Abstract
Telomeres, nucleoprotein structures at the ends of linear eukaryotic chromosomes, are important for the maintenance of genomic stability. Telomeres were considered as typical heterochromatic regions, but in light of recent results, this view should be reconsidered. Asymmetrically located cytosines in plant telomeric DNA repeats may be substrates for a DNA methyltransferase enzyme and indeed, it was shown that these repeats are methylated. Here, we analyse the methylation of telomeric cytosines and the length of telomeres in Arabidopsis thaliana methylation mutants (met 1-3 and ddm 1-8), and in their wild-type siblings that were germinated in the presence of hypomethylation drugs. Our results show that cytosine methylation in telomeric repeats depends on the activity of MET1 and DDM1 enzymes. Significantly shortened telomeres occur in later generations of methylation mutants as well as in plants germinated in the presence of hypomethylation drugs, and this phenotype is stably transmitted to the next plant generation. A possible role of compromised in vivo telomerase action in the observed telomere shortening is hypothesized based on telomere analysis of hypomethylated telomerase knockout plants. Results are discussed in connection with previous data in this field obtained using different model systems.
INTRODUCTION
Telomeres are specialized nucleoprotein structures, usually formed by minisatellite DNA repeat sequences located at the ends of linear eukaryotic chromosomes. Telomeres are essential for maintenance of genomic integrity, compensating for the replicative loss of DNA at chromosomal termini and distinguishing natural chromosome ends from chromosome breaks [reviewed in (1)]. The basic functions of telomeres are performed in chromatin context. Telomere repeats and adjacent subtelomeric regions are associated with histones and non-histone proteins, including a number of telomere-specific proteins.
Regulation of gene expression and chromatin structure via epigenetic mechanisms has been convincingly documented using many model organisms [reviewed in (2)]. Two kinds of modifications of macromolecules are crucial for epigenetic regulation: DNA methylation and modifications of histone proteins [reviewed in (3)]. Furthermore, regulatory roles of small and non-coding RNAs have been established (4). These mechanisms modulate the dynamics of chromatin structure governing activation/repression of resident genes, e.g. in response to developmental and environmental stimuli. The general epigenetic landscape in plants is considerably more varied compared with that in animals. (i) Cytosines located in CG, CHG (H = A, T, C) and asymmetrical CHH sequences can be methylated in plant genomes, while predominantly CG methylation and a lower level of non-CG methylation during specific developmental phases were reported in animal cells [(5), reviewed in (6)]. Methylation of asymmetrically localized cytosines in plant telomeric CCCTAAA sequences was reported in Arabidopsis thaliana (7,8) and in tobacco (9). (ii) Genes encoding enzymes catalysing DNA demethylation during specific developmental phases and at specific genomic loci were identified in Arabidopsis (10,11), while in mammalian cells, demethylation is linked to base excision repair processes (12). (iii) The plant-specific RNA polymerases IV and V (13,14) catalyse the synthesis of RNA molecules involved in RNA-directed DNA methylation pathways.
The involvement of epigenetic mechanisms in the regulation of telomere homoeostasis is a popular research topic and studies in this field have been carried out using various animal models. In these cells, telomeric tracts, as well as adjacent subtelomeric regions, are maintained in a heterochromatic state associated with heterochromatin-specific histone modification. Nevertheless, in recent studies using mouse embryonic fibroblasts, association of telomeres with both the heterochromatin-specific (H3K9me3) and the euchromatin-specific (H3K4me3) epigenetic marks were reported, although the H3K4me3 loading was lower than that of H3K9me3 (15), and the level of heterochromatic marks was surprisingly low at telomeres in human fibroblasts (16) and T-cells (17). While mammalian telomeres lack CG sequences, the natural substrate of known mammalian DNA methyltransferases, in human somatic cells, the subtelomeric repeats are CG-rich and methylated (18,19). The importance of subtelomeric DNA methylation and heterochromatin-specific modifications of telomeric histones for telomere homoeostasis was reported in both human and mouse cells, where loss of heterochromatin-specific modifications led to significant lengthening of telomeres and in some cases, to an increase in telomere recombination (20–25). In contrast to these studies, Roberts et al. (26) reported that telomere lengths were not affected in mouse epigenetic mutants, challenging the idea of epigenetic control of telomere homoeostasis in mammalian cells.
In the classic model, and similarly as in mammalian cells, plant telomeres were viewed as heterochromatic loci (27). A more recent study, however, characterized A. thaliana telomeric chromatin as an intermediate heterochromatin possessing both hetero- and euchromatin-specific histone modifications (8). Vaquero-Sedas et al. (28) even concluded that telomeres of A. thaliana exhibit predominantly euchromatic features, while subtelomeres and interstitial telomeric sequences are of a heterochromatic nature.
Other factors possibly involved in telomere homoeostasis are telomeric transcripts [telomeric repeat containing RNA (TERRA)], the discovery of which has challenged the long-standing opinion that telomeres are transcriptionally inert (29). Although intensively studied, connections between TERRA and telomerase activity/telomere homoeostasis are far from clear. TERRA and transcription of telomeres do not affect telomere lengths, and TERRA does not inhibit telomerase activity in vivo in human cancer cells (30). Conversely, telomere length-dependent inhibition of telomerase activity in vitro by TERRA (31), binding of TERRA to the RNA subunit of telomerase (hTR) and partially to the telomerase catalytic subunit (hTERT) in human cells (32), and telomerase-independent telomere shortening induced by up-regulation of TERRA in Saccharomyces cerevisiae (33), have been reported. In plants, the presence of TERRA transcripts has been reported in Arabidopsis (8) and in tobacco BY-2 cells (9).
Here, we examine the length and methylation of telomeres in A. thaliana methylation mutants (met 1-3 and ddm 1-8), and in wild-type (wt) plants that were germinated in the presence of hypomethylation agents. The MET1 gene encodes the DNA methyltransferase (34) responsible for maintaining methylation of cytosines located in CG sequences. Proper MET1 gene function is crucial for the maintenance of the general epigenetic pattern, including CG and non-CG methylation, histone modifications and chromatin structure (35). The DDM1 gene encodes a protein with similarities to the SWI/SNF family of chromatin remodelling factors (36). A mutation in this gene leads to a significant decrease in the overall level of DNA methylation (37).
Zebularine [1-(β-d-ribofuranosyl)-1,2-dihydropyrimidine-2-one, ZEB] is a cytidine analogue (38) that is stable in aqueous solution and has relatively low toxicity [reviewed in (39)]. Its hypomethylation effect is similar to that of the traditionally used cytidine analogues, 5-azacytidine and 5-aza-2′-deoxycytidine (40). These compounds are incorporated into DNA during replication and form stable complexes with DNA methyltransferases, thereby reducing their methylation activity. In the model plants A. thaliana (41), and Nicotiana tabacum (9), ZEB-induced loss of DNA methylation was described.
DHPA [(S)-9-(2,3-dihydroxypropyl)adenine] (42) is a competitive inhibitor of S-adenosyl-l-homocysteine hydrolase, the enzyme that degrades S-adenosyl-l-homocysteine, a by-product of transmethylation reactions and a potent inhibitor of all methyltransferases, to homocysteine and adenosine. A number of experimental studies have been carried out using this drug for epigenetic modifications, including its application in plant systems (9,43–46).
We observe distinct telomere shortening in chemically hypomethylated plants and in later generations of methylation mutants. The shortening of telomeres neither correlates with the change in transcription of the telomerase reverse transcriptase (AtTERT) gene nor with telomerase activity assayed in vitro. Hypotheses considering in vivo processes involved in telomere shortening and the role of telomerase in the maintenance of shortened telomeres are presented.
