2. RFs include proteins individually required to maintain embryonic stem (ES) cells in a pluripotent state [notably: octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), and NANOG3,4,5,6], as well as proteins [e.g. CMYC, Kruppel-like factor 4 (KLF4) and LIN28] that promote self-renewal and suppress cellular differentiation7,8. Combinations of RFs with highest iPSC activity (e.g. OCT4 and SOX2 plus either KLF4 and CMYC or alternatively, NANOG and LIN28) were discovered from libraries of candidate RFs by combinatorial screens1,2.
In principle, patient-derived iPSCs could be used for autologous stem cell therapies without the ethical and graft rejection problems associated with using embryo-derived stem cells. In practice, the application of iPSCs to human regenerative medicine will require efficient methods to introduce RFs into somatic cells combined with ways to guard against dysregulated RF activity and vector-induced mutations. Unfortunately, somatic cell reprogramming is relatively inefficient and thus requires relatively efficient methods to regulate RF activity. Although retroviruses can achieve sufficient gene transfer efficiencies, any vector that integrates into the genome is potentially mutagenic. Moreover, RFs that enhance iPSC formation may undermine subsequent clinical applications, as illustrated by CMYC, which collaborates with other RFs to enhance iPSC formation but induces tumors in iPSC-derived tissues9. Similarly, ectopic OCT4 and KLF4 promote epithelial dysplasias10,11. For these reasons, various strategies have been developed to produce transgene-free iPSCs, including: loxP flanked vectors12, excisable transposons13, adenovirus14,15 and Sendai virus16 vectors and non-integrating episomal vectors17,18. Other approaches have moved completely away from DNA-based expression vectors, including: synthetic modified RNA19, epigenetic regulation by chemical compounds (see20 for a review) and direct uptake of cell-penetrating RF proteins21,22. In principle, the latter approaches not only avoid major drawbacks (genetic damage and gene dysregulation) associated with gene-based vectors, they also provide better quantitative and temporal control of stem cell reprogramming.
In the present report, we investigated the use of macromolecule intracellular transduction technology (MITT) to deliver biologically active RFs into human skin fibroblasts. MITT was used previously to deliver peptides and proteins to a variety of tissues (notably liver, lung, pancreas and lymphoid tissues), resulting in dramatic protection against lethal inflammatory diseases23,24,25,26,27, suppression of pulmonary metastases28 and inhibition of subcutaneous tumor xenografts29,30. The technology exploits the ability of hydrophobic macromolecule transduction domains (MTDs) to promote bidirectional transfer of peptides and proteins across the plasma membrane29,31,32. By contrast, cationic protein transduction domains (PTDs, e.g. those derived from HIV Tat and Antennapedia) enhance protein uptake predominately through absorptive endocytosis and macropinocytosis, which sequester significant amounts of protein into membrane-bound and endosomal compartments33,34. In practice, uptake mechanisms are strongly influenced by the cargo, and must be investigated on a case-by-case basis33,34,35. In the present study, we experienced difficulties expressing recombinant RF proteins in soluble form. The problem was solved, in part, by testing multiple MTD sequences. The resulting cell-permeable RFs induced colonies with extended capacity for self-renewal that resembled ES cells with regard to morphology and biomarker expression.
Results
Cell-permeable reprogramming factors
Protein sequences derived from the leader peptides of secreted proteins, designated macromolecule transduction domains, have been shown to enhance the cellular uptake of recombinant proteins both in vitro and in vivo. These domains were identified from a screen of 1,500 potential hydrophobic signal peptides for sequences with protein transduction activity as assessed using an EGFP reporter protein28,29. However, achieving practical levels of cytosolic protein delivery requires solving problems relating to protein yield, solubility, structure/activity, route of entry, and sequestration. In practice, the process is largely empirical.
To deliver recombinant transcription factors for the purpose of reprogramming differentiated somatic cells, candidate MTDs were modified and analyzed for the ability to promote the uptake of FITC-labeled proteins by cultured cells. The resulting MTDs (MTD47, MTD52, MTD84, MTD86, MTD132, MTD173 and MTD181; Supplementary Tables S1 A and B) displayed higher protein transduction activity than either a random peptide (SANVEPLERL: S) or the membrane translocating sequence from fibroblast growth factor 4 (FGF4 MTS: AAVLLPVLLAAP: Mm) (Supplementary Figure S1 and S2).
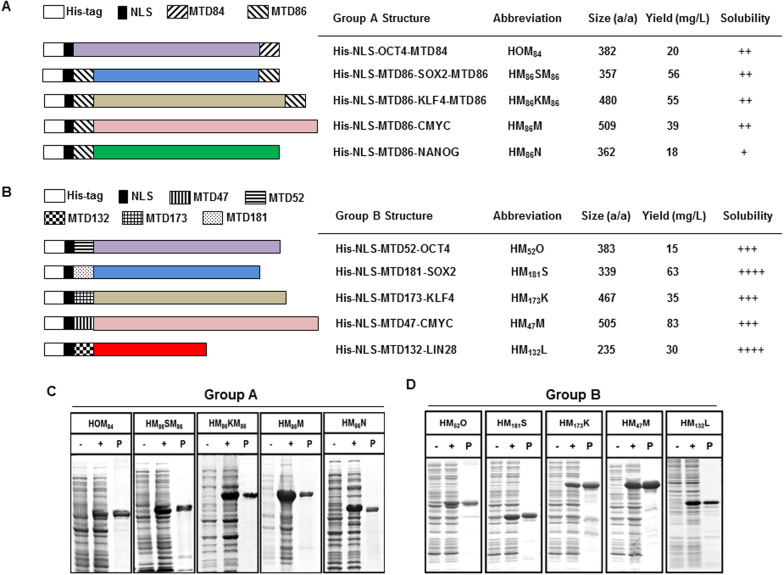
Recombinant fusion proteins bearing various MTD sequences were designed to deliver two different collections of reprogramming factors (RFs) into somatic cells. Group A RFs [OCT4 (O), SOX2 (S), KLF4 (K), CMYC (M), and NANOG (N)] contained MTD84 or MTD86 attached to the N-terminus (HM86M and HM86N), to the C-terminus (HOM84) or to both ends (HM86SM86 and HM86KM86) of the protein (Figure 1A); whereas, the Group B RFs [OCT4 (O), SOX2 (S), KLF4 (K), CMYC (M), and LIN28 (L)] contained MTD47, MTD52, MTD132, MTD173 or MTD181 affixed to the N-terminus (Figure 1B). Each Group A and Group B RF employed the MTD that was empirically determined to produce the fusion protein with the greatest solubility (data not shown). A 6-histidine tag (6xHis: H) and a nuclear localization sequence (NLS) derived from SV40 large T-antigen (KKKRK) were also fused to the N-terminus of RFs to facilitate single-step affinity chromatography and cellular nuclear localization, respectively. Each protein was expressed in E. coli, purified under denaturing conditions (Figure 1 A and B) and refolded as described previously28,29. The individual CP-RFs were similarly capable of activating the expression of cognate reporter genes (Supplementary Figure S3 and data not shown). Control proteins (HO, HS, HM, HK, HL and HN) that lacked a MTD sequence were also prepared (data not shown). Finally, plasmids used to express Group A RFs utilized mammalian gene sequences; whereas, coding sequences for Group B reprogramming factors were modified to conform to E. coli codon preferences.
