Abstract
The regulatory networks of differentiation programs and the molecular mechanisms of lineage-specific gene regulation in mammalian embryos remain only partially defined. We document differential expression and temporal switching of BRG1-associated factor (BAF) subunits, core pluripotency factors and cardiac-specific genes during post-implantation development and subsequent early organogenesis. Using affinity purification of BRG1 ATPase coupled to mass spectrometry, we characterized the cardiac-enriched remodeling complexes present in E8.5 mouse embryos. The relative abundance and combinatorial assembly of the BAF subunits provides functional specificity to Switch/Sucrose NonFermentable (SWI/SNF) complexes resulting in a unique gene expression profile in the developing heart. Remarkably, the specific depletion of the BAF250a subunit demonstrated differential effects on cardiac-specific gene expression and resulted in arrhythmic contracting cardiomyocytes in vitro. Indeed, the BAF250a physically interacts and functionally cooperates with Nucleosome Remodeling and Histone Deacetylase (NURD) complex subunits to repressively regulate chromatin structure of the cardiac genes by switching open and poised chromatin marks associated with active and repressed gene expression. Finally, BAF250a expression modulates BRG1 occupancy at the loci of cardiac genes regulatory regions in P19 cell differentiation. These findings reveal specialized and novel cardiac-enriched SWI/SNF chromatin-remodeling complexes, which are required for heart formation and critical for cardiac gene expression regulation at the early stages of heart development.
INTRODUCTION
The heart arises initially as the cardiac crescent from the lateral plate mesoderm at the late gastrula stage. Shortly thereafter, cardiac crescent cells migrate medially to form the first identifiable cardiac structure: the linear heart tube (1–3). As the linear heart tube grows, it loops out into the pericardial space, subsequently leading to the four-chambered heart formation. Gene expression patterns during cellular differentiation and development are orchestrated by highly dynamic chromatin structure that controls the ability of the transcriptional machinery to gain access to gene loci. Epigenetic regulation of gene expression, in part via chromatin-remodeling factors to change nucleosome positioning, plays a vital role in the elegant process of cardiac specification and development (4).
Adenosine triphosphate (ATP)-dependent chromatin-remodeling complexes (along with histone modifying enzymes) regulate transcriptional activity by changing chromatin structure, resulting in activation and repression of genes within cells (5–7). The canonical ∼1500 kDa multisubunit Switch/Sucrose NonFermentable (SWI/SNF) complexes consist of 12 protein subunits. Among these subunits, Brahma Related Gene (BRG) and Brahma (BRM) are alternative ATPases, each of which is sufficient to remodel nucleosome arrays in vitro (8). Conversely, the roles of the other SWI/SNF subunits are less well established. It has been suggested that BAF155 and BAF170 provide scaffolding functions for other SWI/SNF subunits as well as regulating their protein levels (9,10). Furthermore, structural domains of BRG1-associated factors (BAFs) involved in protein–protein interaction and contain sequence-dependent and sequence-independent DNA-binding domains (11,12). In addition, the SWI/SNF complexes are dynamic, which results in formation of a varied set of mammalian SWI/SNF-like chromatin-remodeling complexes (13). These complexes are believed to play vital roles in tissue and cell type specification by regulating specific gene expression patterns (5,14–19).
Genetic deletion of BRG1, BAF155, BAF47 or BAF250a in mice leads to early embryonic lethality (20–23). However the closely related ATPase, BRM, is dispensable for development (24). Depletion of BAF60c and BAF180 leads to defects in heart development and causes late embryonic death (25,26). Loss of BAF250a results in a lack of mesoderm in the mouse embryo and the upregulated genes required for early development and organogenesis (23). Characterization of BAF60c and other BAF complex subunits that co-assembled to form cell type-specific complexes has established BAF complexes as intrinsic factors in differentiation, rather than simply chromatin-remodeling machines (13,19,25,27). These specific BAF complexes act as a driving force toward lineage specification and tissue precursor differentiation. More recent studies have shown associations between cardiac transcription factors and SW/SNF chromatin-remodeling complexes that are critical for heart development and modulate gene expression during fetal heart formation (28–30). However, details of combinatorial assemblies of SWI/SNF complex subunits responsible for altering chromatin and mechanisms underlying role of BAF complexes in heart formation remain unclear. An intriguing possibility is that stoichiometrically distinct BAF complexes may differentially influence chromatin accessibility, thereby controlling ubiquitously expressed transcriptional regulators to spatially and temporally control gene expression during early development.
In this study we demonstrate that the SWI/SNF complex subunits exhibit differential patterns of expression in early development and that a subset of BAFs was elevated in early heart compared with head and trunk. Proteomic analysis uncovered distinct SWI/SNF-like complexes in the heart, head and trunk of the E8.5 embryos. BRG1/BRM, BAF250a/b, 60 b/c and BRD7 are quantitatively enriched subunits of the mSWI/SNF or BAF complexes that are present in the developing heart. Depletion of BAF250a resulted in elevated expression level of a discrete set of cardiovascular genes, impaired differentiation and in an arrhythmic contraction phenotype in an in vitro model of differentiated cardiomyocytes. BAF250a-containing BAF complexes occupied regulatory regions of cardiac-specific genes in the developing hearts consistent with a critical role in transcriptional regulation during cardiac development. In support of this regulatory concept, depletion of BAF250a specifically affected the levels of histone H3K4me1 and H3K27ac at putative enhancer regions. Additionally, BAF250a modulated chromatin structure and limit recruitment of transcriptional activation machinery. Intriguingly, BRG1-associated factor 250a (BAF250a) and Nucleosome Remodeling and Histone Deacetylase (NURD) complex subunits physically interact and functionally cooperate to repress the expression of cardiac-related genes. These results suggest that BAF250a plays a regulatory role in early heart formation by the direct repression of cardiac-specific gene expression.
MATERIALS AND METHODS
Mouse embryo collection
The date of observation of a vaginal plug is set as embryonic day E0.5 by convention. Embryonic development was confirmed by examination of embryo morphology and somite number. Pregnant CD1mice were killed by CO2 at E5.5, E6.5, E7.5, E8.5, E9.5 and E10.5 and embryos were dissected. The head, heart and trunk were surgically separated from E8.5, E9.5 and E10.5 embryos. All mouse experiments were performed in accordance with NIEHS/NIH guidelines covering the humane care and use of animals in research.
Quantitative reverse transcriptase-polymerase chain reaction analysis
RNA was isolated from early embryos using the Arcturus PicoPure RNA Isolation Kit and Invitrogen RNA Isolation Kit; from the heart, head and trunk separately; cDNA was synthesized using the SuperScript First-Strand Synthesis System (Invitrogen) with OligodT primers. Quantitative real-time polymerase chain reaction (qRT–PCR) measurements of individual cDNAs were performed with the SYBR green real-time PCR detection system. Gene-specific primers for BAF subunits, pluripotency factors and cardiac-specific genes were designed for encoded gene transcript available at NCBI database using Primer Express. The rodent glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The primer sequence can be provided upon request. All measurements were performed in triplicate. Values were normalized to GAPDH using the 2−DDCt methods and expressed as ±SD. All mouse experiments were performed in accordance with NIEHS/NIH guidelines covering the humane care and use of animals in research.
Whole mount RNA in situ hybridization
To generate the antisense probe, BAF250a transcript was amplified using the specific primers (Supplementary Table S2.) and cloned into Zero Blunt TOPO vector. The expressed sequence tag (EST) clone was linearized with PstI and used as a template for in vitro transcription with T7 RNA polymerase. Whole mount RNA in situ hybridization was performed as described previously (31).
Western immunoblot analysis
Whole-cell lysate and nuclear extract was prepared using Active Motif kit and protocol. The protein was quantified using the Bradford assay. Proteins were size-separated in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The gels were blotted onto a Nitrocellulose Membrane (Invitrogen), blocked with 5% non-fat dry milk. The blots were reacted with antibodies for BAF subunits and cardiac genes mentioned below followed by horseradish peroxide (HRP)-conjugated anti-mouse IgG or HRP-conjugated anti-rabbit IgG (GE Healthcare Bio-Sciences). Chemiluminescence was detected with enhanced chemiluminescence (ECL) western blot detection kits (GE Healthcare Bio-Sciences), according to the supplier’s recommendations.
Affinity purification and mass spectrometry
Affinity purification and mass spectrometry (MS) of endogenous BAF complexes were performed as previously described (16). In brief, ∼300 litters of E8.5 embryos (∼3000) from pregnant CD1 mice were dissected in chilled phosphate buffered saline (PBS). Heart, head and trunk were surgically separated followed by preparation of heart, head and trunk nuclear extracts for affinity purification and MS analysis. The Brg-1 Antibody (G-7): sc-17796 (Santa Cruz Biotechnology Inc.) was used to immunopurify the SWI/SNF complexes.