MATERIALS AND METHODS
Plant material
Arabidopsis thaliana seeds of the Columbia-0 ecotype, DDM1 (At5g66750) mutant (ddm1-8 strain, SALK000590) and TERT (At5g16850) mutant (tert−/−, SALK061434) were purchased from the Nottingham Arabidopsis Stock Centre (47); seeds of the mutant plant with a T-DNA insertion in the MET1 gene [At5g49160, met1-3 strain, (48)] were kindly provided by Dr Ales Pecinka (GMI, Vienna, Austria). Primers for genotyping are shown in Supplementary Table S1. Seeds were placed on half strength Murashige–Skoog (½ MS, Duchefa) agar plates and grown under cycles of 8 h light (illumination 100 mmol m−2 s−1), 21°C and 16 h dark, 19°C. Plants were grown in soil under the same light/dark conditions favouring leaf growth; 1-month-old plants were then cultivated using 16 h/8 h light/dark cycles, accelerating flowering and seed development. Leaves were harvested from 2-month-old plants.
Arabidopsis thaliana Columbia-0 and tert−/− seeds were germinated for 7 days on agar plates containing ½ MS medium supplemented with ZEB (Sigma) or DHPA at concentrations of 100 and 250 µM. Plants were then grown in soil as described earlier.
Analysis of telomere lengths by the terminal restriction fragment method
Analysis of telomere length by the terminal restriction fragment (TRF) method is based on the digestion of genomic DNA by a frequently cutting restriction endonuclease without a recognition site in the G-rich telomeric sequences. After hybridization with a radioactively labelled telomeric oligonucleotide probe, the signal corresponds to non-digested telomeric tracts (plus subtelomeric regions up to the first restriction site upstream of the telomeres). Analyses were performed as previously described (49,50).
Analysis of cytosine methylation in telomeric repeats
Bisulfite conversion of genomic DNA was carried out using the EpiTect Bisulfite Kit (Qiagen). During the bisulfite treatment, non-methylated cytosines are converted to uracils and amplified as thymines in the subsequent PCR, while 5-methylcytosines are resistant to this reaction (51). To analyse the methylation of all cytosines located in telomeric repeats, we followed the protocol described in (8), with modifications (9). Bisulfite-modified DNA (300 ng) was transferred to a Hybond XL membrane (GE Healthcare) by vacuum dot blotting (Bio-Rad Dot Blot). Membranes were hybridized overnight at 42°C with [32P] labelled oligonucleotide probes in ULTRAhybTM–Oligo Hybridization Buffer (Ambion). A ‘loading’ probe (CCCTAAA)4 was used as a loading normalizer, and the ‘degener’ probe (TTAGRRT)4, where R = A or G, detected telomeres where the third cytosine of the (ACCCTAA)n repeat was methylated, and other cytosines were either methylated or non-methylated. After washing under low stringency conditions (twice at 50°C for 30 min in 2× SSC, 0.1% SDS), hybridization signals were visualized using the FLA7000 phosphoimager (FujiFilm) and analysed using the MultiGauge software (FujiFilm).
Methylation of telomeric cytosines located in the proximal part of the 1L chromosome arm telomere was analysed as described in (8). PCR primer sequences for amplification of the fragment containing telomeric repeats are shown in Supplementary Table S1. PCR was carried out using DyNAzymeII DNA Polymerase (Finnzymes) in a cycle consisting of initial denaturation (2 min), 30 cycles of 30 s at 94°C, 30 s at 56°C and 30 s at 72°C, followed by final extension (72°C/8 min). PCR products were cloned using a TOPO TA Cloning Kit (Invitrogen) and sequenced (Macrogene). Methylation of cytosines was analysed by CyMATE software (52) in lengths of ∼390 bp, where the sequencing signals were clear. Statistical evaluation was done by one-way weighted analysis of variance (ANOVA), a statistical tool used for simultaneous analysis of differences between two or more means. Seventeen clones from three Columbia plants, 11 clones from two ddm1-8 G2 plants, 12 clones from two met1-3 G2 plants and 4 clones from a segregated wt plant were analysed. Samples were taken as independent.
To analyse relative methylation of cytosines located in terminal and internal telomeric repeats, high-molecular weight DNA was isolated and digested by Bal31 nuclease as described (53,54). DNA in agarose plugs was either digested by Tru1I (MseI) restriction endonuclease (Fermentas/Thermo Fisher Scientific) and analysed by TRF, or isolated by QIAEX II Gel extraction Kit (Qiagen), modified by sodium bisulfite, spotted by vacuum blotting onto nylon membrane and hybridized with radioactively labelled ‘loading’ and ‘degener’ probes as described earlier.
RESULTS
Methylation of cytosines in telomeric repeats is significantly lower in hypomethylated plants
Telomere length and methylation of cytosines in telomeric repeats were assayed in hypomethylated A. thaliana var. Columbia-0 plants in which hypomethylation was induced by (i) loss of function of the gene essential for the maintenance of a stable methylation pattern in met1-3 and ddm1-8 mutants and (ii) germinating seeds of wt plants in the presence of the hypomethylation drugs, ZEB or DHPA (Figure 1). To verify the loss of DNA methylation in mutant plants and in seedlings germinated in the presence of hypomethylation drugs, methylation of cytosines in a 180-bp centromeric repeat was investigated using the methylation-sensitive restriction endonuclease HpaII (see Supplementary Methods). Methylation was significantly lower in leaves of mutant plants and in seedlings germinated in the presence of hypomethylation drugs (Supplementary Figure S1), as previously reported (41,48). In leaves of wt plants segregated from the met1-3 mutant background, the methylation was almost comparable to samples without a mutant history, although in some individuals bands evidencing the non-methylated cytosines in the CCGG sequence motif were visible (Supplementary Figure S1A). In mature leaves of DHPA-treated plants that were grown in soil without the drugs, methylation returned to control levels (Supplementary Figure S1B), consistent with the transient hypomethylation effect described for this drug (44). Variable patterns of centromeric repeat methylations were observed in plants germinated in the presence of ZEB and even in the progenies of plants affected by 250 µM ZEB, although in these cases, the levels of non-methylated cytosines in CCGG sequences were significantly lower than those in ZEB-treated seedlings (Supplementary Figure S1B).