Figure 1. Structure and expression of recombinant cell-permeable reprograming factors.
(A–B) Group A reprogramming factors contain a 6× His tag (white), SV40 nuclear localization sequence (black) and MTD84 or MTD86 sequence (hatched) positioned (A). Numbered MTD sequences are positioned at the amino terminal end of human sequences—codon optimized for expression in E. coli (B). Solubility was scored on a 4 point scale ranging from highly soluble proteins with little tendency to precipitate (++++) to largely insoluble proteins (+). (C–D) Recombinant cell-permeable reprogramming factor proteins (group A: C; Group B: D) expressed in E. coli before (−) and after (+) induction with IPTG, and purification by Ni2+ affinity chromatography (P). Each group of the recombinant proteins (group A and group B) was run on the gels (C and D) under the same experimental conditions.
Group A recombinant proteins with MTD13 (LAAAALAVLPL), MTD57 (LIALLAAPLA), MTD85 (LLAAAAALLLA) and MTD108 (ALLAALLAP) in place of MTD84 and MTD86 were also evaluated but the proteins were less soluble, produced lower yields when expressed in E. coli and entered cells less efficiently (data not shown); therefore, these proteins were not evaluated further.
Cellular reprogramming by group A reprogramming factors
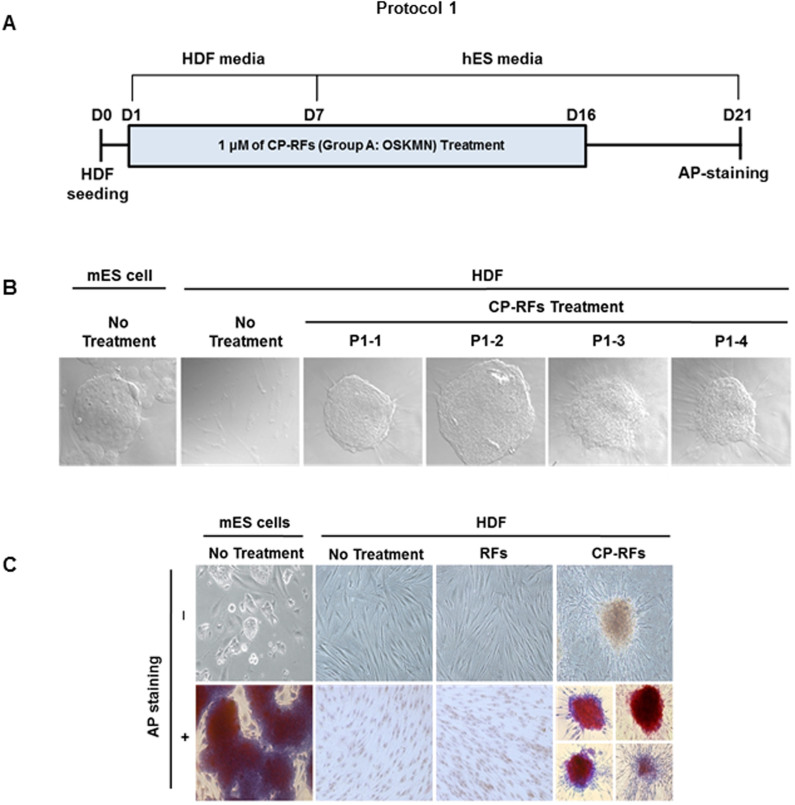
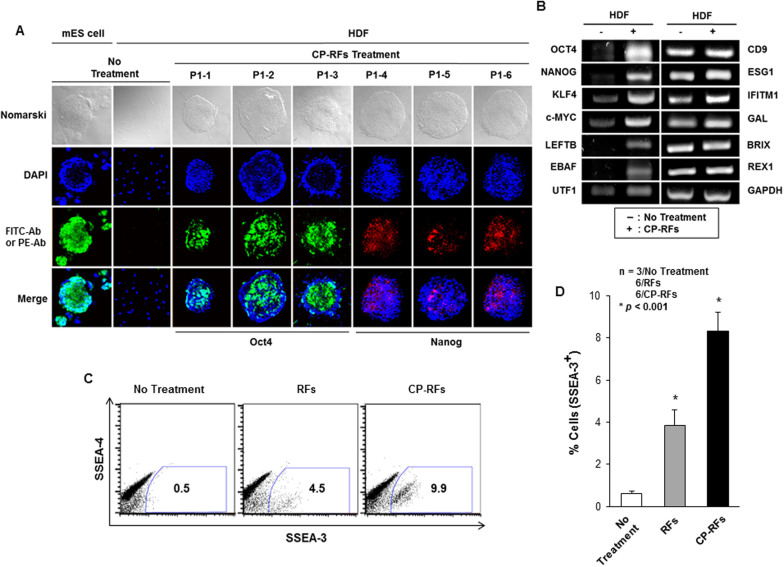
We first assessed the ability of human dermal fibroblasts (HDFs) to be reprogrammed by a cocktail of Group A RFs (OSKMN). 2 × 105 HDFs, maintained in HDF media, were treated with Group A CP-RFs (1 μM each) for 16 days. Media and recombinant proteins were replaced daily and the cultures were switched to human embryonic stem (hES) cell media after 7 days (Protocol 1, Figure 2A). Twenty-three granulated colonies emerged in cultures treated with MTD-containing RFs between day 5 and day 8; whereas, no colonies were observed in cultures treated with control proteins that lacked MTD sequence. The colonies morphologically resembled mouse embryonic stem (mES) cells (Figure 2B), and expressed alkaline phosphatase, as assessed by TRA-2-49/6E antibody staining 21 days into the experiment (Figure 2C). The 21-day colonies also expressed higher levels of stem cell-specific biomarkers36 as compared to cells from untreated controls, including OCT4 and NANOG as assessed by indirect immunofluorescence (Figure 3A) and OCT4, NANOG, KLF4 CMYC, LEFTB, EBAF and UTF1 as determined by reverse transcriptase PCR (Figure 3B). By contrast, expression of all non-stem cell differentiation markers examined (CD9, ESG1, IFITM1, GAL, BRIX, REX1 and GAPDH) was unchanged (Figure 3B).