Spectrum counting and quantitative analysis of MS data
Each lane from the polyacrylamide gel was manually cut into 24 equal pieces and digested with trypsin (Promega) for 8 h in an automated fashion with a Progest robotic digester from Genomic Solutions. Briefly, minced gel bands were incubated twice for 15 min in 100 µl of 25 mM ammonium bicarbonate and 50% (v/v) acetonitrile. The gel was then dehydrated by a 20-min incubation in 100 µl of acetonitrile followed by drying under a nitrogen stream. About 250 ng of trypsin (Promega) were added, followed by an 8-h incubation at 37°C. The resulting peptides were extracted by first collecting the digest supernatant, then incubating the gel with 50 µl of water for 20 min and collecting the resulting supernatant and finally, collecting the supernatants from two independent 20-min incubations of the gel in 50 µl of 5% (v/v) formic acid and 50% (v/v) acetonitrile. All supernatants were pooled during the collection process and were lyophilized to dryness. The lyophilized samples were suspended in 35 µl of 0.1% formic acid. Liquid Chromatography- Electrospray ionization-Mass Spectrometry (nanoLC-ESI-MS/MS) analyses were then performed using an Agilent 1100 nanoLC system online with an Agilent XCT Ultra ion trap mass spectrometer with the Chip Cube Interface. About 20 µl of the peptide mixture from the in-gel digest were loaded onto an Agilent C18 chip (75 µm × 43 mm) followed by a 15-min wash of 5% acetonitrile, 0.1% formic acid. Peptides were eluted by applying a linear gradient from 5% acetonitrile, 0.1% formic acid to 50% acetonitrile, 0.1% formic acid to the column over 45 min. This was followed by a 5-min gradient from 50% acetonitrile, 0.1% formic acid to 95% acetonitrile, 0.1 % formic acid and then a 10-min hold at 95% acetonitrile, 0.1% formic acid. The mass spectrometer was used in the positive ion, standard-enhanced mode and included settings of a mass range from 200 to 2200 m/z, an ionization potential of 2.1 kV, an ICC smart target of 100 000 or 200 ms of accumulation and a 1.0 v fragmentation amplitude. MS/MS data were acquired using a data-dependent acquisition format with the six most abundant ions from each MS scan further interrogated by MS/MS. The automated switching for MS/MS required a threshold of 10 000 counts. A single peak list was generated from the data obtained from each of the 24 nanoLC-ESI-MS/MS analyses using the Data Extractor feature of the SpectrumMill software from Agilent. The Data Extractor settings included limiting the data search to deconvolved ions observed between 300 and 5000 Da and a retention time between 10 and 60 min. MS scans with the same precursor mass (±1.5 m/z) and retention time within 30 s were merged. Moreover, of the remaining MS/MS spectra, only spectra that contained sequence tag information greater than two residues were submitted for database searching. The resulting extracted data were then searched against the NCBInr database using the MS/MS Search function in the SpectrumMill software. Search settings included a trypsin specificity with one missed cleavage allowed, a precursor ion mass tolerance of 2.0 Da, a product ion mass tolerance of 1.0 Da, variable methionine oxidation and a minimum matched spectral intensity of 70%. Proteins identified with a distinct summed MS/MS search score >20 were tabulated. At this threshold, the false-positive rate was essentially 0% as determined by searching against a reversed sequence database. Moreover, sequence assignments of MS/MS spectra were manually validated.
Co-immunoprecipitation
Co-immunoprecipitation was performed using the Active Motif Co-IP Kit. The lysates were incubated with BRG1 antibody NB100-2594, (NOVUS), BAF250 (PSG3)- sc-32761; Santa Cruz, CHD4 (ab70469); abcam, Histone deacetylase 1 (HDAC1) (ab7028); abcam. The immunocomplexes were recovered. SDS buffer was added to the precipitate and boiled for 10 min. Proteins were size-separated in SDS–PAGE. The gels were blotted onto a Nitrocellulose Membrane (Invitrogen), blocked with 5% non-fat dry milk and incubated with the antibodies; CHD4 cat # A301-081 A (BETHYL LAB), BAF53a cat # A301-391 A (BETHYL LAB), BAF155 (DXD7)-sc-32763, Santa Cruz and DNMT1 cat # 39204 (Active Motif). HRP-conjugated secondary antibodies (GE Healthcare) were used for detection using the ECL method (GE Healthcare Bio-Sciences).
Histology and Immunostaining
Histological analysis and immunostaining were performed as described earlier (32). E7.5 and E8.5 embryos were fixed in 4% paraformaldehyde, dehydrated through graded alcohol and embedded in paraffin. Sections (5 µm) were deparaffinized and stained with the following antibodies: BAF250 (PSG3)- sc-32761, Santa Cruz, ARID1A/BAF250 (A301-040 A), BETHYL, BAF53A (AB3882) abcam, Anti-SMARCB1 (HPA018248), SIGMA, BAF155 (DXD7)-sc-32763, Santa Cruz, BRG1-(G7)-sc-17796, Santa Cruz, BAF170 (E-6), sc-17838, Santa Cruz, Myocardin (Myocardin H-300, sc-32766, Santa Cruz), cTnT (ab8295, Abcam), Myl3 (ab680-100 Abcam) and secondary anti-mouse, anti-rabbit and DAPI from Invitrogen (Molecular probe).
Small interfering RNA cell transfection
Small interfering RNA (siRNA) duplexes were synthesized by Dharmacon targeting the mRNA coding region of BAF250a siGENOME SMARTpool cat# M-040694-01-0005 and previously described BAF250-3, 5′-CGACAUGAUUCCUAUGGCAdTdT-3′ (33), BRG1- siGENOME SMARTpool SMARCA4 cat# M-041135-01-0005, BAF155- ON-TARGET plus siRNA SMARCC1 cat# J-044249-06-0005, BAF53a cat# J-044249-07-0005, CHD4 siGENOME cat # D-052142-02-0010 and CHD4 siGENOME cat # D-052142-01-0010 and on-target plus cat # D-001810-0X non-target-control (NTC) (used as a negative control of transcriptional activation knockdown). Cell culture and nucleofection were performed according to the Lonza Optimized Protocol for P19 cell line (ATCC). The cells were extracted 48–72 h post transfection. The extents of BAF250a, BRG1, BAF155, BAF53a and CHD4 protein knockdown and cardiac gene expression were determined by immunoblotting and qRT–PCR, respectively.
Chromatin immunoprecipitation
Hearts from ∼20 litter of E9.5 CD1 mice were dissected in chilled PBS. Chromatin immunoprecipitation (ChIP) was performed following the instruction manual of the Low Cell # ChIP Kit, Diagenode. Chromatin was immunoprecipitated using BAF250 (PSG3) × TansCruz - sc-32761 antibody, Santa Cruz, BRG1 NB100-2594; NOVUS, CHD4 (ab70469); abcam, HDAC1 (ab7028); abcam, PolII (ab5131); abcam, H3K27ac (ab4729); abcam, H3K4me1 (ab8895); abcam, BAF53a (ab3882), abcam. Immunoprecipitated chromatin was PCR-amplified for 35 cycles in embryonic heart ChIP analysis. ChIP analysis was also performed with the Stratagene Mx300P and Brilliant SYBR Green quantitative PCR (qPCR) master mix. Average cycle threshold amplification values and percentage of sample input were calculated. PCR primers are available in Supplementary Table S2. The primer positions are denoted relative to the transcriptional start site (+).
Reporter assay
Gata4 Promoter Luciferase Reporter vector was purchased from SWITCH GEAR GENOMICS. These vectors as well as an episomal Renilla-luciferase plasmid (pREP7_RL) to normalize for transfection efficiency and a siRNA of BAF250a and non-target-control, were transfected into P19 cells using Fugene-6 (Roche). The cells were cultured for 2 days and harvested for luciferase assay using the dual luciferase assay kit (Promega).
DNase sensitivity assays
P19 cells were cultured as previously described. The cells were extracted 48–72 hrs post transfection to make embryoid bodies treated with Dimethyl sulfoxide (DMSO). At Day 4 cells aggregates were then collected in cold PBS. Nucleus isolation and DNase digestion were performed as previously described, with modifications (34). Nuclei were isolated by incubation for 10 min on ice with 5 ml Dounce buffer [20 mm Tris–HCl (pH 7.4), 3 mm CaCl2, 2 mm MgCl2, 0.3% Nonidet P-40, 1x Sigma protease inhibitor cocktail, 0.15 mM spermine, 0.5 mM spermidine) followed by Dounce homogenization. Nuclei were pelleted by centrifugation at 200g for 7 min. Nuclei were washed twice in resuspension buffer [10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, 0.15 mM spermine and 0.5 mM spermidine] and pelleted by centrifugation at 200g for 7 min. Nuclei were then resuspended in 1 ml resuspension buffer and nuclei were counted. Additional resuspension buffer was used to generate equal concentrations of nuclei between samples. Nuclei were aliquoted into microcentrifuge tubes and incubated at 37°C for 5 min. About 1 U of DNase was added to each of the aliquoted samples with the exception of the uncut control and the samples were incubated at 37°C. Digestion was stopped by addition of an equal volume of termination buffer [20 mM Tris (pH 7.4), 200 mM NaCl, 2 mM EDTA, 1% SDS, 200 μg/ml proteinase K]. Reaction mixtures were incubated overnight at 37°C, followed by two phenol–chloroform extractions and one chloroform extraction of the DNA. DNA was quantified on a Qubit analyzer (Invitrogen) and equal amounts of DNA were analyzed by real-time PCR using the appropriate primer sets.