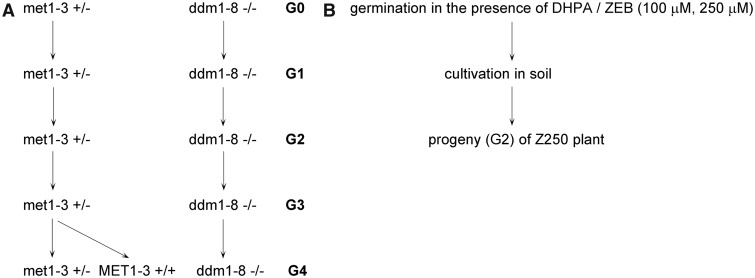
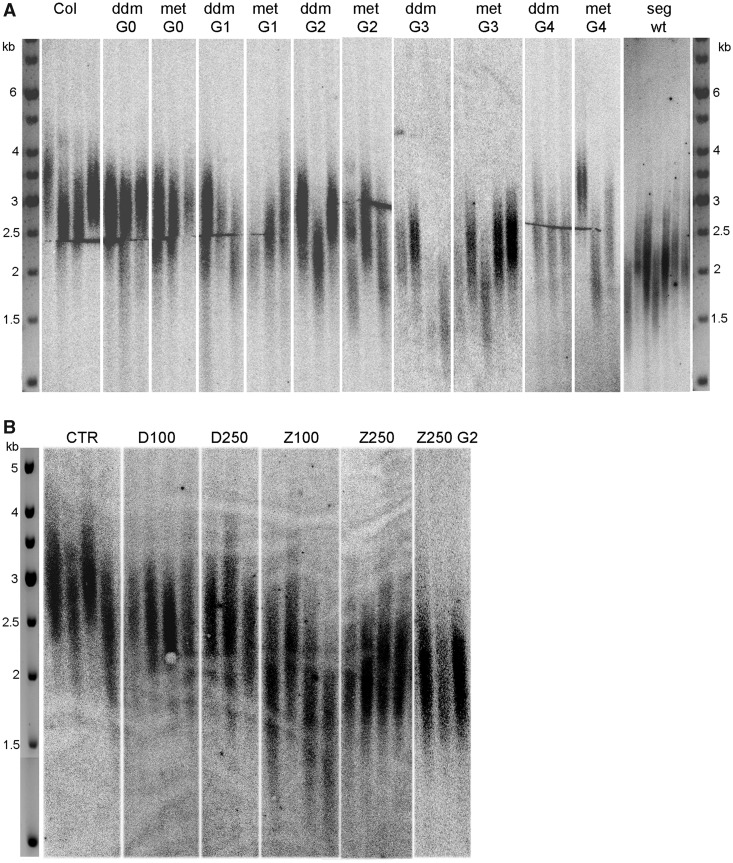
Figure 1.
Schematic picture of the experimental strategy. (A) met1-3 and ddm1-8 mutant plants were propagated and genotyped in each generation. Material for analyses was collected from ddm1-8−/− homozygous individuals and met1-3+/− heterozygous plants; met1-3−/− homozygous mutants were selected with extremely low frequency and did not grow to the reproductive stage (48). G0 of ddm1-8 plants represents the T3 progeny of the original accession. In the fourth generation (G4), segregated wt plants (MET1-3+/+) were selected for analysis of telomere length. (B) Arabidopsis thaliana seedlings were germinated in the presence of hypomethylation drugs DHPA or ZEB for 7 days. Plants were then cultivated in soil, and after 9 weeks, leaves were collected for analysis. Progenies (G2) of plants that had been treated with 250 µM ZEB (Z250) were grown for telomere length analysis.
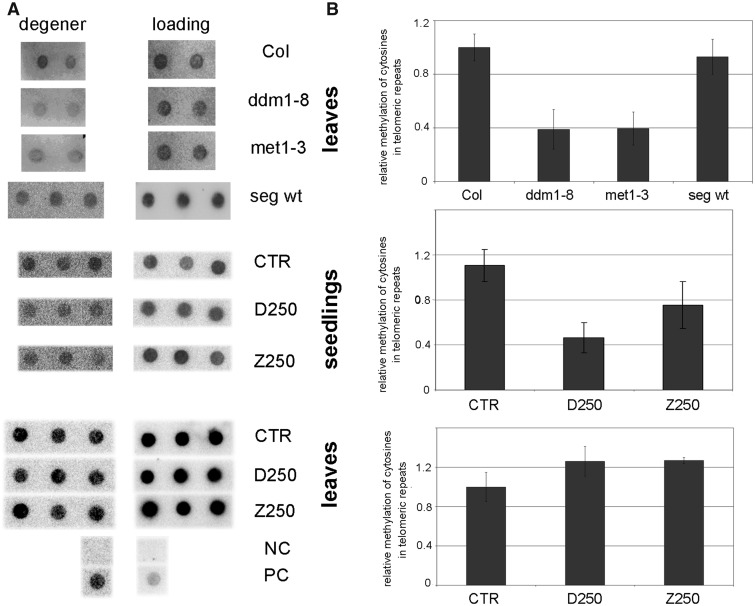
To analyse cytosine methylation in telomeric repeats regardless of their chromosomal position, bisulfite-converted DNA was spotted onto nylon membrane and hybridized with a radioactively labelled probe that detected methylated and partially methylated telomeric repeats (‘degener’ probe). Relative differences between signals from A. thaliana wt plants compared with methylation mutants demonstrated a loss of methylated telomeric cytosines within the mutant background (Figure 2). A similar pattern was observed when populations of seedlings germinated in the presence of 250 µM DHPA and 250 µM ZEB were analysed—a distinctly lower hybridization signal using the ‘degener’ probe in hypomethylated seedlings as compared with the control non-treated sample. Hypomethylation did not persist in mature leaves of drug-germinated plants or in wt plants segregated from the met1-3 mutant background; in these samples, methylation of telomeric cytosines recovered to the level of control/wt plants (Figure 2).
Figure 2.
Analysis of general methylation of telomeric cytosines. (A) About 300 ng of bisulfite modified DNA isolated from plant tissues was spotted onto the membrane and hybridized with the radioactively labelled ‘degener’ probe (TTAGRRT)4, R = A or G, which generated a signal from methylated and partially methylated fractions, and the ‘loading’ probe (CCCTAAA)4 complementary to the G-strand of telomeres, which was used for normalization. Top panel: for analysis of ddm1-8 and met1-3 methylation mutants, leaves of G2 plants were collected from 10 individuals, two representatives are presented; 10 Columbia wt (Col; two presented) and 7 wt plants segregated from the met1-3 mutant (seg wt; three presented) were analysed. Middle panel: analysis of seedlings grown on control medium (CTR), medium supplemented with 250 µM DHPA (D250) and 250 µM ZEB (Z250); three biological replicates were analysed. Bottom panel: analysis of mature leaves from three representative plants grown from drug-treated seedlings. NC, negative control, DNA from the plasmid pUC19; PC, positive control, non-converted genomic DNA isolated from A. thaliana leaves. (B) Relative methylation of telomeric cytosines in hypomethylated A. thaliana tissues. Hybridization signals were evaluated by the MultiGauge software (FujiFilm), and expressed as the ‘degener’/‘loading’ ratio. Signals of Columbia leaves and control non-treated samples (CTR) were arbitrary taken as 1 in respective analyses.