Figure 2. Cell-permeable OCT4, SOX2, KLF4, CMYC and NANOG induce iSC colonies (Protocol 1).
(A) Experimental timeline. Human dermal fibroblasts (HDF) were plated (day zero, D0), allowed to attach overnight and were treated daily with HDF media alone (no treatment), with 1 μM (each) recombinant OCT4, SOX2, KLF4, CMYC and NANOG proteins (RFs) or the same proteins containing MTD sequences to promote intracellular delivery (CP-RFs). The media was changed to hES media on day 7 (D7) and protein treatments (24 hrs/day) ended on day 16 (D16). (B) Colony morphologies. Photomicrographs of 4 representative iSC colonies induced by CP-RF treatment compared to a murine embryonic stem (mES) cell colony. (C) CP-RF induced colonies express alkaline phosphatase.
Figure 3. Cell-permeable reprogramming factor-induced colonies express stem cell markers.
(A) OCT4 and NANOG Immunostaining. Murine ES cell colony, MEF feeder layer cells (MEF) and colonies induced by treatment with CP-RFs (Protocol 1: 1 μM each protein, OSKMN, daily for 16 days) were stained. (B) RT-PCR analysis of stem cell biomarker expression. (C–D) Flow cytometric analysis of SSEA-3 and SSEA-4. Cells were stained with fluorescent antibodies against stage specific embryonic antigen 3 (SSEA-3) and 4 (SSEA-4), and measured by FACS analysis (C) and quantified (D). The data are presented as means ± s.d. (n = 3/No Treatment; 6/RFs; 6/CP-RFs). * p < 0.001 as determined by a Student's unpaired t-test.
These results indicate that cell-permeable Group A RFs can induce colonies with enhanced capacity for self-renewal and these colonies display morphologies and patterns of biomarker expression consistent with somatic to stem cell reprogramming. While the reprogramming was inefficient (0.01%), no colonies were observed (0%) following treatment with RF proteins lacking a MTD sequence. Since the colonies were selected based on enhanced self-renewal, unselected cells in the cultures were also tested for evidence of cellular reprogramming after treatment with CP-RF. For this we monitored the expression of stem cell-specific surface markers, SSEA-3 and SSEA-4, by flow cytometry as shown with a representative plots (Figure 3C) and analyzed statistically (Figure 3D, p < 0.001). Levels of SSEA-3 (and to a lesser extent SSEA-4) expression suggest a greater level of cellular reprogramming in the population as a whole than was inferred by colony outgrowth.
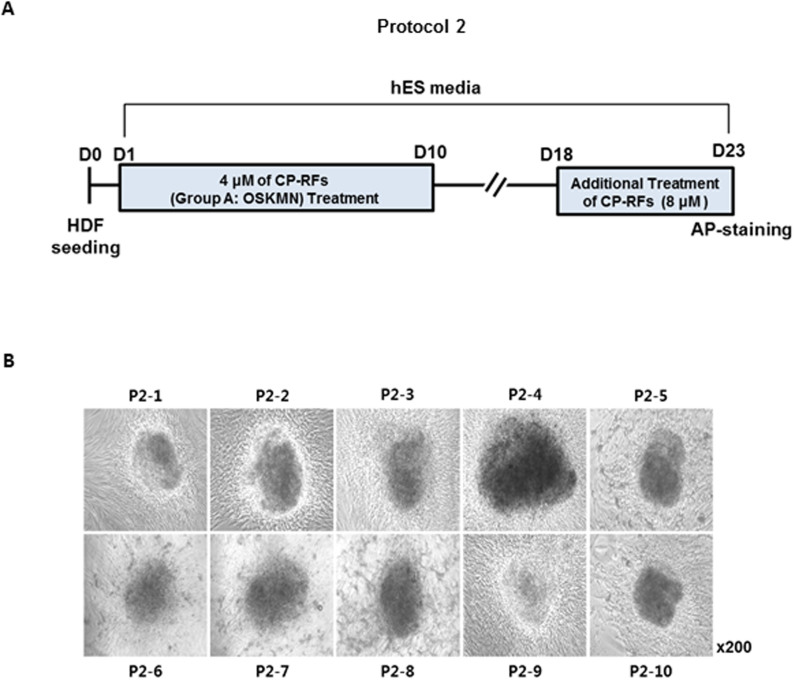
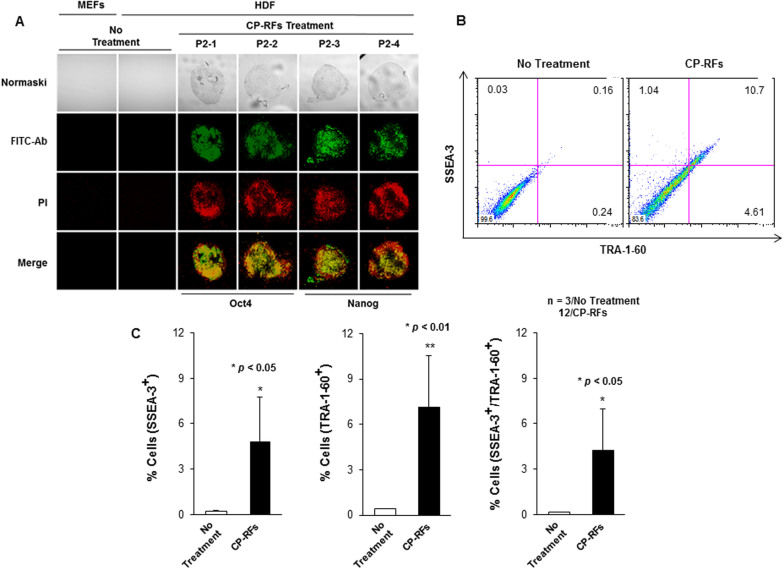
Group A CP-RFs were further tested using a variety of conditions, as illustrated by Protocol 2 (Figure 4A). Maintaining cells in hES media from the start had no effect (data not shown); whereas, increasing the concentration of CP-RFs (4 μM each) increased the frequency of colony formation by ~2.5 fold (0.025%) compared to Protocol 1 (0.01%) which used lower concentration of CP-RFs (1 μM each). As before, the colonies emerged between day 5 and 8. Prolonging the treatment interval had no effect on colony formation (data not shown), nor did a second application of 8 μM CP-RFs administered between days 18 and 23 (Figure 4A). The colonies morphologically resembled ES cells, although this was less apparent at day 23 (Figure 4B), when the colonies had become over-grown. As before, the colonies expressed alkaline phosphatase (data not shown), OCT4 and NANOG (Figure 5A) and the cell surface markers TRA-1-60 and SSEA-3 detected by flow cytometry as shown with a representative plots (Figure 5B) and analyzed statistically (Figure 5C, * p < 0.05 or ** p < 0.01). The latter were expressed at levels 9 to 12 and 8 to 33 fold higher than in untreated HDFs (Figure 5 B and C, and data not shown).
