Abstract
Replication Protein A (RPA) is a heterotrimeric protein complex that binds single-stranded DNA. In plants, multiple genes encode the three RPA subunits (RPA1, RPA2 and RPA3), including five RPA1-like genes in Arabidopsis. Phylogenetic analysis suggests two distinct groups composed of RPA1A, RPA1C, RPA1E (ACE group) and RPA1B, RPA1D (BD group). ACE-group members are transcriptionally induced by ionizing radiation, while BD-group members show higher basal transcription and are not induced by ionizing radiation. Analysis of rpa1 T-DNA insertion mutants demonstrates that although each mutant line is likely null, all mutant lines are viable and display normal vegetative growth. The rpa1c and rpa1e single mutants however display hypersensitivity to ionizing radiation, and combination of rpa1c and rpa1e results in additive hypersensitivity to a variety of DNA damaging agents. Combination of the partially sterile rpa1a with rpa1c results in complete sterility, incomplete synapsis and meiotic chromosome fragmentation, suggesting an early role for RPA1C in promoting homologous recombination. Combination of either rpa1c and/or rpa1e with atr revealed additive hypersensitivity phenotypes consistent with each functioning in unique repair pathways. In contrast, rpa1b rpa1d double mutant plants display slow growth and developmental defects under non-damaging conditions. We show these defects in the rpa1b rpa1d mutant are likely the result of defective DNA replication leading to reduction in cell division.
INTRODUCTION
Replication Protein A (RPA) is a eukaryotic, single-stranded DNA (ssDNA)-binding protein composed of three associated subunits, RPA1 (∼70 kDa), RPA2 (∼32 kDa) and RPA3 (∼14 kDa). The primary biochemical function of the heterotrimeric RPA complex (referred to hereafter as RPA) is to protect and preserve ssDNA from nucleolytic degradation and hairpin formation, similar to SSB (single-stranded binding protein) in prokaryotes (1,2). Consistent with this function, RPA plays essential roles in almost all DNA metabolic pathways including S-phase genome replication, DNA recombination and DNA excision repair.
Importantly, RPA plays a key role in the activation and maintenance of cellular responses to DNA damage. Downstream cellular responses to detected DNA damage include regulation of cell-cycle transitions (checkpoints), induction of DNA repair, changes in gene transcription and in some cases apoptosis (programmed cell death). These responses are ultimately mediated through the two closely related protein kinases, ATM (Ataxia Telangiectasia mutated) and ATR (ATM and Rad3-related) (3–5). While ATM is activated primarily by double-strand breaks, ATR is activated by a wide variety of lesions that result in stalled replication forks, such as DNA breaks, UV photoproducts and DNA crosslinks. These stalled replication forks, as well as DNA excision activities involved in repairing the lesions, induce functional uncoupling (physical disassociation) of helicase and polymerase activities resulting in the persistence of abnormally long stretches of ssDNA (6–8). Studies in yeasts and animal cells suggest that RPA-coating of these ssDNA stretches act as a molecular signal to activate ATR-dependent downstream phosphorylation, primarily through an associated protein called ATRIP (9–11). An ATRIP ortholog has recently been described in plants, and mutants in ATRIP display a nearly identical phenotype to atr mutants when challenged with replication blocking agents (12,13). This suggests plants encode a similar system of RPA-dependent activation of ATR in the DNA-damage response.
Interestingly, RPA itself is a target of phosphorylation by ATM, ATR, and the related kinase DNA-PK (found only in animals) in response to DNA damage. During the unperturbed cell cycle, RPA activity is regulated through cyclin-dependent kinase phosphorylation of the RPA2 subunit during DNA replication and mitosis, and dephosphorylation as cells progress into G1 (14,15). Cyclin-dependent kinase phosphorylation can ‘prime’ RPA2 for additional phosphorylation by ATR, ATM and DNA-PK in response to DNA damage (1). These phosphorylation changes to RPA2 can have effects on RPA activity during DNA repair and replication (16–21). For example, RPA hyper-phosphorylation mimetic mutants, engineered with multiple negative amino acids at known phosphorylation sites within RPA32 are unable to interact with replication centers (20). Thus, models have been proposed whereby phosphorylation of RPA2 by ATR/ATM/DNA-PK acts as a switch to modulate active DNA replication if DNA damage persists within the cell.