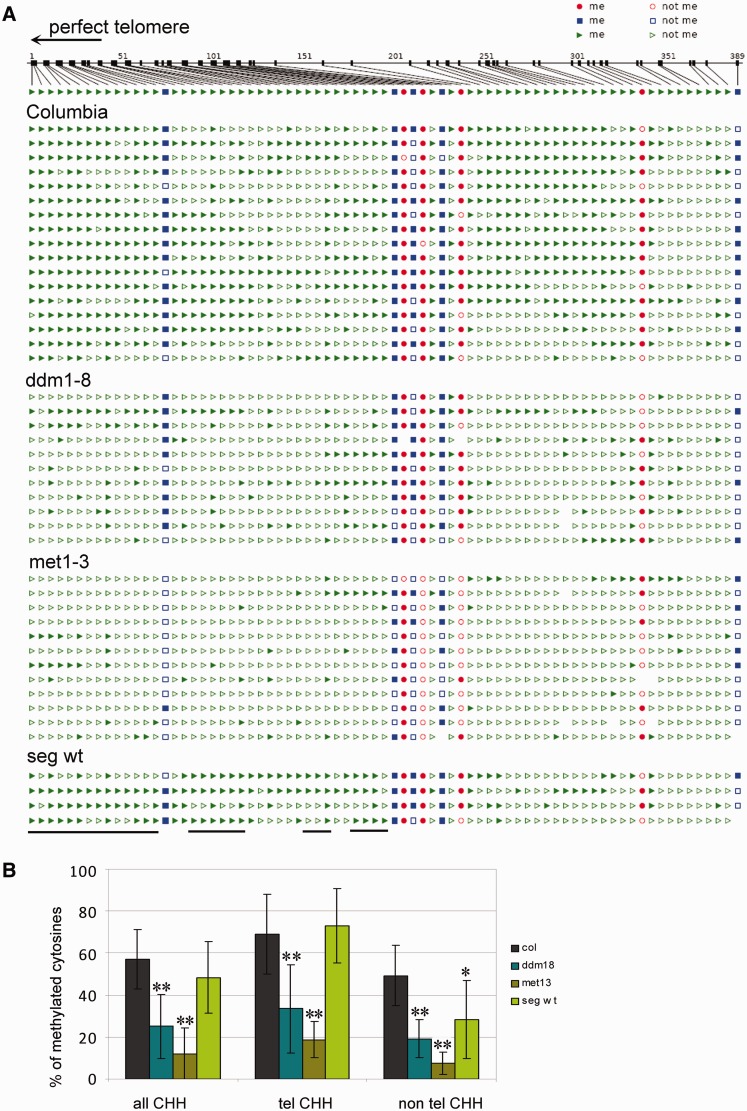
Results obtained from the above analyses provide information about methylation of cytosines in telomeric repeats in terminal (in telomeres) and internal (in ITSs) chromosomal locations within the A. thaliana genome. Because ITSs represent a relatively large proportion of all telomeric repeats [20–70%, depending on the methodology used and sequence accuracy, (55,56)], we followed the approach described in (8) and carried out a detailed analysis of methylation in a specific telomeric region; the proximal part of the 1L chromosome arm telomere. According to bisulfite genomic sequencing data from wt Columbia leaves, almost 60% of all asymmetrically located cytosines and >60% of cytosines located in perfect telomeric repeats are methylated (Figure 3A and B). These data demonstrate, at least for this specific region, the presence of methylated cytosines in A. thaliana telomeres. In both met1-3 and ddm1-8 methylation mutants, the number of methylated cytosines in asymmetrical sequences, within and outside of telomeric repeats, is significantly lower compared with Columbia wt leaves (Figure 3). In wt segregated from the met1-3 mutant, methylation recovered almost to the Columbia wt level, although a reduced level of methylation was observed in cytosines located outside of perfect telomeric repeats. Based on these data, methylation of cytosines in telomeric repeats appears to be dependent on the activity of enzymes that maintain a stable methylation pattern.
Figure 3.
Analysis of cytosine methylation in the proximal part of the 1L telomere arm. (A) Distribution of methylated cytosines along the 389-bp region, which is delimited by primers derived from the subtelomeric region and a specific insertion into the 1L telomere (8). Seventeen clones from three Columbia plants, 11 clones from two ddm1-8 G2 plants, 12 clones from two met1-3 G2 plants and 4 clones from a segregated wt plant (seg wt) were analysed. Red circles, CG methylation; blue squares, CHG methylation; green triangles, CHH methylation; filled symbols, methylated cytosine; empty symbols, non-methylated cytosine. Positions of cytosines located in perfect telomeric repeats are delimited by the black lines below the figure. The arrowhead determines the direction to the perfect telomere. (B) Graphical representation of the level of all methylated cytosines located in non-asymmetrical sequences (all CHH), in perfect telomeric repeats (tel CHH) and outside these repeats (non-tel CHH). Standard deviations reflect variability between clones. The level of methylated cytosines is significantly lower in methylation mutant clones (**P < 0.01 for all sequence contexts). In the segregated wt, methylation of cytosines outside of telomeric repeats (non-tel CHH) is reduced (*P < 0.05).
Although dot-blot analyses revealed cytosine hypomethylation in telomeric repeats of DHPA- and ZEB-treated seedlings (Figure 2, middle panel), no such trend was observed for cytosines located in the proximal 1L chromosome arm telomere; methylation patterns of asymmetrically located cytosines within and outside of perfect telomeric repeats, in treated seedlings (Supplementary Figure S2) and in leaves of plants germinated in the presence of drugs (not shown), were similar to controls.
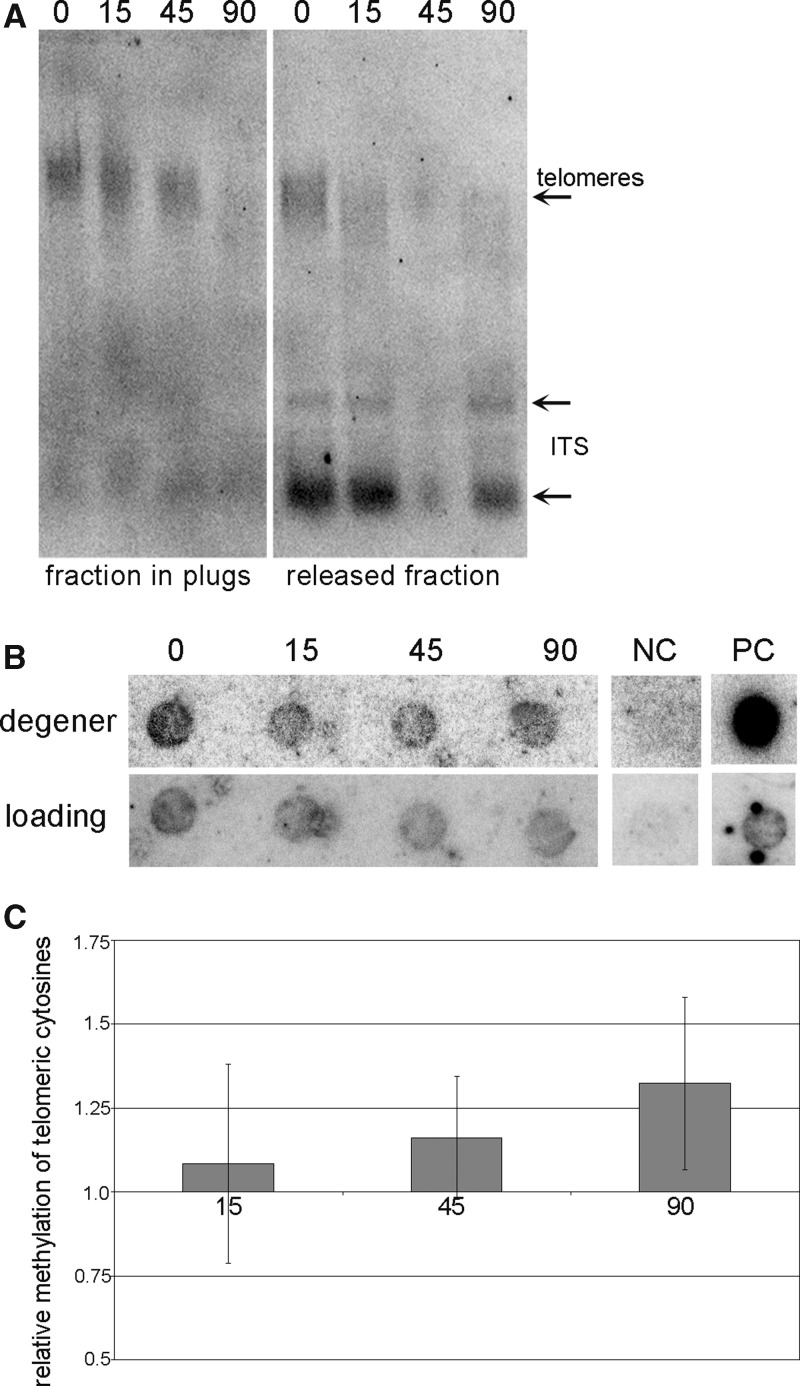
Bearing in mind previous contradictory data revealing the presence (7,8) or absence (28) of methylation of telomeric cytosines in A. thaliana, we aimed to determine the relative level of methylation at terminally located telomeric repeats. To selectively degrade telomeres, high-molecular weight DNA from A. thaliana leaves was digested by Bal31 exonuclease. Telomere shortening by Bal31 was verified by TRF analysis (Figure 4A); a significant loss of the telomere-specific signal during Bal31 digestion was observed, while signals from ITSs were comparable in Bal31-treated and non-treated samples. DNA was isolated from agarose plugs, modified by sodium bisulfite, spotted onto nylon membrane and hybridized with radioactively labelled ‘loading’ and ‘degener’ probes (Figure 4B). The reduction in the telomere-specific signal from the loading probe in Bal31-digested samples (to about one half as compared with the non-digested control) demonstrated the loss of terminal telomeric repeats. Comparison of the hybridization signals obtained using the ‘degener’ probe relative to the ‘loading’ probe did not reveal any statistically significant differences and showed a relatively stable methylation of telomeric cytosines during BAL31 digestion (Figure 4C); this demonstrated that methylation of cytosines in telomeres and in interstitial telomeric repeats was similar.