Figure 4. Cell-permeable OCT4, SOX2, KLF4, CMYC and NANOG induce iSC colonies (Protocol 2).
(A) Experimental timeline. Human dermal fibroblasts (HDF) were plated (day zero, D0), allowed to attach overnight and were treated daily with HDF media alone (no treatment), with 4 μM or 8 μM (each) recombinant proteins (RFs) or the same proteins containing MTD sequences to promote intracellular delivery (CP-RFs). (B) Colony morphologies. P2 stands for Protocol 2 followed by the serial ID number for each colony.
Figure 5. Cell-permeable reprogramming factor-induced colonies (Protocol 2) express stem cell markers.
(A) OCT4 and NANOG Immunostaining. Untreated HDFs and colonies induced by treatment with CP-RF (Protocol 2) were stained. (B–C) TRA-1-60 and SSEA-3 expression (B) and relative numbers of single and double positive cells (C). The data are presented as means ± s.d. (n = 3/No Treatment; 12/CP-RFs). * p < 0.05 and ** p < 0.01 as determined by a Student's unpaired t-test.
Since colonies induced by Group A RFs resembled iPSCs with regard to enhanced capacity for self renewal, colony morphology, and biomarker expression, we repeatedly tried to expand the cells in order to assess their differentiation potential. However, we were unable to expand the colonies either by picking individual colonies or by passaging all cells on plates that contained multiple colonies. These unsuccessful efforts employed a variety of growth conditions including those (lot selected FBS, LIF, mouse embryo feeder layers and collagen-treated culture dishes) routinely used to establish murine ES cell lines from pre-implantation embryos.
Cellular reprogramming by group B reprogramming factors
The Group A RFs tended to precipitate after being added to cells maintained either in human dermal fibroblast (HDF) or human embryonic stem cell (hESC) media, and CP-NANOG (HM86N) had the worst solubility problems. We therefore developed a second group of CP-RFs, designated Group B, in which NANOG was replaced with LIN28. LIN28 has been reported to promote cell proliferation by regulating mRNA stability of various cell cycle regulators in ESCs. Each Group B RF also employed a different MTD (Supplementary Table S1B), empirically selected to produce fusion proteins with the greatest relative solubility.
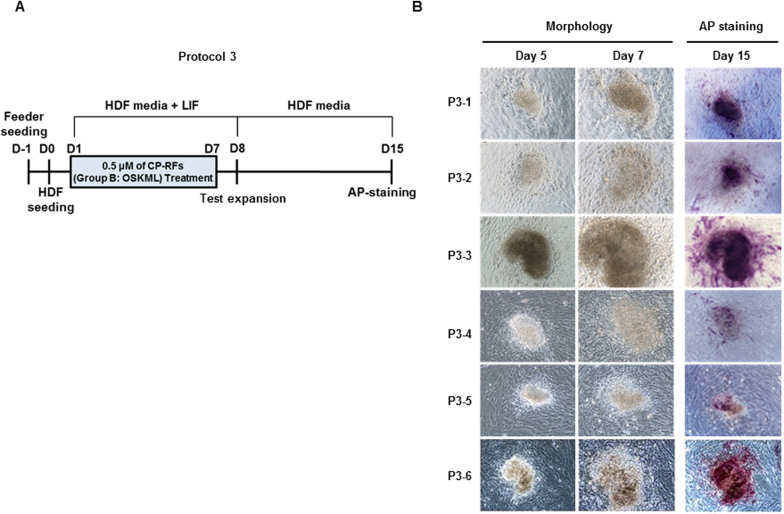
In Protocol 3 (Figure 6A), 1 × 105 HDFs maintained in HDF media including LIF were plated on irradiated mouse embryo fibroblast feeder layers and treated daily for seven days with 0.5 μM of Group B RFs for 3 hrs. Colonies emerged between days 3 to 8 (7 colonies at day 3, 9 at day 5 and 3 at day 8) at an overall frequency of 0.02% and were tested for the ability to be expanded in mass culture on day 8. After adding fresh feeder cells, the colonies were maintained for another 7 days in the absence of RFs and stained with alkaline phosphatase (Figure 6B). Control cultures (i.e. feeder cells alone) treated with RFs did not produce colonies, indicating the colonies were derived from HDFs, which based on cell morphology, enhanced capacity for self-renewal and alkaline phosphatase staining had undergone somatic to stem-cell reprogramming.
Figure 6. Cell-permeable OCT4, SOX2, KLF4, CMYC and LIN28 induce iSC colonies (Protocol 3).
(A) Experimental timeline. Irradiated murine embryonic fibroblasts were plated at one day before (D-1) human dermal fibroblasts (HDF) (day zero, D0). Cultures were treated daily for 7 days with HDF media alone (no treatment), for 3 hours daily with 0.5 μM (each) codon-optimized recombinant proteins (RFs) or the same proteins containing MTD sequences to promote intracellular uptake (CP-RFs). Test expansion indicates when an effort was made to expand colonies in mass culture. (B) Colony morphologies and alkaline phosphatase staining.
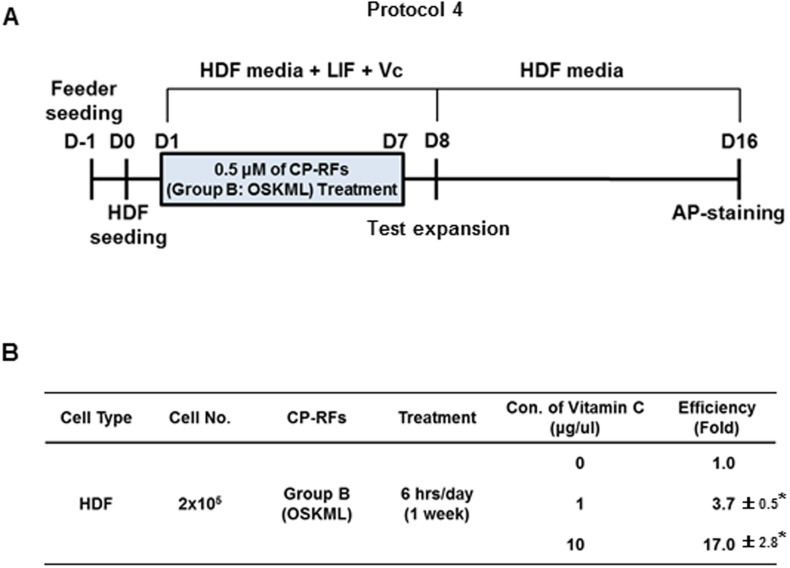
It has been reported that vitamin C could improve the reprogramming efficiency and promote the transition of partially reprogrammed iPSCs to a fully reprogrammed state37. The effect was attributed to anti-oxidant properties of ascorbic acid and suppression of replicative senescence. Therefore, we decided to use vitamin C as an adjuvant for reprogramming. Increasing the exposure to Group B RFs from 3 to 6 hours per day had no effect (data not shown) on colony formation (Protocol 4, Figure 7A). However, the addition of 1 and 10 mg/ml vitamin C to the media increased the number of colonies by 3.7- (0.074%) and 17-fold (0.34%), respectively (Figure 7B, p < 0.001). These results may suggest that the combination treatment of optimized CP-RFs and an adjuvant molecule could increase the efficiency of iSC generation. However, we were again unable to expand these colonies either individually or in mass culture using a variety of growth conditions including those routinely used to establish murine ES cell lines.