RESULTS
BAF subunits show differential spatiotemporal expression patterns in early development
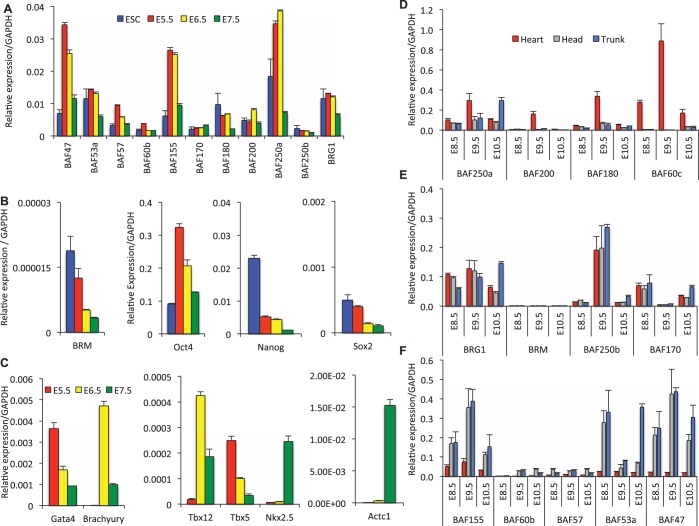
To begin to determine the temporal functions of BAF subunits during early development we examined mRNA expression levels. We found that BAF250a was highly expressed in E5.5 and E6.5 wild-type embryos and reduced in E7.5, relative to levels in Embryonic Stem Cells (ES cells) (Figure 1A), while BAFs 155, 57 and 47 were elevated at Days E5.5 and E6.5. In contrast, other BAF subunits displayed only minor changes in the expression pattern through early development relative to ES cells (Figure 1A). Intriguingly, the pluripotency genes, Sox2 and Nanog, are downregulated from E5.5–7.5 (Figure 1B), while Oct4 was elevated at Days E5.5 and E6.5 (Figure 1B). We next examined a limited number of cardiac genes and observed that Gata4 and Tbx5 expression is greater in the E5.5 and E6.5 embryos and reduced in E7.5 embryos. Whereas, the mesoderm-specific Brachyury and Tbx12 genes were expressed at low levels in E5.5 and elevated at E6.5 before exhibiting reduced expression at E7.5. In contrast the Nk2.5 and Actc1 were most highly expressed in E7.5 embryos (Figure 1C).
Figure 1.
Differential expression and temporal switching of BAF subunits and core pluripotency genes during early development. Quantitative RT–PCR analysis of (A) BAF subunits, (B) core pluripotency factors and (C) cardiac-specific genes in embryonic stem cells and at indicated (E5.5–7.5) developmental stages (D), (E), (F) BAF subunits and (G) cardiac genes exhibit differential expression pattern and spatiotemporal switching during early (E8.5–10.5) organogenesis. Relative expressions were normalized to GAPDH and each experiment was repeated twice. The data are expressed as mean ± SD.
To determine if these expression differences persisted during later development, we compared the quantitative temporal and spatial expression patterns separately in heart, head and trunk tissues of E8.5, E9.5 and E10.5 embryos. We found that BAF subunits show differential spatiotemporal expression patterns that switch during development. BAF60c is exclusively expressed in the heart and is much reduced in the head and trunk; consistent with an earlier report (25) (Figure 1D). Along with BAF250a, BAF180 and BAF200 were also elevated in the E9.5 heart compared with the head and trunk (Figure 1D), consistent with a more prominent role for these subunits in heart development. However, expression levels of BRG1, BAF250b and BAF170 were similar in heart, head and trunk at E8.5 and E9.5 (Figure 1E). In contrast, BAFs 155, 60b, 57, 53a and 47 expression levels were higher in E8.5, 9.5 and 10.5 head and trunk compared with that in the heart (Figure 1F). During Days E5.5–E7.5, BRM expression levels were substantially lower (Figure 1B) than that seen for BRG1 and the BAFs at Days E8.5–E10.5 (Figure 1E). Intriguingly, cardiac-related genes were highly expressed in E8.5 and E9.5 heart relative to head and trunk, a profile reminiscent of the BAF250a, BAF180, BAF200 and BAF60c profile (Figure 1D and G). Finally, in the adult animals the expression of the BAF subunits was much lower in the heart compared with the ovary, brain and kidney (Supplementary Figure S1). These findings led us to investigate if BAF subunits BAF250a, 60c, 180 and 200 exhibit differential spatiotemporal expression patterns that might be informative for specific biological processes such as heart development.
Characterization SWI/SNF-like complexes from early mouse embryos
The expression of SWI/SNF complexes is dynamic during development and the combinatorial assemblies of BAF subunits have the capacity to produce distinct functional complexes as in the case of a specialized SWI/SNF complexes found in mouse embryonic stem cells (esBAF) that lack BAF170 (35). To characterize the in vivo SWI/SNF complexes and interacting partners that could specify tissue lineage differentiation, we affinity-purified the endogenous BAF complexes from surgically dissected heart, head and trunk tissues of CD1 E8.5 mouse embryos using an antibody against BRG1 and analyzed these complexes by MS (Supplementary Figure S2). Subsequent proteomic analysis yielded high quality spectra for 112 proteins from the heart, 112 proteins from the head and 71 proteins from the trunk that were co-immunoprecipitated with BRG1 with statistical probabilities exceeding 95% as determined by ProteinProphet (36) (Supplementary Figure S2). Among these proteins, 47 contributed >8% of the total spectra obtained and were found to be common in the heart, head and trunk, including both known and novel components of BAF complexes; 29 proteins were common in the heart and head, 7 proteins were common in the heart and trunk, 12 proteins were common in the head and trunk. In total, 29 proteins were unique to the heart; 24 proteins were unique to the head and 5 proteins were unique to the trunk (Supplementary Figure S2). The predominant composition of known components of the complex was determined by comparison of spectrum counts for the proteins of each sample. The number of spectra acquired for each protein is reflective of protein abundance and has been normalized for the total number of spectra in each dataset. The abundance and composition of the predominant BAF complexes differed significantly in each tissue type (Table 1). Thus, affinity purification revealed spatially distinct types of SWI/SNF-like complexes within the E8.5 heart, head and trunk, which might regulate chromatin during organogenesis.
Table 1.
Details of assembly of the distinct SWI/SNF-like or BAF chromatin-remodeling complexes that exist in the developing E8.5 (embryonic day 8.5) mouse embryos
| Brg1-associated factors (BAF) | Cardiac-enriched BAF |
Head Tissue BAF |
Trunk Tissue BAF |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of spectra in heart | AA coverage (%) in heart | No. of distinct peptides in heart | MS/MS score in heart | No. of spectra in head | AA coverage (%) in head | No. of distinct peptides in head | MS/MS score in head | No. of spectra in trunk | AA coverage (%) in trunk | No. of distinct peptides in trunk | MS/MS score in trunk | |
| BRG1 | 85 | 25 | 33 | 484.87 | 44 | 34 | 21 | 327.69 | 44 | 20 | 26 | 353.92 |
| BAF250a(ARID1A) | 60 | 20 | 30 | 408 | 3 | 1 | 2 | 29.99 | 3 | 1 | 2 | 27.67 |
| BAF155(SMARCC1) | 66 | 23 | 18 | 253.51 | 16 | 8 | 7 | 88.73 | 17 | 8 | 7 | 101.75 |
| BAF60a(SMARCD1) | 37 | 35 | 15 | 198.87 | 10 | 21 | 9 | 97.27 | 6 | 13 | 6 | 56.10 |
| BRM(SMARCA2) | 31 | 8 | 12 | 180.40 | 19 | 6 | 10 | 126.55 | 18 | 7 | 10 | 124.10 |
| BAF170(SMARCC2) | 37 | 8 | 9 | 130.28 | 10 | 5 | 5 | 71.11 | 6 | 3 | 3 | 38.22 |
| ACTG1(β ACTIN) | 32 | 17 | 9 | 127.23 | 23 | 41 | 9 | 120.28 | 18 | 23 | 8 | 103.72 |
| BAF180 (PBRM1) | 15 | 7 | 9 | 110.91 | 11 | 6 | 8 | 101.09 | 8 | 3 | 5 | 68.62 |
| BAF60b(SMARCD2) | 12 | 17 | 7 | 92.11 | – | – | – | – | – | – | – | – |
| BAF47(SMARCB1) | 18 | 21 | 7 | 84.77 | 11 | 19 | 6 | 81.28 | 9 | 13 | 5 | 68.26 |
| BAF57(SMARCE1) | 9 | 19 | 6 | 79.69 | 2 | 8 | 2 | 18.21 | 5 | 10 | 3 | 49.27 |
| BAF250b(ARID1B) | 14 | 1 | 4 | 60.23 | – | – | – | – | – | – | – | – |
| BAF60c(SMARCD3) | 7 | 3 | 2 | 30.56 | 3 | 4 | 2 | 20.01 | – | – | – | – |
| BAF45D(DPF2D4) | 2 | 7 | 2 | 30.19 | 1 | 3 | 1 | 16.86 | 7 | 17 | 5 | 65.46 |
| BRD7 | 2 | 5 | 2 | 21.81 | – | – | – | – | – | – | – | – |
| BAF53(ACTL6A) | – | – | – | – | 7 | 14 | 6 | 79.6 | 9 | 23 | 8 | 109.51 |
| BAF53b(ACTL6B) | – | – | – | – | 2 | 5 | 2 | 22.25 | 2 | 5 | 2 | 25.95 |
| BAF200(ARID2) | 2 | – | 2 | 21.7 | 2 | 1 | 2 | 21.57 | 2 | 1 | 2 | 18.76 |
Distinct SWI/SNF-like complexes exist in the developing embryo. Summary of the BAF (BRG1-associated factors) peptides identified by mass spectrometry in the developing heart, head and trunk of the E8.5 (embryonic day 8.5) mouse embryos referred to as cardiac-enriched BAF (ceBAF), head tissue BAF and trunk tissue BAF. Values in bold are indicating the enrichment of specific subunits which are highly variable in heart, head and trunk.