Figure 4.
Analysis of relative methylation of cytosines in telomeric repeats. (A) High-molecular weight DNA in agarose plugs was digested by Bal31 exonuclease and analysed by TRF. The fraction retained in agarose plugs and the fraction released into the solution were analysed separately. The time of Bal31 digestion (in min) is marked above the lines. The positions of telomeres and ITSs are depicted by arrowheads. (B) DNA isolated from agarose plugs was modified by sodium bisulfite and analysed by Southern hybridization with radioactively labelled ‘loading’ and ‘degener’ probes. The time of BAL31 digestion (in min) is depicted. NC, negative control, DNA from the plasmid pUC19; PC, positive control, non-modified genomic DNA isolated from A. thaliana leaves. (C) Relative methylation of telomeric cytosines in Bal31-digested samples. Hybridization signals were evaluated by the MultiGauge software (FujiFilm), and expressed as the ‘degener’/‘loading’ ratio. The ratio in BAL31 non-digested samples was arbitrary taken as 1. Five independent experiments were evaluated.
Telomere shortening in hypomethylated A. thaliana plants and in plants with a history of hypomethylation
Stable telomere length is important for genomic stability. We determined the lengths of telomeric repeats to establish whether A. thaliana genomic hypomethylation influences telomere homoeostasis. In Columbia wt plants, telomeres were ∼3 kb long with some variations between individuals (Figure 5). Up to the second generation (G2) of methylation mutants, telomeres were maintained at about wt lengths, although some individuals with shorter telomeres were observed, even in G1 and G2. In G3, telomeres of mutants were markedly shortened by ∼500–700 bp in met1-3 plants and even more than 1 kb in some ddm1-8 individuals. Telomere shortening continued into the next generation (G4) although in this case, plants with shortened telomeres as distinct as in G3 were not observed (Figure 5A).
Figure 5.
Telomere lengths in hypomethylated A. thaliana plants. Lengths of telomeres were analysed by the TRF method in methylation mutants (A) and in plants germinated in the presence of hypomethylation drugs (B). (A) G0–G4 generations of ddm1-8 and met1-3 mutant plants and plants without T-DNA insertion segregated from met1-3 G3 mutants (seg wt, for details, see Figure 1) were analysed. (B) CTR, control non-treated A. thaliana plants; D100, plants germinated in the presence of 100 µM DHPA; D250, plants germinated in the presence of 250 µM DHPA; Z100, plants germinated in the presence of 100 µM ZEB; Z250, plants germinated in the presence of 250 µM ZEB; Z250 G2, progenies of plants germinated in the presence of 250 µM ZEB.
Telomeres in plants that had been treated by hypomethylation drugs during germination were also distinctly shorter, and the shortening was more pronounced in ZEB-treated plants. While telomeres of plants germinated in the presence of DHPA were ∼500 bp shorter compared with non-treated controls, in ZEB-treated individuals, shortening by more than 1 kb was observed (Figure 5B). Telomere shortenings in plants that were grown in the presence of different concentrations of methylation inhibitors (100 and 250 µM) were similar, thus independent of the drug concentrations used.
To investigate possible recovery from telomere shortening induced by genome hypomethylation, telomere lengths were analysed in wt plants segregated from the met1-3 mutant background, and in the progenies of plants treated with 250 µM ZEB (Figure 1). Telomeres in both groups remained markedly shorter (Figure 5A and B), demonstrating meiotic stability of this phenotype.
Involvement of telomerase in telomere shortening of hypomethylated A. thaliana plants
Telomerase is an enzyme responsible for the elongation of telomeres, and loss of its function leads to progressive telomere shortening during subsequent plant generations (50,57). To determine whether telomerase dysfunction is involved in the process of telomere shortening observed in later generations of met1-3 and ddm1-8 mutants, and in plants germinated in the presence of hypomethylation drugs, telomerase activity and transcription of the gene encoding the telomerase protein subunit (AtTERT) were analysed in buds, i.e. in tissue with high telomerase activity (58). Telomerase activity and AtTERT gene transcription were assayed in generations G2 and G4 of met1-3 and ddm1-8 relative to the wt (see Supplementary Methods). Neither telomerase activity nor AtTERT transcription was significantly affected in any of the mutants (Supplementary Figure S3). Correspondingly, in plants germinated in the presence of hypomethylation drugs, telomerase activity and transcription were at control levels in telomerase-positive tissues investigated, in 7-day seedlings and in buds (data not shown). Therefore, in A. thaliana plants, neither AtTERT transcription nor telomerase activity (as examined by assay in vitro) was sensitive to hypomethylation.
In addition to telomerase, other proteins involved in telomere homoeostasis in A. thaliana have been described. Nevertheless, transcription of six selected genes coding for such proteins [telomerase RNA subunits AtTER1, AtTER2 (59), AtPOT1a (60), ATPOT1b (61,62), AtTRB1 (63), AtCTC1 (60)] was at wt levels in buds collected from G2 and G4 generations of met1-3 and ddm1-8 mutants, as well as in buds of plants treated with hypomethylation drugs (data not shown). In addition, analysis of TERRA transcripts (see Supplementary Methods) using primers specific for 2R, 3L and 5L chromosome arms (8) did not reveal significant differences between individuals germinated in the presence of 250 µM DHPA or 250 µM ZEB, except for slightly elevated levels of TERRA derived from the 3L chromosome arm (Supplementary Figure S4).