Figure 7. Vitamin C enhances iSC reprogramming (Protocol 4).
(A) Experimental timeline. Radiation-inactivated murine embryonic fibroblasts were plated at one day before (D-1) human dermal fibroblasts (HDF) were plated (day zero, D0). Cultures were treated as described in Figure 6, except the cultures were treated with proteins for 6 hours each day in HDF media containing 0, 1 or 10 mg/ml L-ascorbic acid. (B) Vitamin C enhances iSC colony formation. The percent of treated cells that gave rise to iSCs is tabulated based on the means of three separate experiments ± s.d. (n = 3/group). * p < 0.001 as determined by a Student's unpaired t-test.
Discussion
The reprogramming of somatic cells into stem cells has tremendous medical potential as a way to generate patient-specific stem cells for tissue regeneration and repair. In the present study we used macromolecular intracellular transduction technology to develop cell-permeable OCT4, SOX2, KLF4, CMYC, NANOG and LIN28 transcription factors and tested mixtures of these proteins for the ability to reprogram primary human dermal fibroblasts. Protein cocktails consisting of cell-permeable OCT4, SOX2, KLF4, CMYC and either NANOG or LIN28 induced stem cell-like colonies (iSCs) with frequencies ranging from 0.01 to 0.34% as summarized in Table 1. The earliest colonies emerged within three days after starting protein treatment (0.5 or 4 μM each; added daily for 3 or 24 hours over 7 days) and resembled stem cells in terms of morphology, extended capacity of self-renewal, and expression of stem cell-specific markers, notably OCT4, NANOG, KLF4, CMYC, LEFTB, EBAF, UTF1, SSEA3 and alkaline phosphatase. By contrast, control proteins lacking MTD sequences did not induce stem cell colonies. However, the response to reprogramming factors appeared to be transient (TRA-2-49/6E+; TRA-1-60+; SSEA-3+; SSEA-4low), as we were unable to expand the colonies further and therefore were unable to assess their differentiation potential.
Table 1. Summary of conditions used for human dermal fibroblast reprogramming.
| Conditions | Protocol 1 | Protocol 2 | Protocol 3 | Protocol 4 |
|---|---|---|---|---|
| Feeder | No | No | Yes | Yes |
| Supplements | No | No | LIF | LIF + Vitamin C |
| Minimum Period of Treatment | 5 days | 5 days | 3 days | 3 days |
| Concentration | 1 μM each | 4 μM each | 0.5 μM each | 0.5 μM each |
| Efficiency | 0.01% | 0.025% | 0.02% | 0.34% |
| CP-RFs | Group A (OSKMN) | Group B (OSKML) | ||
Treatment protocols utilized CP-RF (Group A: OCT4, SOX4, KLF4, CMYC and NANOG; OSKMN) or codon-optimized CP-RF (Group B: OCT4, SOX4, KLF4, CMYC and LIN28; OSKML). Reprogramming efficiencies were calculated as the percent of HDFs giving rise to alkaline phosphatase positive colonies.
In principle, protein transduction technologies provide a direct way to manipulate biochemical processes in living cells under non steady-state conditions. The approach exploits the ability of specific basic, amphipathic and hydrophobic (or amphipathic depending on cargo) protein sequences to enhance the uptake of proteins and other macromolecules by mammalian cells33,34. The 7 MTDs described in the present study add to a growing list of signal peptide-derived sequences that enhance protein uptake by cultured cells. Features of the MTD responsible for protein transduction activity are clearly shared by a variety of peptide sequences raging from 7 to 12 amino acids in size. We are currently trying to identify optimal sequence and/or structural determinants for cytosolic and tissue delivery/uptake and to assess potential contributions by cargo sequences. Our efforts to develop cell-permeable reprogramming factors were hampered by poor solubility of the recombinant proteins in culture media. The problem was improved, but not completely eliminated by extensively testing a variety of MTD sequences with each cargo protein.
The Yamanaka group reported a gradual reprogramming process of fibroblasts, in which granulated colonies appeared approximately two weeks after retroviral transduction of 4 RFs, and distinct types of colonies that were flat and resembled hESCs were observed around day 2538,39. In Protocol 1, we used Group A-5 CP-RFs and observed many granulated colonies (efficiency: 0.01%) within a week, and the colonies maintained in ES cell media displayed ESC-like morphology and robust expression of stem cell-specific markers (ex, SSEA-3, p < 0.001). The efficiency was increased to 0.025% with expression of biomarkers (ex, TRA-1-60, p < 0.05; SSEA-3, p < 0.01) when the cells were treated with a higher concentration of CP-RFs in Protocol 2, and in both cases colony formation was not observed with RFs without MTD sequences. These results suggest that the reprogramming process depended on intracellular levels of RF proteins.
In Protocols 3 and 4, we reduced the duration of daily treatment of CP-RFs from 24 hrs/day to 3 hrs/day for a week, and we reduced the concentration of protein in cells from 1–4 μM of Group A CP-RFs (OSKMN) to less than 0.5 μM of Group B (OSKML). Reprogramming efficiencies maintained (0.02%) even though the duration and the concentration were significantly reduced (both 8-fold) compared with Protocols 1 and 2.
Compared to previous reports using CP-RFs, we observed higher reprogramming frequencies (0.025% for 3 days to 0.001% for 56 days21 and 0.002% for 35 days22, respectively) using human dermal fibroblasts as reported with newborn human fibroblasts21 and mouse embryo fibroblasts22. Moreover, these efficiencies were achieved using purified recombinant RF proteins, unlike previous studies that used unpurified RFs present in cell lysates21 or RFs supplemented with valproic acid, a histone deacetylase inhibitor, to enhance reprogramming22. Thus, our study is the first to achieve stem cell reprogramming with purified CP-RFs alone.