Unique composition of BAF components triggers heart formation in developing E8.5 embryo
The predominant BAF complexes in each tissue type differ considerably in composition, particularly when BAF-associated proteins from heart are compared with those from head and trunk tissues (Table 1). A total number of 536 peptides were determined and 762 unique spectra were obtained from the heart. The purification of BRG/BRM complexes yielded most (16 out of 18) of the family members of the core subunits of BAF complexes in heart, specifically, BRG1/BRM, BAF250a/b, BAF180, BAF155, BAF170, BAF60a/b/c, BAF57, BAF47, BRD7, β-Actin, BAF200 and BAF45d (Table 1). An examination of the relative abundance and combinatorial assembly of the BAF subunits in the heart revealed a number of intriguing observations. Firstly, BRG1, BAF250a, BAF155 and BAF60a, b, c were highly enriched in cardiac-enriched BAF complexes. Indeed, whereas BAF250b, along with BRD7 and BAF60b, was only detected in the heart purified complexes, BAF250a was present at substantially greater representation than BAF250b reflecting their relative expression levels as described earlier (Figure 1). Enrichment of BAF250a might be a key factor in determining role of cardiac-enriched BAF complexes in heart formation. In contrast, the BAF250b subunit is greatly reduced or undetectable in E8.5 heart, head and trunk BAF complexes. This result is consistent with the lower expression of BAF250b in early embryo (Figure 1). Surprisingly, the data in Table 1 show an ∼3:1 ratio of BRG1 to BRM peptides indicating that both catalytic subunits, which have been shown to be mutually exclusive in the complex, are present and abundant. This raises the unexpected possibility that the human Brg-1 antibody might cross-react with murine BRM. This observation may also provide an intriguing insight into the relative abundance of BRG1 and BRM in the early mouse embryo as well as the respective developmental importance of BRG1 and BRM as exemplified by the viability of the BRM-/- mouse (24). Interestingly, the ‘cardiac-specific’ subunit BAF60c was found at lower levels in heart compared with BAF60a/b, and also is present in head BAF complexes, consistent with previous studies, which show presence of BAF60c in npBAF complexes (19,25). Consequently, the presence of BAF60b specifically in the cardiac BAF complexes indicates that several types of complexes can co-exist in the developing embryonic heart (Table 1 and Supplementary Figure S2). Peptides from polybromo-associated BAF (PBAF) BAF180 and BAF200 were detected in heart, head and trunk BAF complexes. However, BRD7 was exclusively obtained in the heart and undetectable in head and trunk by MS analysis, suggesting that BRD7 is a cardiac-enriched subunit of the BAF complexes (Table 1).
The neural-specific subunits, BAF53a and BAF53b are undetectable by MS in intact cardiac-enriched BAF complexes. In the developing embryo, Baf53a/b is mostly expressed in proliferating neural progenitors and differentiated cells. BAF53a/b was enriched in head and trunk tissues compared to heart. Six peptides (BAF53a) and two peptides (BAF53b) were identified in head and eight peptides (BAF53a) and two peptides (BAF53b) were identified in trunk BAF complexes, consistent with their specialized role in the central nervous system. No unique peptides from the BAF45a, 45b and 45c subunits could be detected, whereas five unique peptides, from a total of eight different peptides, were noted for BAF45d in heart, head and trunk. BAF45a, 45b and 45c were at very low to undetectable levels in intact BAF complexes shown at this stage of early mammalian embryo.
Our data thus far suggest the presence of distinct cardiac-enriched SWI/SNF-like complexes defined by quantitative enrichment, combinatorial assembly and of diverse BAF subunits that may be unique from those found in head, trunk and other previously characterized cell lines (17,35). We, therefore, hypothesized that combinatorial assembly and relative abundance of BAFs can provide the scaffold for cardiac-enriched BAF complexes in the developing heart and may provide a basis for fetal cardiac gene regulation for early lineage specification and heart formation. Our findings are consistent with the view that due to cardiac tissue heterogeneity in the early embryonic heart, multiple distinct SWI/SNF complexes co-exist in the developing heart, which we refer to as cardiac-enriched BAF complexes. As BAF250a is the largest subunit and belongs to BAF-A complexes, which is highly enriched in cardiac-enriched SWI/SNF-like complexes, we decided to explore further the role of the BAF250a-conating BAF complexes in cardiac gene regulation during cardiac differentiation.
BAF250a is vital in heart development and diversely co-localizes with BAFs in early embryo
The preceding gene expression and proteomics analyses revealed that BAF250a is a highly expressed, major subunit of cardiac-enriched BAF complexes. Whole mount RNA in situ hybridization and immunofluorescence localization studies confirmed that BAF250a mRNA and protein are strongly expressed in the cardiac crescent as early as E7.5 followed by the heart and head at E8.5 embryo (Supplementary Figure S3A–G). Cross-sectional analysis of the E8.5 embryos used for whole mount RNA in situ hybridization revealed that BAF250a mRNA was located in multiple cell types of the heart (Supplementary Figure S3H–K). These results suggest that BAF250a is an important subunit of the cardiac-enriched BAF-A complexes for early heart development.
Given the prominent expression of BAF250a in heart and the abundance in the cardiac BAF complexes, we examined the co-localization of BAF250a with BRG1 and other BAF protein subunits in the E8.5 embryo. Immunostaining revealed that BAF proteins were extensively stained at multiple sites in the E8.5 embryo. BAF250a is often co-localized with BRG1 in the head, mesenchyme and somites and elaborates punctuate staining pattern in heart, consistent with a spatial association between these subunits in early development (Supplementary Figure S4C). However, BAF155 and BAF250a were strongly co-localized in the dorsal aorta and vitelline vein and less so in the heart, which suggests a strong association in vasculogenesis (Supplementary Figure S4F). Similarly, BAF170 and BAF250a were also co-localized in the dorsal aorta (Supplementary Figure S4I). In contrast, BAF53a and BAF47 were strongly co-localized with BAF250a in the optical vesicle, head mesenchyme and somites, and less in the heart (Supplementary Figure S4L and O). This result suggests that spatial association of BAF250a with BAF155 and BAF170 may be relevant for vasculogenesis, whereas co-localization of BAF250a with BRG1 within the cardiac field may contribute to heart formation. We also found that BAF250a-containing BAF complexes co-localize with cardiac-specific proteins in the developing heart. Co-immunostaining shows that BAF250a and a number of cardiac-specific proteins are found together in developing heart (Supplementary Figure S5).
Chromatin modifying enzymes co-immunoprecipitaed with BAF250a
To explore molecular mechanisms mediated by BAF complexes in early development, we examined expression of BRG1 subunits in cultured P19 cells, which exhibit characteristics of ES cells (37). We found 84 proteins from P19 cells co-immunoprecipitated with Brg1 with probability exceeding 95% as determined by ProteinProphet (36). The 394 unique spectra obtained correspond to 280 unique peptides including both novel and known components of BRG1 complexes (Supplementary Table S1). We found 145 proteins from P19 nuclear extract that were co-immunoprecipitated with BAF250a including known and unique proteins of BAF complexes with probabilities exceeding 95% determined by ProteinProhet (36) (Supplementary Table S1). The number of proteins co-immunoprecipitated with BRG1 and BAF250a involved in various biological processes including developmental and cellular process such as Dnmt1, HDAC1, HDAC2, CHD3, Pold 1, Nucleolin, Mybbp1a, Annexin2, SFPQ and Actc1. We found specifically that BAFs and a number of chromatin modifying enzymes co-immunoprecipited with BAF250a in P19 cells and in heart, head and trunk of developing embryo (Supplementary Table S1 and Supplementary Figure S6). This result suggests that BAF250a-containing BAF complexes could achieve its specificity by combining with other chromatin modifying enzymes in context-dependent manner.
BAF subunits differently regulate heart-specific genes
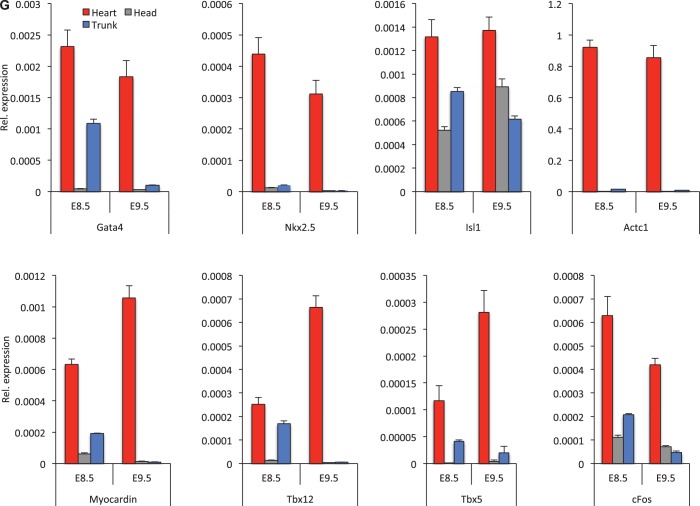
To analyze the functional significance of the cardiac-enriched BAF components on cardiac gene expression, we independently knocked down BAF250a, BRG1, BAF155 and BAF53a proteins in P19 cells by siRNA (Supplementary Figure S7A, B, C and D) in cultured P19 cells. Expression analysis demonstrated that silencing different BAF subunits resulted in different transcriptional profiles of cardiac genes. Interestingly, depletion of BAF250a augmented the expression level of most of the 40 genes examined; 18 of them were upregulated more than 2-fold, when analyzed in P19 cells (Figure 2A). Myh11, cFos, Isl1, Gja5 and Gata6 expression levels were increased >5-fold, whereas those of Snail, Actc1, Ndrg1, Gata4, Hand1, Nkx2.5, Wnt11, Acta2, Ctgf, Pecam, cTnT, Srf and Myocardin were upregulated >2-fold. These results support the hypothesis that BAF250a primarily represses expression of cardiac-specific genes. To test this idea, we performed the reciprocal experiment by over-expressing of BAF250a in P19 cells and analyzed cardiac gene expression. As predicted, most of the targets that were upregulated with the depletion of BAF250a were downregulated following BAF250a induction (Figure 2H). Interestingly, while BAF250a induction repressed most examined genes, Wnt11, Ndrg1 and Gata6 expression was unchanged. The results from the loss and gain of function analysis of BAF250a are consistent with repressive action on cardiac genes.