Although plant and animal DNA metabolism and DNA-repair responses are highly conserved in most aspects, RPA regulation in plants appears surprisingly different. In contrast to the single RPA1, RPA2 and RPA3 subunits found in yeasts and mammals [excluding humans where there are two RPA2-like genes (22)], plants encode multiple RPA1, RPA2 and RPA3 subunits. Rice contains three RPA1 paralogs, three RPA2 paralogs and one RPA3 homolog (23,24). Interaction studies suggest the subunits form at least three heterotrimeric complexes (25). Although studies in pea suggest that an RPA32 subunit is phosphorylated at certain developmental stages (26), in vitro studies in rice indicate that rice RPA2-1 (RPA32-1) is not hyper-phosphorylated in response to DNA damage, and protein levels of RPA2-1 decrease following DNA damage (27)
In Arabidopsis, we find (see below) five paralogs of RPA1, two of RPA2 and two of RPA3, consistent with earlier genomic analyses (24,25,28). A previous study of the T-DNA insertion mutants rpa1a and rpa1b suggested that rpa1a is lethal while rpa1b displays hypersensitivity to DNA-damaging agents (25). However, a recent study of a viable (T-DNA insertion) rpa1a mutant suggests that RPA1A is required for class I crossovers in meiosis, but does not appear to play a significant role in (SPO11-dependent) meiotic double-strand break repair (28). In addition, an Arabidopsis mutant in the RPA2A subunit (termed ROR1 for Repressor of Silencing 1), suggests a role for RPA2A in transcriptional gene silencing and meristem maintenance (29).
Employing T-DNA insertion mutants of all five of the Arabidopsis RPA1 subunit genes, we present here evidence that the RPA1 gene family has diverged into two functionally distinct groups. One group appears responsible for promoting genomic replication, and another group appears responsible for promoting DNA repair and recombination. Furthermore, we show that within the repair/recombination group individual RPA1 subunits display unique functions in response to DNA damage. Based on these results, we hypothesize that the functional differences found within the RPA1 gene family in Arabidopsis represents a unique mechanism of RPA regulation common to plants.
MATERIALS AND METHODS
Plant materials and growth
All Salk T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center (ABRC). The Salk ID for each mutant is as follows: rpa1a, Salk_017580 (25,28); rpa1b, Salk_088429 (25); rpa1c, Salk_085556; rpa1d, Salk_140762; rpa1e, Salk_120368; atr-2, Salk_032841 (30); atm-2, Salk_006953 (31). Lines homozygous for the T-DNA insert were isolated by PCR using gene-specific and T-DNA-specific primers (http://signal.salk.edu/tdnaprimers.2.html). The sequences of the gene-specific primers are as follows: Salk_017580 (F), 5′-CTTAGTTTTCTAGTGATCTCTG-3′, Salk_017580 (R), 5′-GAATCTCCCCTCCATCATAGTC-3′; Salk_088429 (F), 5′-GTACATACGTGAATCA-3′, Salk_088429 (R), 5′-AAGTGTTTTGAAGTAC-3′; Salk_085556 (F), 5′-GAGAACAACAGCACCACTGATGTA-3′, Salk_085556 (R), 5′-GTGTCTCTAGTTCCTGAGGTTCCA-3′; and Salk_140762 (F), 5′-TCTCACGGCTTTTAGTTTTCAC-3′, Salk_140762 (R), 5′-AGATCTCTTCTATCATAGAGTC-3′. Salk_120368 (F), 5′-TTGGTATTGTGTCATCTATCA-3′. Salk_120368 (R), 5′-CAACCTTACGGATGATATCTTC-3′. Salk_006953 (F), 5′-GGTTGGGCAGTTCCAAAGATGA-3′, Salk_006953 (R), 5′-TCTCTCCTTGTTTCAAGCTCTG-3′. Salk_032841 (F), 5′-CAAGGGTTCCGATGTTCAAAGTG-3′. Salk_032841 (R), 5′-CAATCAGCAGGAAAAGACAATCA-3′. All characterized lines segregated as a single Mendelian locus, and are recessive. PCR-genotyping information for rpa1c-2 (SALK_139567), rpa1d-2 (SALK_149669) and rpa1e-2 (SALK_077939) are available upon request.