However, we were unable to expand iSC colonies as expected for iPSC or ES cell lines and therefore were unable to assess cellular differentiation potentials. Our experience differs from previous reports using gene transfer to induce pluripotent stem cell reprogramming. This suggests cell permeable reprogramming factors have a greater propensity as compared to gene transfer approaches toward non-productive or partial reprogramming, even when similar cell culture conditions were used. Future studies will test whether protein-based approaches will benefit from changes in culture conditions known to enhance iPSC reprogramming following gene transfer. Furthermore, testing different protein formulations and optimizing the levels and timing of their application may be required to completely reprogram human somatic cells into pluripotent stem cells with the expected capacity for self-renewal.
Vitamin C has been reported to stimulate iPSC reprogramming of human dermal fibroblasts40. Similarly, we observed 3.7 to 17 more iSCs induced by CP-RFs with the addition of 1 and 10 mg/ml vitamin C, respectively. The effect has been attributed to anti-oxidant properties of ascorbic acid and suppression of replicative senescence; however, the other anti-oxidants tested did not enhance iPSC formation40. Alternatively, the effects of vitamin C--which has long been known to stimulate the proliferation, lifespan and multi-cellular depth of human dermal fibroblasts--are as readily attributed to enhanced collagen production41,42,43,44.
Although we showed that iSC colonies resembled embryonic stem cells with regard to morphology, biomarker expression, and extended capacity for self-renewal, we could not expand the colonies to assess their differentiation potential in vivo. The iSC colonies formed more rapidly (3 to 5 days) than previous studies describing successful iPSC reprogramming. Therefore, the speed of initial colony outgrowth appears to be a negative indication with regard to iPSC reprogramming. This experience is expected to help other groups interested in using CP-RFs to generate functional iPSCs.
While we could not confirm the pluripotency of the colonies generated from the terminally differentiated cells treated with CP-RFs, CP-RFs incorporating macromolecule transduction domains should result in a better quantitative and temporal control of the reprogramming. After somatic cells are induced to form iSC colonies, the transcription factors turnover seamlessly, with no trace left behind.
This study is the first to achieve partial stem cell reprogramming with purified CP-RFs alone through MITT. The approach circumvents safety concerns that preclude clinical applications of retroviral and other gene-based delivery systems. However, we were unable to expand iSC colonies as expected for iPSC or ES cell lines and therefore were unable to assess cellular multilineage differentiation potentials. This experience is expected to help other groups interested in using CP-RFs to generate functional iPSCs.
Methods
Construction of expression vectors for recombinant proteins
MTDs were identified by screening candidate signal peptides for hydrophobic sequences with protein transduction activity (Jo et al., manuscript in preparation). MTD84 and MTD86 for Group A recombinant proteins, and MTD47, MTD52, MTD132, MTD173 and MTD181 for Group B proteins were derived from signal sequences from AAK63068 and NP_629842 from Phytophthora cactorum and Streptomyces coelicolor, respectively, and NP_627512, NP_775628, NP_628377, NP_624384 and CAB84257 from StrePtomyces coelicolor, Homo sapiens, Streptomyces coelicolor, Streptomyces coelicolor and Neisseria meningitides, respectively.
Coding sequences for EGFP (E) and reprogramming factors (OCT4, SOX2, KLF4, CMYC, NANOG and LIN28; abbreviated O, S, K, M, N, and L, respectively) were cloned into pET-28a(+) (Novagen) from PCR-amplified DNA segments (Supplementary Table S2).
For Group A recombinant RF proteins, the 2 MTDs were used for development of 35 recombinant proteins by attaching to the N-terminus, to the C-terminus or to both ends of each factor (2 MTDs × 5 RFs × 3 different structures = 30 MTD-fused proteins + 5 MTD-free control proteins). Among them, we chose CMYC and NANOG with N-terminal fused to MTD86, OCT4 with C-terminal fused to MTD84 and SOX2 and KLF4 with both ends fused to MTD86 because these proteins were the most soluble and produced the highest when expressed in E. coli.
Group B proteins (OCT4, SOX2, KLF4, CMYC and LIN28) used N-terminal MTDs (MTD47, MTD52, MTD132, MTD173 and MTD181), empirically chosen based on their relative cell permeability (1.8; 1.6; 1.3; 2.3 and 3.5 fold) compared to a reference domain--membrane translocating sequence from Fibroblast Growth Factor 4 (FGF4 MTS: AAVLLPVLLAAP) assessed using an EGFP reporter protein. pET-28a(+)-based vectors expressing O, S, K, M, N, and L were further optimized by incorporating E.coli codon preferences. Expression constructs were sequenced prior to use.
Cell permeability and intracellular localization
Recombinant proteins were purified from E. coli BL21-CodonPlus (DE3) cells, purified under denaturing conditions and refolded as described previously28,29,30. Recombinant proteins were conjugated to FITC according to the manufacturer's instructions (Pierce Chemical, Rockford, IL). RAW 264.7 cells (Korean Cell Bank, Seoul, Korea) were treated with 10 μM of FITC-labeled proteins for 1 hr at 37°C and washed with PBS three times. To remove cell-surface bound FITC-labeled protein, the cells were treated with proteinase K (10 μg/ml) for 20 min at 37°C. Quantitative cell permeability was determined by FACS analysis (FACS Calibur; Becton Dickinson, Franklin Lakes, NJ). To visualize intracellular localization of the proteins, NIH3T3 cells (Korean Cell Bank, Seoul, Korea) were treated with 10 μM of FITC-conjugated recombinant proteins for 1 hr at 37°C, washed with PBS three times, and treated with proteinase K (10 μg/ml) for 20 min at 37°C. Treated cells were counterstained with the nuclear fluorescent stain propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) at a concentration of 1 μg/ml and washed with cold PBS three times. The intracellular localization was determined using confocal laser scanning microscopy. Each experiment was conducted at least three times.