Figure 2.
Expression pattern of cardiac-specific genes are shown in P19 cells after individual depletion of (A) BAF250a, (B) BRG1, (C) BAF155, (D) BAF53a and (E), (F), (G) CHD4 and (H) after transient overexpression of BAF250a. In each panel, black bars show non-target control and gray bars show BAF subunits and CHD4. All transcript levels were normalized to GAPDH. Data are mean ± SD; n = 3; *P < 0.05. The data represent three biological replicates and each experiment was repeated twice.
Knockdown of BRG1, BAF155 and BAF53a resulted in a range of expression patterns for the cardiac-specific genes analyzed (Figure 2B, C and D). Myocardin, Pecam, Wnt11, Gata4, Actc1 snail, Gata6, cFos and Myh11 appeared to be relatively unaffected. Isl1, Gja5, Ndrg1, Hand1, Nkx2.5, cTnT and Srf were upregulated (≥2-fold) in P19 cells upon inhibition of BRG1 (Figure 2B). BAF155 is an abundant subunit of the cardiac-enriched BAF complexes. However, depletion of BAF155 has relatively mild effects—a >2-fold increase of expression of Ctgf, Wnt11, Acta2, Nkx2.5, Actc1, Snail, Gja5, Isl1 and ≤2-fold increase of expression of Hand1, Gata4, Ndrg1, Gata6 and cFos. There was little effect on Myocardin, Srf, CtnT, Pecam and Myh11 expression (Figure 2C). The reduction of BAF53a expression resulted in increased expression of Isl1, Snail, cTnT and Srf (>5-fold) and a more modest increase (>2-fold) of expression of Gja5, Gata6 and Hand1 (Figure 2D). Myocardin, Pecam, Wnt11, Ctgf, Gata4, Ndgr1, Actc1 and Myh11 were relatively unaffected. Thus, in agreement with our proteomics data, suggesting distinct complexes with quantitative enrichment of specific BAF subunits in the developing heart, our knockdown analysis indicate that specific BAF subunits may deliver functional specificity and suggest that BAF250a-containing complexes influence a broad spectrum of cardiac gene expression.
BAF-A complex is required for fully functional cardiac differentiation in vitro and repressively regulate cardiac gene expression
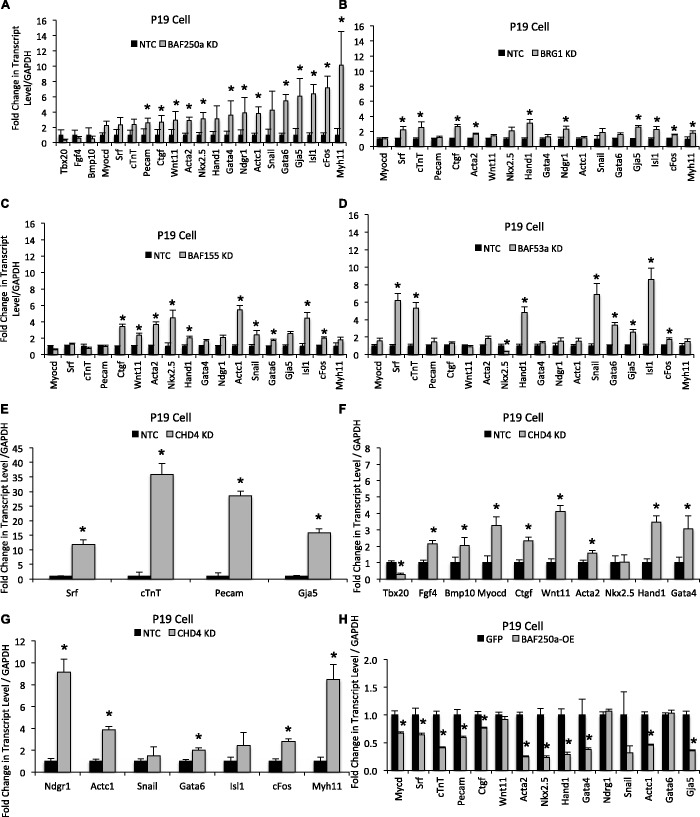
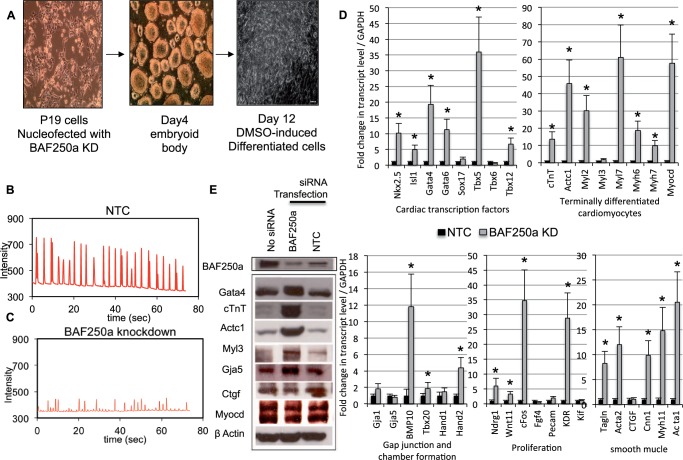
To determine if BAF250a depletion has an effect on cardiac differentiation and cardiac gene expression in differentiated cardiomyocytes, we depleted BAF250a protein in P19 cells and evaluated the ability of these cells to differentiate into cardiomyocytes upon induction with DMSO (38) using the treatment schedule depicted in Figure 3A. When P19 cells were transfected with non-target control siRNA, treatment of cell aggregates with DMSO induced the formation of rhythmically beating cells resembling primary cardiomyocytes. These cells appear to have an embryonic phenotype (Figure 3B and Supplementary Movie S1). In contrast, we observed fewer cardiomyocytes in differentiated cultures of BAF250a-depleted P19 cells, and those cardiomyocytes that were present exhibited an arrhythmic contraction phenotype. Interestingly, Action Potential (AP) and Action Duration were severely altered in BAF250a knockdown; differentiated cardiomyocytes compare with control cardiomyocytes (Figure 3C and Supplementary Movie S2). This result suggests that BAF250a depletion severely compromised the cardiogenic action of DMSO-induced differentiation. We then expanded our analysis by examining the expression of a specific set of key regulators of cardiovascular development, the expression of which is potentially mediated by BAF250a during cardiac differentiation. Cardiac morphogenesis and differentiation are governed by a set of transcription factors that regulate the expression of genes implicated in morphogenesis and patterning during heart development as well as genes required for cardiac terminal differentiation (39). Our qRT–PCR analysis showed that several important genes belonging to the core cardiac transcriptional machinery, such as Nkx2.5, Isl1, Gata4, Gata6, Tbx5 and Tbx12, were enhanced in BAF250a-depleted cells. In addition to cardiac transcription factors, BAF250a loss also increased the expression of genes specific for terminally differentiated cardiomyocyte, proliferation, gap junction and chamber formation in addition to smooth muscle cells as determined by qRT–PCR analysis (Figure 3D). Protein levels were also elevated among members of a subset of analyzed cardiac-specific proteins—Gata4, cardiac troponin, cardiac actin, Myl3 and Gja5—compared with the control cells (Figure 3E). BAF proteins were not changed relatively in BAF250a-depleted cells (Supplementary Figure S8).
Figure 3.
Cardiac-specific genes augmented in cardiomyocytes after differentiation of cultured P19 cells upon inhibition of BAF250a. (A) DMSO-induced differentiation of P19 cells. Micrograph shows Day 12 differentiated cardiomyocyte derivatives. (B), (C) DMSO-induced differentiation of P19 cells in conjunction with BAF250a knockdown results in impaired ‘myocardial function’ of P19 cell differentiated cardiomyocytes as demonstrated by spontaneous arrhythmic contraction. Panel B shows control; panel C shows BAF250a knockdown. (D) Quantitative mRNA expression of cardiac-specific genes in DMSO induced differentiated P19 cells upon BAF250a knockdown compared with non-target control. Transcript level was normalized to GAPDH. The data are expressed as mean ± SD *P < 0.05. The data represent three biological replicates and each experiment was repeated twice. (E) Western blot analysis of DMSO-induced differentiated P19 cells either not transfected, transfected with BAF250a siRNA or transfected with a non-target control siRNA. Antibodies specific for BAF250A, β-Actin and the indicated cardiac-specific proteins were used to determine their relative levels.