The wild-type control Arabidopsis (Arabidopsis thaliana) accession was Columbia (Col-0). For plate experiments, seeds were surface sterilized by soaking in a solution of 10% bleach for five minutes and then rinsing three times with double-distilled sterile water. Seeds were sown on nutrient phytoagar plates containing 1X MS salts (PlantMedia, Dublin, Ohio, USA) pH 5.7, 0.05 g/L MES and 1.0% (w/v) phytoagar (PlantMedia, Dublin, Ohio, USA). Seeds were stratified at 4°C for 2 days in the dark before being placed vertically in a growth chamber under cool-white lamps filtered through Mylar (Golden State Plastics, Sacramento, CA, USA) at an intensity of 100–150 μmol/m2/sec at 22°C, and a photoperiod of 16 h light/8 h dark. For soil experiments, seeds were stratified, sown and germinated on 1X MS phytoagar plates in the same condition as described above and on the fifth day transferred to soil growing medium (SUNGRO Horticulture, Seba Beach, Canada) in pots. Plants were irrigated once in 3 days with a solution of water and Miracle-Gro® plant fertilizer, 0.45 g/Liter (Scotts Miracle-Gro products inc., Marysville, OH, USA).
For DNA damage sensitivity assays, 40–50 surface-sterilized wild-type and mutant seeds were sown on plates containing 1X MS phytoagar media with or without DNA-damaging agents: hydroxyurea (HU), camptothecin (CPT), mitomycin-C (MMC) (all from SIGMA, St. Louis, MO, USA) and aphidicolin (APH) [USBiological, Swampscott, MA, USA or A.G. Scientific Inc., San Diego, CA, USA]. For gamma-radiation assays, Arabidopsis seeds and plants were irradiated using a Cs137 source (Massachusetts Institute of Technology, Cambridge, MA, USA), dose rate ∼60 radiations/minute. Sterilized seeds were imbibed in water at 4°C for 2 days, irradiated and then immediately placed on 1X MS phytoagar plates for germination in the growth chamber. For irradiation of seedlings, plate grown 5-day-old seedlings were irradiated and then immediately returned to the growth chamber. For UV-B treatment 1X MS phytoagar plate grown 5-day-old seedlings were irradiated with UV lamps (Spectronics, Estbury, NY, USA) filtered through cellulose acetate (SABIC Polymershapes, Devens, MA, USA) to eliminate UV-C for different time periods and then returned to the growth chamber. Three replicate plates were used per treatment, and for root-length measurement plates were first photographed (9–11 days after germination) with a digital camera (Kodak, China) and measured using ImageJ software (32).
For relative root-growth comparisons of the rpa1b rpa1d double mutant, plants were prepared and grown as described above for 4 days and transferred to new 1X MS phytoagar plates with or without 22.5 nM CPT or 2 μg/ml MMC. For UV-B treatment, 4-day-old plants were transferred to new 1X MS phytoagar plates and treated with 0.5 J/sec/m2 UV-B for 24 h or left untreated. To calculate the relative percent growth reduction of the treated plants compared to the untreated plants we used the formula:
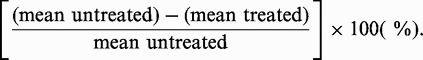 |
Histochemical staining
PcyclinB1;1:GUS promoter/reporter staining was performed as described previously (33,34) with minor modifications. The GUS reporter gene produces β-glucuronidase enzyme and enzymatically converts 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) into a visible blue stain in plant tissues. Seedlings were placed in 50 mm NaPO4, pH 7.2, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6 and 2 mm X-Gluc (Gold Biotechnology, St. Louis, MO, USA) and then incubated at 37°C overnight. Tissues were then washed in 70% EtOH for 1 h and then directly imaged under microscope.