Treatment with cell-permeable reprogramming factors
Human dermal fibroblasts prepared from the facial skin of a healthy Western European descent woman in her thirties were purchased (CELLnTEC, Stauffacherstr, Switzerland) and maintained according to the manufacturer's instructions. Human resource (human dermal fibroblasts) handling and experimental procedures were approved and performed in accordance with the guidelines of Institutional Review Board (IRB) of the ProCell R&D Institute, ProCell Therapeutics, Inc. The cells were cultured at 37°C in a humidified atmosphere of 5% CO2 in a HDF medium (M106 (Cascade Biologic, Carlsbad, CA) + LSGS (Cascade Biologic, Carlsbad, CA)). The HDF medium was exchanged with a hESC medium (DMEM/F12 (HyClone, Waltham, MA) supplemented with 20% KSR, 2 mM L-glutamine, 2 mM MEM Non-Essential Amino Acid, 0.1 mM β-mercaptoethanol, and 0.1% penicillin/streptomycin (all from GIBCO, Carlsbad, CA)). Mouse embryonic stem (mES) cells were also cultured1. All plates we used were coated with 0.1% gelatin. The cells were treated with the recombinant proteins at a given concentration and duration as followed; Protocol 1: HDFs (2 × 105) were treated with Group A-5 CP-RFs (OSKMN, 1 μM each) for 16 days. The cells in presence of proteins were incubated in HDF media for the first 7 days and then in hESC media for 9 days. During the treatment, recombinant proteins were replaced daily (24 hrs/day). After protein treatment, the cells were maintained in hESC media for 5 days. Protocol 2: HDFs (5 × 104) were treated with Group A-5 CP-RFs (4 μM each) for 10 days and maintained in the absence of proteins for 8 days, following additional protein treatment (8 μM each) in hESC media for 5 days. Recombinant proteins were replaced every day (24 hrs/day). Protocol 3: HDFs (1 × 105) maintained in HDF media including LIF (1 μg/ml) on feeder cells derived from mouse embryos were treated with Group B–5 CP-RFs (OSKML, 0.5 μM each) for 7 days (3 hrs/day). The colonies were transferred on the fresh feeder cells at day 8 and were maintained for another 7 days in the absence of proteins in hES media. Protocol 4: HDFs (2 × 105) were treated like Protocol 3 except the addition of vitamin C (0, 1 or 10 μg/μl) and daily treatment (6 hrs/day).
Alkaline phosphatase staining
A capsule of Fast Blue RR salt in the alkaline phosphatase staining kit (Sigma Aldrich, St. Louis, MO) was dissolved in water to prepare a diazonium salt solution. Naphthol AS-MX phosphate alkaline solution was added to the diazonium salt solution to prepare an alkaline staining mixture. The cells in the 6-well plate were fixed for about 30 seconds and rinsed gently with water for 45 seconds. After addition of the alkaline staining mixture, the cells were incubated at room temperature for 30 minutes (avoiding sunlight), washed with water, counterstained with a Mayer's hematoxylin solution for 10 minutes and rinsed with water for 2 minutes.
Immunocytochemistry
The colonies were scraped with a capillary glass tube (bent into a ring shape) and allowed to adhere onto 8-well chamber slides (chamber slide, NUNC, Waltham, MA). The next day chamber slides were rinsed twice with PBS, and the colonies were fixed in 2% paraformaldehyde (JUNSEI, Chuo-ku, Tokyo) for 20 minutes, washed twice with PBS, treated with 0.1% Triton X-100 for 5 minutes, rinsed twice with PBS, and treated with 2% BSA (Sigma Aldrich, St. Louis, MO)/PBS at room temperature for 1 hour. Thereafter, the colonies were treated with goat polyclonal anti-OCT4 or anti-hNANOG antibodies (diluted 1:1000 in 2% BSA/PBS, Abcam, Cambridge, MA) overnight at 4°C, washed twice with PBS, treated with Alexa Fluor 488 rabbit anti-goat IgG (Invitrogen, Carlsbad, CA) or Alexa Fluor 546 rabbit anti-goat IgG secondary antibodies (diluted 1:500 with 2% BSA/PBS) at room temperature for 1 hour. After rinsing with PBS three times, nuclei were stained with 300 nM DAPI (4,6-diamidino-2-phenylindole, Sigma-Aldrich, St. Louis, MO) at room temperature for 5 minutes in the dark. The colonies were washed with PBS three times, mounting medium (Vector Labs, Burlingame, CA) and coverslips were applied, and after 15 minutes, the cells were examined by a laser scanning confocal microscope equipped with Nomarski optics.
Reverse transcriptase polymerase chain reaction (RT-PCR)
RNA was extracted from cells using Trizol Reagent (Invitrogen, Carlsbad, CA), and 5 μg of RNA was used for reverse transcription reaction with SuperScript II (Invitrogen, Carlsbad, CA) and oligo-dT primer, according to the manufacturer's instructions. PCR cycles (30–35) were performed with a temperature profile consisting of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
Flow Cytometry
Cells and colonies were treated with trypsin/EDTA (Invitrogen, Carlsbad, CA) at 37°C for 3 minutes to detach and dissociate, washed twice with PBS supplemented with 0.1% BSA, treated with Fc Blocker CD16/32 (Fc Blocker, BD Pharmingen, San Jose, CA) for 10 minutes to block non-specific antibody binding to cell surface Fc receptors, washed twice with PBS supplemented with 0.1% BSA and stained on ice with tumor rejection antigen (TRA)-1-60 (BD Bioscience, San Jose, CA) at a concentration of 1 μg/1 × 106 for 20 minutes or with Phycoerythrin (PE)-labeled SSEA-3 antibody (BD Bioscience, San Jose, CA). The cells were washed twice with PBS plus 0.1% BSA. The prepared cells were analyzed with FACS Calibur (Beckton-Dickinson, San Jose, CA) using the CellQuest Pro cytometric analysis software (CellQuest Pro, BD, San Jose, CA).
Luciferase assay
To test the trans-activational activity of 4 CP-RFs (HM52O, HM181S, HM173K and HM47M), the luciferase expression vectors containing a transcriptional regulatory element (TRE) of each transcription factor in its promoter region were used. The luciferase expression vectors systems containing reporter vector expressing firefly luciferase and normalization vector expressing renilla luciferase were also used (SABiosceinces, Valencia, CA). HEK293 cells at 70% confluence in 24-well plates were transfected with 400 ng of the luciferase expression vector mixture using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturers' instructions. At 12 hrs post-transfection, the cells were treated with 1 μM of CP-RFs in serum free DMEM for 6 hrs and transferred to serum-containing (10% FBS) DMEM for 24 hrs. Transactivation activity of CP-RFs was determined with the Dual-Glo Reporter Assay System (Promega, Fitchburg, WI).
Statistical analysis
All experimental data obtained are expressed as the means ± S.D. Statistical significance was evaluated using a one-tailed Student's t-test. Statistical significance was established at p < 0.05.
Author Contributions
D.J. conceived and designed the experiments. J.L., J.K. and J.K. performed the experiments. D.J. analyzed the data and wrote the paper.