BAF250a-containing SWI/SNF complexes directly regulates cardiac-specific genes in the developing heart
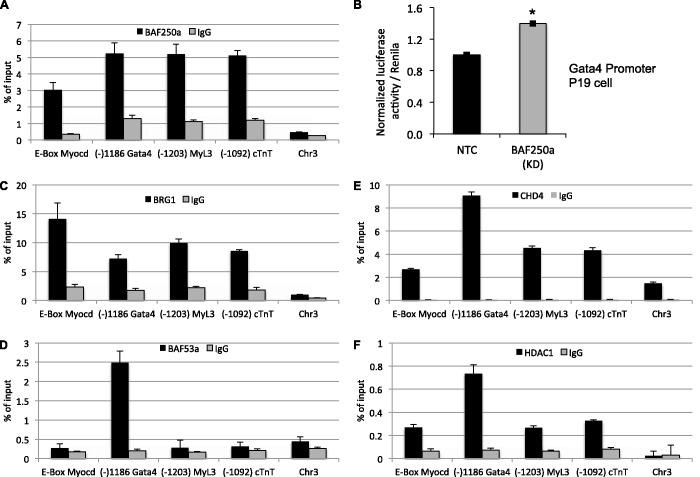
We next turned to ChIP assays to investigate a direct role for BAF250a in cardiac gene expression in the developing heart. We began our analysis of embryonic Day 9.5 hearts by examining enrichment of BAF250a on the enhancer/promoter region of cardiac-specific genes. ChIP analysis revealed BAF250a binding on DNA sequences upstream from the transcription start site (TSS) of myocardin, Gata4, Myl3 and cTnT in the developing early heart (Figure 4A) . Interestingly, the strongest enrichment was found within the genomic region previously identified as the cardiac and smooth muscle enhancer that drives endogenous myocardin expression in developing embryo (data not shown) (40). To evaluate further the functional consequences of BAF250a binding on transcriptional activity, we transfected the Gata4 promoter-luciferase construct and BAF250a siRNA into P19 cells. We found that inhibition of BAF250a caused an ∼1.5-fold induction in the Gata4 reporter activity (Figure 4B). We next evaluated the occupancy of BRG1 and BAF53a at the same targets to determine if their occupancy overlapped that of BAF250a at the same loci. BRG1 enrichment was detected on the BAF250a binding regions of the upstream DNA sequence of the all four cardiac-specific genes (Figure 4C), while BAF53a did not bind on the DNA sequence of cardiac genes except Gata4 (Figure 4D), suggesting that BAF250a-containing SWI/SNF-like complexes can directly regulate gene expression in the developing heart and support a direct repression of Gata4 by BAF-A complexes.
Figure 4.
BAF250a-containing BAF complexes regulate chromatin in early heart. (A) ChIP–qPCR showing BAF250a occupancy compared with immunoglobulin-G (IgG) in E9.5 hearts. (B) Luciferase reporter assay in P19 cells introduced with Gata4 promoter reporter construct and BAF250a siRNA. Fold changes (1.5) are shown compared with NTC (non-target control) *P < 0.001. The data are expressed as mean ± SD. (C), (D), (E), (F) Chromatin from the developing hearts was immunoprecipitated with BRG1, BAF53a, CHD4 and HDAC1 antibodies. DNA was analyzed by qPCR to determine if BRG1, BAF53a, CHD4 and HDAC1 occupy on the BAF250a binding DNA of cardiac genes. A non-genic fragment of chromosome 3 was used as negative genomic control. Data were plotted as the percentage of total input or chromatin bound. Each experiment was repeated twice.
It is likely that BAF250a-containing BAF complexes contribute to the exquisite regulation of cardiac gene expression by establishment of a repressive chromatin structure during cardiogenesis. We hypothesized that known repressor proteins such as HDAC1 and CHD4 might be recruited to the BAF250a binding region of cardiac gene enhancer/promoters in the developing heart. ChIP analyses showed that HDAC1 and CHD4 were clearly detected on the regulatory DNA of myocardin, Gata4, Myl3 and cardiac troponin-T in the developing heart (Figure 4E and F). To analyze the functional significance of CHD4 on cardiac gene expression, we knocked down CHD4 in P19 cells by siRNA (Supplementary Figure S7E) in cultured P19 cells. As predicted, CHD4 depletion in P19 cells results in upregulation of cardiac-related genes (Figure 2E, F and G), which correlates with BAF250a function. Srf, cTnT, Pecam and Gja5 expression levels were increased >10-fold (Figure 2E), whereas Fgf4, Bmp10, Myocd, Ctgf, Wnt11, Acta2, Hand1, Gata4, Ndrg1, Actc1, Gata6, cFos and Myh11 were increased >2-fold (Figure 2F and G). This supports that BAF250a and CHD4 functionally cooperate to repress cardiac genes in P19 cells. Taken together, these data demonstrate that BAF250a may facilitate an association between BAF and NURD complexes for repressive chromatin on the cardiac genes.
SWI/SNF and NURD complexes physically interact and coordinate in transcriptional regulation
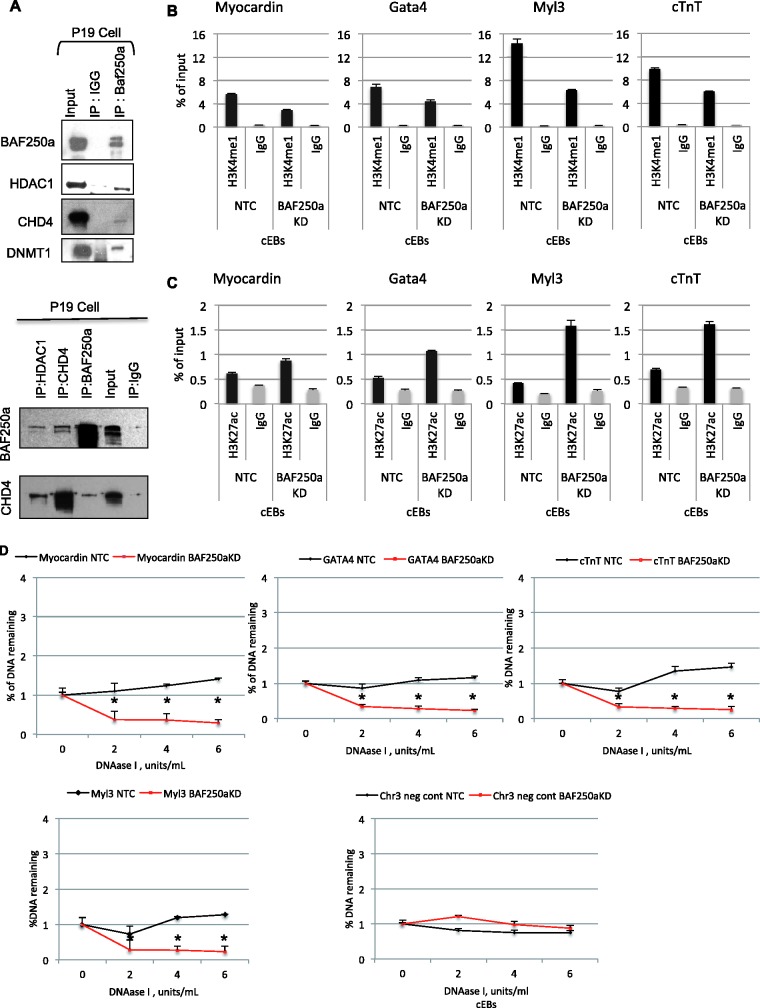
The discovery of the cardiac-enriched chromatin-remodeling complexes suggests that these complexes may provide distinctive composite surfaces for efficient of interactions with regulators that define lineage specification. Indeed, proteins involved in regulating higher order chromatin organization and presumably associated with transcriptional repression, interacted with BAF250a, including CHD3/4, HDAC1, HDAC2 and Dnmt1 (Supplementary Table S1). We were able to detect BAF250a interactions with CHD4, HDAC1 and Dnmt1 by co-immunoprecipitation (Figure 5A) . Interactions of BAF250a-containing BAF complexes with other chromatin regulators may provide additional layer of regulatory mechanisms of heart-specific genes for precise cardiogenesis.
Figure 5.
BAF250a forms a complex containing repressive complex proteins. (A) Representative western blot of immunoprecipitation using antibodies against BAF250a and IgG in P19 cells followed by probing with the antibodies against HDAC1, DNMT1 and CHD4 shows physical interaction with BAF250a. (B) Cardiac-specific genes have decreases H3K4me1 (poised chromatin mark) at the genomic loci of cardiac genes in BAF250a-depleted cell compared with control during cardiac differentiation. (C) H3K27ac (active chromatin mark) enrichment at the enhancer/promoter region of cardiac genes upon BAF250a depletion in excess to control during cardiac differentiation. (D) BAF250a/SWI/SNF modulates chromatin accessibility of target genes during cardiac differentiation. BAF250a is required for chromatin remodeling at cardiac genes promoter region. The data represents three biological replicates and each experiment was repeated twice.