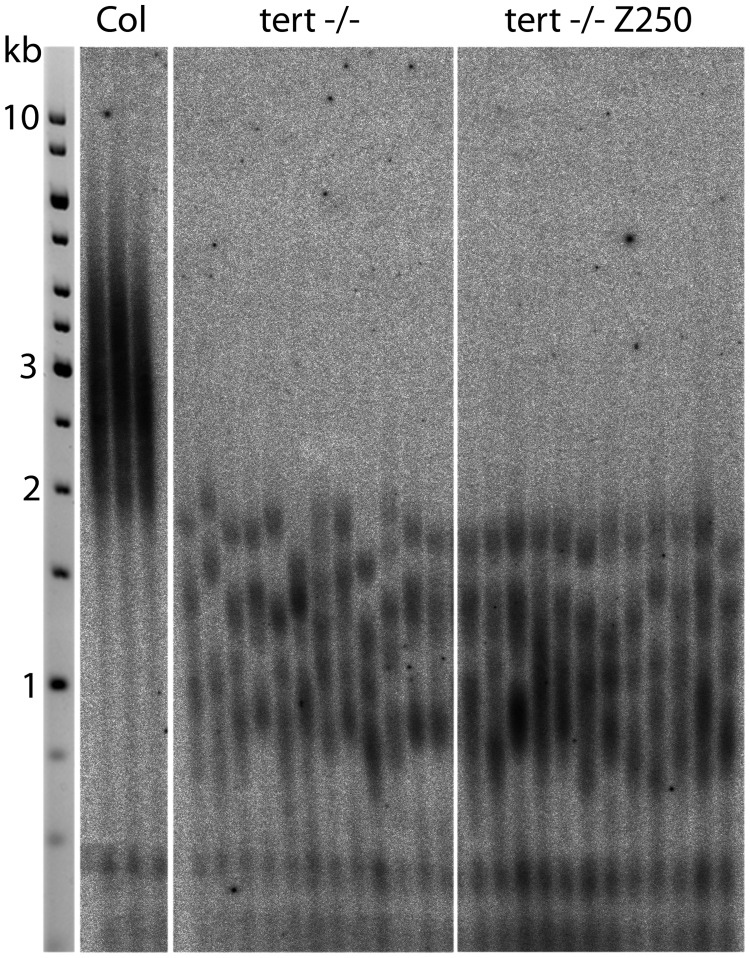
Due to the complexity of the process of telomere maintenance, it is tricky to obtain unambiguous information on possible in vivo involvement of telomerase in telomere shortening. We analysed telomere lengths in plants with loss of telomerase function; in tert−/− mutants (50,57), germinated in the presence and absence of 250 µM ZEB. According to the working hypothesis, if telomere shortening in hypomethylated A. thaliana plants is dependent on telomerase action, then the lengths of telomeres in hypomethylated tert−/− mutants would be similar to those in tert−/− mutants germinated in control medium because active telomerase is entirely absent here and therefore cannot be modified by hypomethylation during germination. If any telomerase-independent factor participates in telomere shortening, both effects will be additive and presumably telomeres will be even shorter in hypomethylated tert−/− mutant plants. As shown in the TRF profiles in Figure 6, telomere lengths in hypomethylated tert−/− plants remained the same as those in mutants germinated in control medium. Although these experiments suggest involvement of telomerase in the observed telomere shortening in hypomethylated plants, they do not identify directly a specific underlying step in telomerase in vivo regulation.
Figure 6.
Telomere lengths in hypomethylated telomerase mutant (tert−/−) plants. Lengths of telomeres were analysed by TRF in leaves of tert−/− plants and tert−/− plants germinated for 7 days in the presence of 250 µM ZEB (tert−/− Z250).
DISCUSSION
The long-established view of plant telomeres as heterochromatic loci was challenged by the recent data demonstrating the presence of both heterochromatin- and euchromatin-specific epigenetic modifications of telomeric histones in A. thaliana (8). Following an independent study by another group (28), telomeric sequences were hypothesized to have a euchromatic character for A. thaliana telomeres and a heterochromatic character for interstitial telomeric sequences. Even in the field of mammalian telomeric chromatin, where data on the importance of heterochromatin-specific modifications for telomere stability were collected mainly using mouse models [reviewed in (64)], the general doctrine of telomeric heterochromatin was challenged by observations in human cells (16,17). Contrary to the universality of the genetic code, there is no unique or generally valid epigenetic code, and data obtained in different model systems are required to shed more light on the epigenetic nature of telomeric chromatin and to distinguish between specific features of a model system and more general principles.
Methylation of telomeric cytosines is dependent on the activity of MET1 a DDM1 enzymes
Arabidopsis thaliana is an excellent model organism for epigenetic studies due to the accessibility of mutant line collections, including lines with loss-of-function of important epigenetic factors. On the other hand, hypomethylation induced by chemical drugs may be a relatively easy way to disturb an established epigenetic landscape in a broad range of model organisms. Such compounds were used in a pilot study analysing the epigenetic regulation of telomeres in cultured cells of N. tabacum (9).
In our experiments, cytosine methylation, including cytosines in telomeric repeats, was significantly lower in mutant plants with loss of function of enzymes (MET1 and DDM1) that are essential for the maintenance of a stable pattern of methylation. After restoration of the wt allele in a met1-3 mutant background the methylation in both symmetrically (centromeric repeat; Supplementary Figure S1A) and asymmetrically (telomeres; Figures 2 and 3) located cytosines returned almost to the wt pattern. This agrees with a recent observation that loss of methylation in genic regions is persistent in segregated wt while methylation in another type of sequence, transposable elements, recovers to wt level, suggesting site-specific regulation of DNA methylation (65). Similarly, treatment of A. thaliana plants with hypomethylation drugs during germination led to a decreased level of telomeric and centromeric cytosine methylation in seedlings, i.e. during the developmental stage when plant tissue is in direct contact with the drug (Figure 2; Supplementary Figure S1B). During cultivation of plants in soil without drugs, DHPA-induced hypomethylation was completely reversed (Figure 2; Supplementary Figure S1B), as was observed in previous experiments using tobacco plants and cell cultures (44,45). Although the hypomethylating effect of ZEB was also reported as transient (41), in our experiments, distinct hypomethylation of cytosines located in CCGG sequences was detectable even in the next plant generation (Supplementary Figure S1B). This may be due to a higher concentration of ZEB or a different A. thaliana ecotype used in our study. Nevertheless, our data agree with that of 5-azacytidine-induced stable hypomethylation of symmetrically located cytosines, as observed in repetitive sequences in tobacco plants (66). This is also consistent with similar hypomethylation mechanisms of both 5-azacytidine and ZEB drugs.
Although in the leaves of methylation mutants loss of methylation of asymmetrically located cytosines in the proximal part of the 1L telomere arm was demonstrated (Figure 3), no changes were detected in drug-treated seedlings (Supplementary Figure S2). This difference may be attributed to different methods of induction of the hypomethylation state (chemical versus genetic).
Even though it was demonstrated that cytosines located in perfect telomeric repeats in the proximal part of the telomere on the 1L chromosome arm are methylated [(8) and Figure 3; Supplementary Figure S2], and that the extent of this methylation is dependent on the activity of MET1 and DDM1 enzymes (Figure 3), questions remain about the level of true (terminal) telomere methylation in A. thaliana (28). Our analyses of the relative methylation of cytosines located in CCCTAAA sequences of Bal-31 digested DNA, i.e. in samples depleted of telomeres (Figure 4), demonstrated that cytosines in telomeres and in interstitial telomeric sequences are methylated to similar extents. The finding that cytosines in A. thaliana true telomeres are methylated is consistent with the data on drug-induced hypomethylation of telomeric cytosines in tobacco (9) where significant levels of intrachromosomal telomeric repeats were not found (67).