Additional information
Ethics statement: Human resource (human dermal fibroblasts) handling and experimental procedures were approved and performed in accordance with the guidelines of Institutional Review Board (IRB) of the ProCell R&D Institute, ProCell Therapeutics, Inc.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. Shinya Yamanaka (Institute for Frontier Medical Sciences, Kyoto University) for providing the human RFs cDNA. We also thank Dr. Earl Ruley (Vanderbilt University School of Medicine) for his critical suggestion/comment and many young scientists who were involved in the early stage of this study for their technical assistance. This work was supported by grants of the Industrial Strategic Technology Development Program of Ministry of Knowledge Economy (10032101 to D.J.) and the Biotechnology Development program of Ministry of Education, Science and Technology (2009-0094004 to D.J.), Republic of Korea.
Footnotes
D. Jo was the founding scientist of ProCell Therapeutics, Inc., and is affiliated to Vanderbilt University at present. J. Lim and J. Kang are currently employees of ProCell Therapeutics, Inc. Hereby; these three authors disclose a financial interest in the company. The other author disclosed no potential conflicts of interest. ProCell Therapeutics, Inc. has filed patent for “establishment of induced pluripotent stem cell using cell-permeable reprogramming transcription factor for customized stem cell therapy” under the name of Daewoong Jo; Junghee Lim; Jungeun Kim; Sooyoung Jeong; Inhee Jung. The relevent application number is PCT/KR2010/001559. There are no further patents, products in development or marketed products to declare.
References
- Takahashi K. & Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- Yu J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- Avilion A. A. et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17, 126–140 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K. et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 (2003). [DOI] [PubMed] [Google Scholar]
- Nichols J. et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998). [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J. & Smith A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24, 372–376 (2000). [DOI] [PubMed] [Google Scholar]
- Cartwright P. et al. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132, 885–896 (2005). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood 105, 635–637 (2005). [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T. & Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 (2007). [DOI] [PubMed] [Google Scholar]
- Foster K. W. et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene 24, 1491–1500 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K., Yamada Y., Beard C. & Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477 (2005). [DOI] [PubMed] [Google Scholar]
- Soldner F. et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 136, 964–977 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K. et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Nagaya M., Utikal J., Weir G. & Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science 322, 945–949 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T. & Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949–953 (2008). [DOI] [PubMed] [Google Scholar]
- Ban H. et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proceedings of the National Academy of Sciences of the United States of America 108, 14234–14239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K. et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458, 771–775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N. & Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell 3, 595–605 (2008). [DOI] [PubMed] [Google Scholar]
- Kim D. et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 4, 472–476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo D., Liu D., Yao S., Collins R. D. & Hawiger J. Intracellular protein therapy with SOCS3 inhibits inflammation and apoptosis. Nat Med 11, 892–898 (2005). [DOI] [PubMed] [Google Scholar]
- Liu D. et al. Suppression of Staphylococcal Enterotoxin B-induced Toxicity by a Nuclear Import Inhibitor. J Biol Chem 279, 19239–19246 (2004). [DOI] [PubMed] [Google Scholar]
- Liu D., Zienkiewicz J., DiGiandomenico A. & Hawiger J. Suppression of acute lung inflammation by intracellular peptide delivery of a nuclear import inhibitor. Mol Ther 17, 796–802 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Y. et al. Peptide-directed suppression of a pro-inflammatory cytokine response. J Biol Chem 275, 16774–16778 (2000). [DOI] [PubMed] [Google Scholar]
- Moore D. J. et al. In vivo islet protection by a nuclear import inhibitor in a mouse model of type 1 diabetes. PLoS One 5, e13235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. et al. Cell-permeable NM23 blocks the maintenance and progression of established pulmonary metastasis. Cancer Res 71, 7216–7225 (2011). [DOI] [PubMed] [Google Scholar]
- Lim J. et al. Antitumor activity of cell-permeable p18(INK4c) with enhanced membrane and tissue penetration. Mol Ther 20, 1540–1549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. et al. Anti-Tumor Activity of Cell-Permeable RUNX3 Protein in Gastric Cancer Cells. Clinical Cancer Research 19, 680–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy A., Kandasamy S. K., Lee D. K., Kidambi S. & Larson R. G. Structure, topology, and tilt of cell-signaling peptides containing nuclear localization sequences in membrane bilayers determined by solid-state NMR and molecular dynamics simulation studies. Biochemistry 46, 965–975 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach R. A. et al. Receptor/transporter-independent targeting of functional peptides across the plasma membrane. J Biol Chem 279, 11425–11431 (2004). [DOI] [PubMed] [Google Scholar]
- Fischer P. M. Cellular uptake mechanisms and potential therapeutic utility of peptidic cell delivery vectors: progress 2001-2006. Med Res Rev 27, 755–795 (2007). [DOI] [PubMed] [Google Scholar]
- Heitz F., Morris M. C. & Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol 157, 195–206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M., Poon G. M. & Gariepy J. The importance of valency in enhancing the import and cell routing potential of protein transduction domain-containing molecules. Biochim Biophys Acta 1758, 355–363 (2006). [DOI] [PubMed] [Google Scholar]
- Adewumi O. et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25, 803–816 (2007). [DOI] [PubMed] [Google Scholar]
- Esteban M. A. et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79 (2010). [DOI] [PubMed] [Google Scholar]
- Takahashi K., Okita K., Nakagawa M. & Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2, 3081–3089 (2007). [DOI] [PubMed] [Google Scholar]
- Takahashi K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- Esteban M. A. et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79. [DOI] [PubMed] [Google Scholar]
- Hata R. & Senoo H. L-ascorbic acid 2-phosphate stimulates collagen accumulation, cell proliferation, and formation of a three-dimensional tissuelike substance by skin fibroblasts. J Cell Physiol 138, 8–16 (1989). [DOI] [PubMed] [Google Scholar]
- Kurata S. & Hata R. Epidermal growth factor inhibits transcription of type I collagen genes and production of type I collagen in cultured human skin fibroblasts in the presence and absence of L-ascorbic acid 2-phosphate, a long-acting vitamin C derivative. J Biol Chem 266, 9997–10003 (1991). [PubMed] [Google Scholar]
- Phillips C. L., Tajima S. & Pinnell S. R. Ascorbic acid and transforming growth factor-beta 1 increase collagen biosynthesis via different mechanisms: coordinate regulation of pro alpha 1(I) and Pro alpha 1(III) collagens. Arch Biochem Biophys 295, 397–403 (1992). [DOI] [PubMed] [Google Scholar]
- Phillips C. L., Combs S. B. & Pinnell S. R. Effects of ascorbic acid on proliferation and collagen synthesis in relation to the donor age of human dermal fibroblasts. J Invest Dermatol 103, 228–232 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information