BAF-A complex switch active/poised chromatin marks at cardiac-specific genes during cardiac lineage specification
We next examined the recruitment of BAF-A-containing complexes to cardiac enhancer/promoter elements during cardiac lineage differentiation by asking whether BAF250a plays a role to shift the chromatin signature of the cardiac-specific genes from a poised state to an active state in Day 4 Cardiac Embryoid Bodies (cEBs) conditioned with DMSO. In BAF250a-depleted cells, the level of myocardin-H3K4me1 was suppressed under a cardiac differentiation conditions (Figure 5B), suggesting that BAF250a is required to maintain the H3K4me1 poised mark in the myocardin enhancer/promoter during cardiac development. These changes in the H3K4me1 levels were not specific to the myocardin gene. BAF250a-depleted P19 cells also displayed suppression of H3K4me1 levels during cardiac differentiation in the enhancer/promoters for Gata4, Myl3 and cTnT, while their H3K27ac levels increased in BAF250a-depleted cEBs (Figure 5C). These results confirm that BAF250a is important in maintaining the H3K4me1 mark in cardiac genes during lineage specification. Interestingly, the myocardin-H3K4me1 levels in the cardiac genes remained higher during cardiac differentiation in the control cEBs, while the myocardin-H3K27ac levels were low under parallel differentiation condition in control cEBs (Figure 5B and C). We also obtained similar data for other cardiac genes (Figure 5B and C). These data suggest that the loss of BAF250a results in a decrease of H3K4me1 and deposition of H3K27ac on the regulatory region of cardiac-specific genes, thus allowing us to conclude that BAF250a maintains the repressive chromatin state in cardiac genes.
We also examined the histone modification status at putative enhancers/promoters. We observed significant levels of H3K4me1 at the Myocardin, Gata4, Myl3 and cTnT enhancers (Figure 5B). Upon depletion of BAF250a there was a reduction of H3K4me1 at all the target sites. In contrast, an examination of H3K27ac demonstrated an elevation of this mark upon depletion of BAF250a at the same target sites (Figure 5C).
BAF-A complex maintain closed chromatin of cardiac lineage-specific genes
As BAF complexes regulate gene expression by modulating the accessibility of DNA regulatory elements, it is possible that BAF250a/BAF-A complex may be required to establish the chromatin structure on the target genes (41). We examined the DNase I hypersensitivity at the promoter of myocardin, Gata4, cTnT and Myl3. In all cases depletion of BAF250a resulted in enhanced DNase I hypersensitivity (Figure 5D), consistent with a relatively open chromatin structure in these genes when BAF250a is depleted. The changes of chromatin structure seen in BAF250a-depleted cells were highly correlated with gene expression patterns and histone modifications (Figures 2A, 3D, 5B and C). These results suggest that BAF250a is required to modulate chromatin structure for the establishment of poised chromatin state at the target promoters during early cardiac differentiation.
BAF250a impedes BRG1 and Pol II occupancy on the genomic loci of cardiac genes
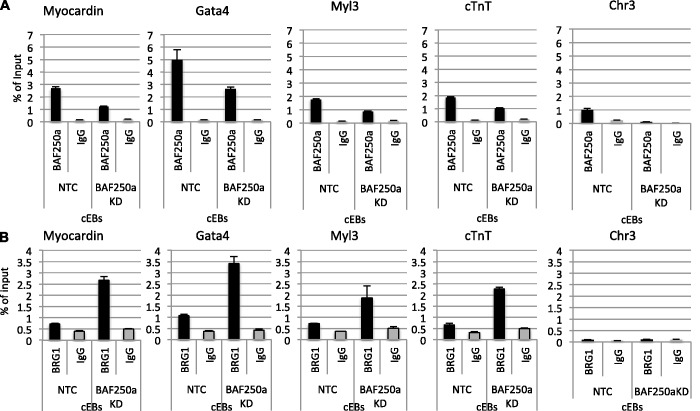
Our earlier observation that depletion of BAF250a and BRG1 had distinct effects on the transcriptional profile of the cardiac specific genes led us to consider that they may have distinct effects on specific genes. Therefore, we sought to determine whether BAF250a depletion might compromise targeting the BRG1-containing SWI/SNF remodeling complexes to cardiac genes. We performed ChIP analysis using Day 4 cEBs treated with DMSO in the presence of BAF250a-specific siRNA duplexes and non-targeting control siRNA. As expected, there was a reduction of BAF250a detected at the genes consistent with the reduction of the BAF250a protein (Figure 6A) . However, upon reduction in BAF250a protein levels and BAF250a at the targets, BRG1 was found to be increased on the genomic region of cardiac genes in BAF250a-depleted cells compared with that observed for the NTC cells, (Figure 6B). As predicted by the increase in gene expression seen previously, RNA polymerase II (Pol II) occupancy was also increased on the upstream DNA sequence of cardiac genes in BAF250a-depleted cells (data not shown). These results suggest that the presence of BAF250a can limit recruitment of transcriptional activation machinery during cardiac differentiation for appropriate gene expression and that BAF250a hampers BRG1 occupancy on the regulatory DNA sequence of cardiac genes during cardiac differentiation.
Figure 6.
BAF250a modulates BRG1 occupancy at cardiac genes. Chip assay with anti-BAF250a and BRG1 antibodies in BAF250a-depleted and non-target control Day 4, embryoid bodies cultured under cardiac differentiation condition. (A) BAF250a binding is decreased in BAF250a-depleted cEBs compare to control. (B) Loss of BAF250a leads to increase in BRG1 occupancy on cardiac genes. The data represent three biological replicates and each experiment was repeated twice.
DISCUSSION
SWI/SNF complexes have substantial transcriptional, compositional and functional diversity and are implicated in wide variety of normal biological processes, such as embryonic development as well as in abnormal processes such as cancer and other diseases (7,13). Ablation of SWI/SNF components in mouse results in defective development, suggesting it is likely required in differentiation-associated remodeling and/or the subsequent engagement of lineage-specifying epigenetic control (23,42). Elucidation of the SWI/NF subunit composition in the developing heart and action of BAF components in cardiac transcriptional regulation remains elusive. The results of this study begin the characterization of the expression patterns and details of the assembly and relative abundance of the SWI/SNF-like complexes that exist in the developing heart and delineates novel mechanisms underlying the role of BAF250a-containg BAF-A complexes in cardiac gene regulation.
Developmental progression requires coordination of chromatin organization and gene expression patterns. Our studies define a previously unknown SWI/SNF complex composition and spatiotemporal expression pattern in early heart development that provides a novel insight into the role of this complex in cardiac lineage decision and subsequent heart formation during early development. Our data show that four specific BAF subunits—BAF250a, BAF60c, BAF180 and BAF200—have expression levels that are higher in the heart compared with head and trunk tissue during early development, suggesting these subunits of the SWI/SNF complex are important for early heart formation. Indeed, depletion of BAF60c and genetic ablation of BAF250a and BAF180 in mouse embryos results cardiac malformation and lethality (25,26,30). We also show that the composition of the BAF complex is quite diverse during development, varying in different tissue types. Indeed, recent studies demonstrated embryonic stem cells and neural cell types contain distinct chromatin-remodeling complexes (16,19,35). Furthermore, we find that combinatorial assembly and relative abundance of the mSWI/SNF-like complexes in the developing heart differ from those in head and trunk tissues. Interestingly, we observed a subset of the SWI/SNF subunits; BAF250a, BAF155, BAF60a, BAF60b and BAF60c are more abundant in the cardiac-enriched BAF complexes, perhaps indicative of their specialized roles in heart development. Somewhat unexpectedly, we discovered that Baf60a and Baf60b were more abundant than Baf60c in the embryonic heart. Indeed it is BAF60b, which appeared to be the cardiac-specific subunit. Importantly, our finding that Baf60c was also present in the head enriched complex is in agreement with the presence of Baf60c in head and previously characterized npBAF complexes (19). Studies of BAF complexes in a context indicate that these complexes undergo progressive changes in subunits composition during transition from a pluripotent stem cell to a multipotent neuronal progenitor cell to a committed neuron (19). These findings are consistent with our data indicating that several types of SWI/SNF complexes may co-exist in the developing heart.
Using siRNA-mediated reduction of BAF proteins, we observed differential effects of BAF subunits on cardiac gene expression. Support of these results can be found in previous analysis of the murine mSWI/SNF (BAF) subunits, which have been shown to play non-redundant roles in development (20,22). As the early developing heart remains primitive and likely to have progenitor cells of multipotent cardiac-related cell types, we speculate that relative enrichment of BAF60 subunits in an intact complex may provide some biological specificity to cardiac cell types and heart formation in early development. These studies indicate that BAF complexes in developing heart have a unique subunit composition that is not seen in head, trunk and other previously characterized cell lines. It is tempting to speculate that the actual composition of the cardiac-enriched BAF complexes may influence how BAF subunits interact with different transcription or chromatin regulators and how a BAF subunit interacts with chromatin on given genomic loci, suggesting that BAF complexes enriched in cardiac tissue may be critical for fetal cardiac gene regulation.