Compromised telomere maintenance in hypomethylated plants raises questions on underlying mechanism
In our previous work, we demonstrated that in Arabidopsis, DNA methylation was not significantly involved in the developmental regulation of transcription of the telomerase catalytic subunit (68). Correspondingly, transcription of the telomerase catalytic subunit and telomerase activity in vitro were not affected in later generations of met1-3 and ddm1-8 mutants nor in plants treated with hypomethylation drugs during germination (Supplementary Figure S3). Analysis of telomerase activity by in vitro TRAP assay only provides information on whether active telomerase is present in the extracted material. The absence of an additive effect of hypomethylation and disruption of telomerase activity (Figure 6) suggests (though indirectly) a possible involvement of telomerase in the observed telomere shortening. Although telomerase activity is essential for telomere elongation, the complex process of telomere maintenance involves numerous factors participating in regulation of telomerase activity in vivo which cannot be reflected by the used in vitro assays. In principle, the genome hypomethylation may affect telomere homoeostasis by three ways: (i) directly via modulation of telomerase transcription (and consequently activity)—which was ruled out [Supplementary Figure S3; (68)]; (ii) by compromised function of telomerase ‘holoenzyme’ complex, which includes, in addition to both core telomerase subunits, a group of telomerase-associated protein factors affecting, e.g. telomerase recruitment, intracellular trafficking and localization and (iii) indirectly by a restricted accessibility of telomere chromatin to telomerase.
In animal models, telomere lengthening and increased recombination of telomeres were observed as consequences of the loss of heterochromatin-specific epigenetic modifications, including DNA methylation [(22) and reviewed in (64)]. On the other hand, the presence of critically short telomeres was correlated with hypomethylation of subtelomeric regions (20) or hypomethylation throughout the genome (69). Varied effects described in different animal model systems, and strikingly, quite opposite effects observed between plant and animal systems, are remarkable. Nevertheless, in telomere/telomerase biology and in epigenetics, considerable differences between plant and animal models do exist at many levels, and epigenetic modulation of telomere length—interlinking these two fields—thus logically reflects organism-specific features of both.
In our previous study, telomere lengths were stable, and telomerase activity was elevated in cultured tobacco cells treated with ZEB or DHPA (9), contrary to the observations in Arabidopsis where telomerase activity in vitro is not changed but telomeres are significantly shorter. These results suggest distinctions in pathways of epigenetic regulation of telomeres and telomerase, even between different model plants. The observed differences may be associated with distinct features of genomes and epigenomes of these plants, in particular a high content of ITSs, a lower global methylation and heterochromatin level (e.g. the absence of subtelomeric chromatin blocks), and a short telomere size in A. thaliana compared with N. tabacum.
Shortened telomeres are maintained over plant generations
Analysis of telomere lengths in wt plants segregated from the met1-3 mutant, and in progenies of plant germinated in the presence of 250 µM ZEB reveals that shorter telomeres are maintained through meiosis. Similar results were obtained using mouse embryonic stem cells impaired in DNA methyltransferases function, but with elongated telomeres (22); a partial increase in subtelomeric DNA methylation was observed after re-introduction of the methyltransferase gene, but the telomeres remained long. These data indicate that epigenetic stress—loss of DNA methylation—is necessary for implementation of telomere length change, but not for its maintenance. Moreover, telomere shortening in hypomethylated A. thaliana is not too dramatic or critical, and very probably does not lead to genomic or chromosomal instabilities (70). Thus telomerase simply maintains the same telomere length as present in parental cells (Figure 7). This scenario is supported by the considerable variability in telomere length, not only between different plant species [i.e. A. thaliana ecotype Columbia 3–5 kb, and N. tabacum 20–150 kb (71)] but also between different ecotypes of A. thaliana; striking variations in telomere length between individuals of the Wassilewkija ecotype have been documented. Crossings between Wassilewskija plants with short and long telomeres showed that telomere length in the progeny was determined by the parental telomere length (72).
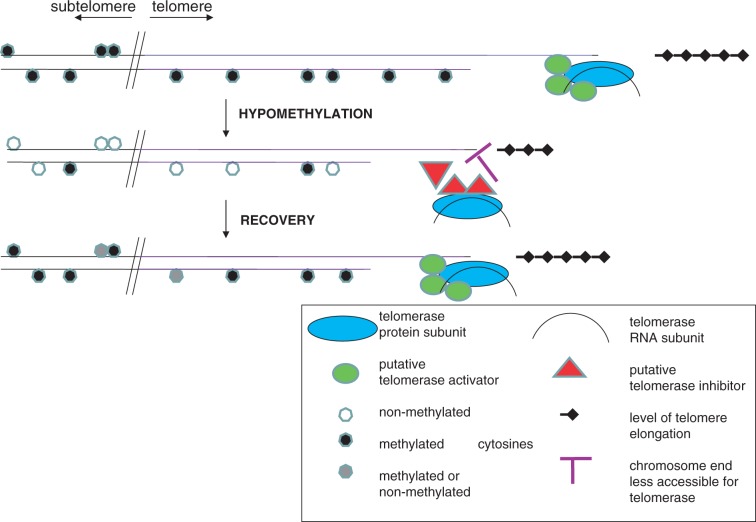
Figure 7.
Schematic depiction of the role of telomerase in the process of telomere shortening and in the subsequent propagation of telomeres. Under normal circumstances (top), telomerase is active in telomerase-positive tissues and maintains the length of telomeres. Complex changes in hypomethylated genomes (middle) lead to compromised telomere maintenance (manifested as telomere shortening), which may be caused either by factors modulating telomerase action in vivo (e.g. the restricted accessibility of chromosome ends or the absence/presence of not yet-identified telomerase activators/inhibitors) or by telomerase-independent process(es). In wt segregated from the met1-3 mutant and in the progenies of plants germinated in the presence of ZEB (bottom), telomeres remain shortened and are maintained at the stable length sufficient for normal cell functions by telomerase. The interface between telomeric and subtelomeric regions is marked by an oblique double line.
Our data demonstrate that similarly to the other repetitive sequences in the plant genome, telomeres are under epigenetic control. Continuation of these studies using different model plants with distinctive telomeric distributions (i.e. A. thaliana with a significant proportion of telomeric repeats in interstitial telomeric sequences versus N. tabacum with a predominant fraction of telomeric repeats located in telomeres) may reveal new knowledge about specific and general features of plant telomeric chromatin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [P501/11/0596], the project ‘CEITEC—Central European Institute of Technology’ [CZ.1.05/1.1.00/02.0068] from the European Regional Development Fund, and the research project RVO61388963 (to Z.J.). Funding for open access charge: the project ‘CEITEC—Central European Institute of Technology’ [CZ.1.05/1.1.00/02.0068].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Technical assistance of Mrs Jana Kapustova is highly appreciated.