The specific subunit compositions of cardiac-enriched SWI/SNF-like complexes are likely to provide a large and specialized surface area that facilitates interaction with other chromatin modifying enzymes during development. Indeed, we found that BAF-A complexes made interactions with repressive chromatin regulators and DNA-methylation machinery (CHD3/4, HDAC1, HDAC2 and DNMT1), suggesting a role in gene silencing and heterochromatin maintenance. The knockdown of BAF250a resulted in fewer differentiated cardiomyocytes in cultured P19 cell aggregates differentiated with DMSO treatment; those cardiomyocytes also exhibited an arrhythmic contraction phenotype and elevated expression of a discrete set of cardiovascular genes. The similar promotion and acceleration of expression of various cardiovascular-related genes suggest that BAF250a depletion induced a change in a broad spectrum of cardiovascular development-related genes rather than simply upregulating specific genes required for cardiac differentiation. Endogenous expression of most BAF proteins shows no change in the BAF250a-depleted cells, suggesting the assembled BRG1 complex is stable in the absence of BAF250a (23,33). These results further suggest that the phenotype observed in BAF250a-depleted cells is not compounded by a change in the BAF subunits, as has been observed in cancer cells in absence of a defined subunit (8,43). These cardiac differentiation defects and dysregulation of cardiovascular genes are varied to those observed in embryos completely lacking BAF250a and embryos completely lacking BAF250a in secondary heart field (SHF) derivatives (via Mef2c-AHF-Cre) (30). As our data suggest that the BAF250a is expressed nearly ubiquitously in early development and can bind to a wide spectrum of both activator and repressive cardiac tissue-specific enhancers/promoters, we propose that the cardiac differentiation defects observed in BAF250a-depleted cells is due to the dysregulation of a wide spectrum of cardiovascular genes during the initial stages of cardiac specification. Deletion of BAF250a in specific cardiac cell type using Mef2c-AHF-CRE; R26-YFP; BAF250f/f cell lines, YFP positive cells were observed at a much later stage of cardiac differentiation at Day 10 and leaving BAF250a intact in the cells during earlier stages shows cardiac differentiation defects, and Mef2c-Cre; BAF250af/f embryos show defects in the developing heart result in embryonic lethality in late gestation development. Our deletion using siRNA uncovers a broader BAF250a-dependent program of gene expression than that observed with Mef2c-AHF-Cre, most likely because of the earlier depletion of BAF250a (30). Alternatively, the divergent results may also reflect the significant differences between ES cells and P19 EC cells in this regard and or the difference in the differentiation protocols employed. We anticipate that deciphering the basis for these differences will be a fruitful area for subsequent investigations.
The changes in expression pattern of BAFs may be critical in modulating chromatin remodeling at specific loci and in buffering gene expression. Thus, functional and compositional specificities of the SWI/SNF complex may govern early development through modulation of the temporal and spatial expression of BAF subunits. In particular, BAF250a and its interacting repressor complex proteins bind regulatory DNA sequences of the cardiac genes in developing heart. Most likely two-mechanisms, DNA-binding transcription factors and the binding to acetylated histone tails, affect recruitment of chromatin remodeling complexes to their targets at specific positions within the genome (44,45). HDACs are recruited to chromatin via transcriptional repressors/co-repressors in the silenced loci. However, recent genome-wide studies show complex binding patterns of HDAC and associated proteins that indicate their recruitment to transcriptionally active loci in metazoans (46,47). HDAC recruited to the 5′ regions of active gene in yeast by Set3 and Set4, results in the recruitment of Rpd3, which suppresses spurious expression within the transcribing gene (48,49). More recently, it has been reported that UpSET is recruited to transcriptionally active genes and interacts with HDAC machinery and its depletion increases histone acetylation levels over transcribed regions (50). Similarly, we have shown that both BAF250a and CHD4 knockdowns accelerate the activation of cardiac genes by deposition of active chromatin marks, increasing chromatin accessibility and reducing poised marks during cardiac differentiation. These epigenetic modification patterns that result from knockdown of BAF250a bear a striking resemblance to those that distinguish active enhancers (H3K27ac+, H3K4me1+/−), which correlate with tissue-specific expression from poised enhancers (K3K4me1+), which correlates with potential gene expression later in development(51–53) during cardiac differentiation, thus further supporting a role for BAF250/BAF complex in repressive cardiac gene regulation during early stages of cardiac differentiation. Consistently, even under conditions of cardiac differentiation, BAF250a and HDAC/NURD complex are recruited to the regulatory region of the cardiac genes and are still marked by the H3K4me1. Additional depletion of BAF250a results in a more open chromatin structure on regulatory region of cardiac genes. Due to the broad expression of BAF250a in multiple tissues, our results suggest that BAF250a can play a general role in the control of multipotent cells by modifying chromatin structure and allowing genes to remain accessible for later activation. Specificity could be achieved by combining BAF subunits with other chromatin modifying enzymes that have more restricted expression, generating complexes of subunits found specifically in the region of the embryo that give rise to the heart. Taken together, these data suggest that BAF250a-containing BAF complexes are crucial in preventing hyperacetylation and maintaining a transcriptionally poised chromatin to prevent spurious gene expression during cardiac differentiation.
The ARID domain of BAF250a binds DNA in a sequence-independent manner and is not required for BRG1 localization (54,55). Unexpectedly, we observed BAF250a depletion increased enrichment of BRG1 occupancy on the regulatory region of cardiac genes under conditions of cardiac differentiation. Recently, it has been shown that the ARID domain of BAF250a implements switch from a BAF-A to a PBAF configuration at target site (56,57). BAF250a may contribute to targeting of SWI/SNF complex to specific genes in context-dependent manner. At the cardiac gene regulatory site, we have shown that BAF250a and repressive complex protein physically interact and BAF250a depletion accumulates BRG1 binding on the regulatory region of cardiac genes. Based on these observations it is possible that repressive nucleosome positioning by BAF250a-containing BAF complexes together with repressive complexes may act as a barrier to BRG1 and Pol II recruitment, perhaps limiting occupancy on target gene loci during development. Such antagonism may be a key regulatory step if the remodeling activity of one complex modulates the activity or recruitment of a different complex, especially considering that several distinct complexes can exist in the developing heart. This premise is supported by integration of multiple chromatin remodelers into the regulatory decision at a given locus. Further genome-wide studies will be necessary to probe the regulatory mechanism of chromatin remodeling complexes that exist in the developing heart.
Our study establishes a biochemical and molecular platform to develop an understanding of the process of heart development at the molecular level and provides an opportunity to identify mechanisms of gene regulation during heart development as well as mechanisms underlying heart developmental abnormalities and disease. Moreover, we have laid the foundation for understanding how the dynamics of chromatin remodeling complexes and the epigenetic landscape integrates transcriptional inputs during normal heart development. These insights will be valuable to develop improved cardiac reprogramming strategies and to elucidate the mechanisms that contribute to congenital heart disease.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Project no. Z01 ES071006-13).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Steven Akiyama for review of the manuscript and Dr Harriet Kinyamu and members of the Archer laboratory for discussion and critical analysis of the experimental approach. We thank Dr Jason Williams for his assistance in the mass spectrometry analysis, Dr Shyamal Peddada for statistical analysis, Jeff Tucker for help with video recordings and analysis of the cardiomyocyte action potential. We also thank the Histology Core Facility and NIEHS animal facility for production and maintenance of the CD1 mice.
REFERENCES
- 1.Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu. Rev. Cell Dev. Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 3.Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 4.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 5.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell Biol. 2002;3:422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 7.Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 8.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol. Cell. Biol. 2005;25:9016–9027. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn DH, Lee KY, Lee C, Oh J, Chung H, Jeon SH, Seong RH. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J. Biol. Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- 11.Simone C. SWI/SNF: the crossroads where extracellular signaling pathways meet chromatin. J. Cell. Physiol. 2006;207:309–314. doi: 10.1002/jcp.20514. [DOI] [PubMed] [Google Scholar]
- 12.Mohrmann L, Verrijzer CP. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 15.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl Acad. Sci. USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 19.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, Muchardt C, Hue L, Pontoglio M, Yaniv M, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 23.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl Acad. Sci. USA. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruneau BG. Chromatin remodeling in heart development. Curr. Opin. Genet. Dev. 2010;20:505–511. doi: 10.1016/j.gde.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei I, Gao X, Sham MH, Wang Z. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J. Biol. Chem. 2012;287:24255–24262. doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miura S, Singh AP, Mishina Y. Bmpr1a is required for proper migration of the AVE through regulation of Dkk1 expression in the pre-streak mouse embryo. Dev. Biol. 2010;341:246–254. doi: 10.1016/j.ydbio.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di-Gregorio A, Sancho M, Stuckey DW, Crompton LA, Godwin J, Mishina Y, Rodriguez TA. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development. 2007;134:3359–3369. doi: 10.1242/dev.005967. [DOI] [PubMed] [Google Scholar]
- 33.Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell. Biol. 2008;28:1413–1426. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burch JB, Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983;33:65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- 35.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl Acad. Sci. USA. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 37.van der Heyden MA, Defize LH. Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc. Res. 2003;58:292–302. doi: 10.1016/s0008-6363(02)00771-x. [DOI] [PubMed] [Google Scholar]
- 38.Paquin J, Danalache BA, Jankowski M, McCann SM, Gutkowska J. Oxytocin induces differentiation of P19 embryonic stem cells to cardiomyocytes. Proc. Natl Acad. Sci. USA. 2002;99:9550–9555. doi: 10.1073/pnas.152302499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- 41.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fryer CJ, Kinyamu HK, Rogatsky I, Garabedian MJ, Archer TK. Selective activation of the glucocorticoid receptor by steroid antagonists in human breast cancer and osteosarcoma cells. J. Biol. Chem. 2000;275:17771–17777. doi: 10.1074/jbc.M908729199. [DOI] [PubMed] [Google Scholar]
- 44.Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 45.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 46.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rincon-Arano H, Halow J, Delrow JJ, Parkhurst SM, Groudine M. UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell. 2012;151:1214–1228. doi: 10.1016/j.cell.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins RT, Furukawa T, Tanese N, Treisman JE. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 1999;18:7029–7040. doi: 10.1093/emboj/18.24.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilsker D, Patsialou A, Zumbrun SD, Kim S, Chen Y, Dallas PB, Moran E. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 2004;32:1345–1353. doi: 10.1093/nar/gkh277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1a-DNA Interactions Are Required for Promoter Occupancy by SWI/SNF. Mol. Cell. Biol. 2013;33:265–280. doi: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 2011;9:e1001206. doi: 10.1371/journal.pbio.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.