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 3.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(Spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X. The epigenetic landscape of plants. Science. 2008;320:489–492. doi: 10.1126/science.1153996. [DOI] [PubMed] [Google Scholar]
- 7.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majerova E, Fojtova M, Mozgova I, Bittova M, Fajkus J. Hypomethylating drugs efficiently decrease cytosine methylation in telomeric DNA and activate telomerase without affecting telomere lengths in tobacco cells. Plant Mol. Biol. 2011;77:371–380. doi: 10.1007/s11103-011-9816-7. [DOI] [PubMed] [Google Scholar]
- 10.Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl Acad. Sci. U.S.A. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao F, Li X, Hiew S, Brady H, Liu Y, Dou Y. Dicer independent small RNAs associate with telomeric heterochromatin. RNA. 2009;15:1274–1281. doi: 10.1261/rna.1423309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brock GJ, Charlton J, Bird A. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999;240:269–277. doi: 10.1016/s0378-1119(99)00442-4. [DOI] [PubMed] [Google Scholar]
- 19.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol. Cell. Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 21.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 23.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J. Cell Biol. 2011;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27:6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 26.Roberts AR, Blewitt ME, Youngson NA, Whitelaw E, Chong S. Reduced dosage of the modifiers of epigenetic reprogramming Dnmt1, Dnmt3L, SmcHD1 and Foxo3a has no detectable effect on mouse telomere length in vivo. Chromosoma. 2011;120:377–385. doi: 10.1007/s00412-011-0318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kavi HH, Xie W, Fernandez HR, Birchler JA. Global analysis of siRNA-mediated transcriptional gene silencing. Bioessays. 2005;27:1209–1212. doi: 10.1002/bies.20328. [DOI] [PubMed] [Google Scholar]
- 28.Vaquero-Sedas MI, Gamez-Arjona FM, Vega-Palas MA. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2010;39:2007–2017. doi: 10.1093/nar/gkq1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 30.Farnung BO, Brun CM, Arora R, Lorenzi LE, Azzalin CM. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS One. 2012;7:e35714. doi: 10.1371/journal.pone.0035714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 32.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeiffer V, Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet. 2012;8:e1002747. doi: 10.1371/journal.pgen.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 35.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 37.Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 38.Kim CH, Marquez VE, Mao DT, Haines DR, McCormack JJ. Synthesis of pyrimidin-2-one nucleosides as acid-stable inhibitors of cytidine deaminase. J. Med. Chem. 1986;29:1374–1380. doi: 10.1021/jm00158a009. [DOI] [PubMed] [Google Scholar]
- 39.Yoo CB, Cheng JC, Jones PA. Zebularine: a new drug for epigenetic therapy. Biochem. Soc. Trans. 2004;32:910–912. doi: 10.1042/BST0320910. [DOI] [PubMed] [Google Scholar]
- 40.Cihak A. Biological effects of 5-azacytidine in eukaryotes. Oncology. 1974;30:405–422. doi: 10.1159/000224981. [DOI] [PubMed] [Google Scholar]
- 41.Baubec T, Pecinka A, Rozhon W, Mittelsten Scheid O. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 2009;57:542–554. doi: 10.1111/j.1365-313X.2008.03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holy A. Aliphatic analogues of nucleosides, nucleotides and oligonucleotides. Collect. Czech. Chem. C. 1975;40:187–214. [Google Scholar]
- 43.Kovarik A, Van Houdt H, Holy A, Depicker A. Drug-induced hypomethylation of a posttranscriptionally silenced transgene locus of tobacco leads to partial release of silencing. FEBS Lett. 2000;467:47–51. doi: 10.1016/s0014-5793(00)01077-2. [DOI] [PubMed] [Google Scholar]
- 44.Koukalova B, Votruba I, Fojtova M, Holy A, Kovarik A. Hypomethylation of CNG targets induced with dihydroxypropyladenine is rapidly reversed in the course of mitotic cell division in tobacco. Theor. Appl. Genet. 2002;105:796–801. doi: 10.1007/s00122-002-0965-6. [DOI] [PubMed] [Google Scholar]
- 45.Fulnecek J, Matyasek R, Votruba I, Holy A, Krizova K, Kovarik A. Inhibition of SAH-hydrolase activity during seed germination leads to deregulation of flowering genes and altered flower morphology in tobacco. Mol. Genet. Genomics. 2011;285:225–236. doi: 10.1007/s00438-011-0601-8. [DOI] [PubMed] [Google Scholar]
- 46.Fojtova M, Kovarik A, Votruba I, Holy A. Evaluation of the impact of S-adenosylhomocysteine metabolic pools on cytosine methylation of the tobacco genome. Eur. J. Biochem. 1998;252:347–352. doi: 10.1046/j.1432-1327.1998.2520347.x. [DOI] [PubMed] [Google Scholar]
- 47.Alonso JM, Stepanova AN. T-DNA mutagenesis in Arabidopsis. Methods Mol. Biol. 2003;236:177–188. doi: 10.1385/1-59259-413-1:177. [DOI] [PubMed] [Google Scholar]
- 48.Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- 49.Jaske K, Mokros P, Mozgova I, Fojtova M, Fajkus J. A telomerase-independent component of telomere loss in chromatin assembly factor 1 mutants of Arabidopsis thaliana. Chromosoma. 2013;122:285–293. doi: 10.1007/s00412-013-0400-6. [DOI] [PubMed] [Google Scholar]
- 50.Ruckova E, Friml J, Prochazkova Schrumpfova P, Fajkus J. Role of alternative telomere lengthening unmasked in telomerase knock-out mutant plants. Plant Mol. Biol. 2008;66:637–646. doi: 10.1007/s11103-008-9295-7. [DOI] [PubMed] [Google Scholar]
- 51.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- 53.Fojtova M, Fulneckova J, Fajkus J, Kovarik A. Recovery of tobacco cells from cadmium stress is accompanied by DNA repair and increased telomerase activity. J. Exp. Bot. 2002;53:2151–2158. doi: 10.1093/jxb/erf080. [DOI] [PubMed] [Google Scholar]
- 54.Sykorova E, Fajkus J, Meznikova M, Lim KY, Neplechova K, Blattner FR, Chase MW, Leitch AR. Minisatellite telomeres occur in the family Alliaceae but are lost in Allium. Am. J. Bot. 2006;93:814–823. doi: 10.3732/ajb.93.6.814. [DOI] [PubMed] [Google Scholar]
- 55.Uchida W, Matsunaga S, Sugiyama R, Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002;77:63–67. doi: 10.1266/ggs.77.63. [DOI] [PubMed] [Google Scholar]
- 56.Gamez-Arjona FM, Lopez-Lopez C, Vaquero-Sedas MI, Vega-Palas MA. On the organization of the nucleosomes associated with telomeric sequences. Biochim. Biophys. Acta. 2010;1803:1058–1061. doi: 10.1016/j.bbamcr.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 57.Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. U.S.A. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proc. Natl Acad. Sci. U.S.A. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl Acad. Sci. U.S.A. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuchar M, Fajkus J. Interactions of putative telomere-binding proteins in Arabidopsis thaliana: identification of functional TRF2 homolog in plants. FEBS Lett. 2004;578:311–315. doi: 10.1016/j.febslet.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 62.Shakirov EV, Surovtseva YV, Osbun N, Shippen DE. The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 2005;25:7725–7733. doi: 10.1128/MCB.25.17.7725-7733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mozgova I, Schrumpfova PP, Hofr C, Fajkus J. Functional characterization of domains in AtTRB1, a putative telomere-binding protein in Arabidopsis thaliana. Phytochemistry. 2008;69:1814–1819. doi: 10.1016/j.phytochem.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 65.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vyskot B, Koukalova B, Kovarik A, Sachambula L, Reynolds D, Bezdek M. Meiotic transmission of a hypomethylated repetitive DNA family in tobacco. Theor. Appl. Genet. 1995;91:659–664. doi: 10.1007/BF00223294. [DOI] [PubMed] [Google Scholar]
- 67.Majerova E, Fojtova M, Mandakova T, Fajkus J. Methylation of plant telomeric DNA: what do the results say? (Vaquero-Sedas, M.I. and Vega-Palas, M.A.: DNA methylation at tobacco telomeric sequences) Plant Mol. Biol. 2011;77:533–536. doi: 10.1007/s11103-011-9833-6. [DOI] [PubMed] [Google Scholar]
- 68.Ogrocka A, Sykorova E, Fajkus J, Fojtova M. Developmental silencing of the AtTERT gene is associated with increased H3K27me3 loading and maintenance of its euchromatic environment. J. Exp. Bot. 2012;63:4233–4241. doi: 10.1093/jxb/ers107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pucci F, Gardano L, Harrington L. Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell. 2013;12:479–486. doi: 10.1016/j.stem.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: plant responses to telomere dysfunction. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- 71.Fajkus J, Kovarik A, Kralovics R, Bezdek M. Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 1995;247:633–638. doi: 10.1007/BF00290355. [DOI] [PubMed] [Google Scholar]
- 72.Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.