Microscopy
GUS stained root tips were imaged with a digital camera mounted inverted light microscope (Olympus CKX41). For Propidium Iodide (PI) staining, wild-type and mutant plants were grown on 1X MS phytoagar plates for 7 days and then stained with 5 μg/ml PI (Calbiochem, La Jolla, CA, USA) for 2 min. Seedling were then mounted with water on a slide and imaged with a Zeiss LSM 510 confocal laser-scanning microscope using the HeNe 543 nm excitation and 475–560 nm emission lines. For DNA-replication assays, wild-type and mutant plants were grown on 1X MS phytoagar plate for 5 days and then incubated in 10 µM 5-Ethynyl-2′-deoxyuridine (EdU), (Invitrogen, Carlsbad, CA, USA) in half-strength MS liquid medium for 30 min. Seedlings were then fixed for 30 min in 4% (w/v) formaldehyde solution in phosphate buffered saline (PBS) with 0.1% Triton X-100. Following 3 × 10 min PBS washes, seedlings were directly incubated for 30 min at room temperature (RT) in EdU detection cocktail (Invitrogen, Click-iT EdU Alexa Fluor 488 HCS assay) followed by a 10-min rinse. Seedlings were then mounted with Vectashield (Vector Laboratories, Burlingame, CA, USA) on a slide and imaged with a Zeiss LSM 510 confocal laser-scanning microscope using the Argon laser 488-nm excitation and 478–553 nm emission lines. To visualize epidermal-cell outlines, the middle region of the third true leaves were dissected from 2-week-old seedlings and mounted with 100% ethanol. The abaxial sides of the leaves were viewed with the Zeiss LSM 510 microscope using transmitted light. Meiotic chromosome spreads were prepared as described (35). Slides were mounted with DAPI (2.5 μg/ml) in Vectashield. Chromosomes were visualized using a fluorescence microscope (OLYMPUS BH-2). Images were then captured using Qimaging (Micropublisher 3.3) camera and processed with Qcapture software.
Phylogenetic analysis
Phylogenetic analysis of the RPA1 protein sequences was performed with the MEGA 4 software package (36). Amino acid sequences were aligned using ClustalW (default parameters) within the MEGA software package, and adjusted manually. The resulting neighbor-joining distance tree was further evaluated employing 1000 bootstrap replicates.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RPA1A (AT2g06510), RPA1B (AT5g08020), RPA1C (AT5g45400), RPA1D (AT5g61000), RPA1E (AT4g19130), ATR (AT5G40820) and ATM (AT3G48190).
RESULTS
The RPA1 gene family is composed of two distinct groups
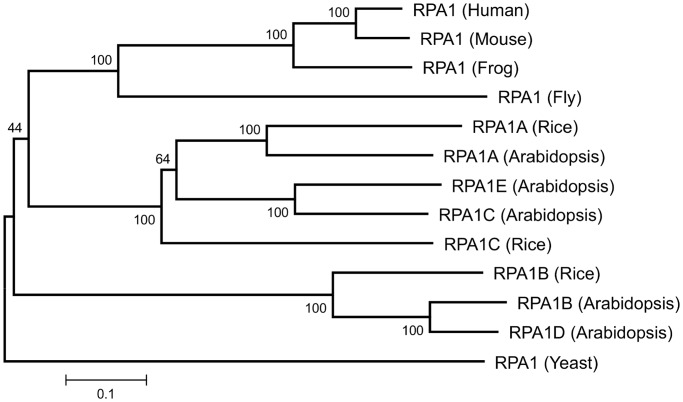
In light of the multitude of RPA functions in yeasts and animals, we wanted to investigate similar functional roles for RPA in Arabidopsis. Searching the Arabidopsis thaliana genome (TAIR10), we identified two putative 14kD small-subunit genes (RPA3A, At3g52630; RPA3B, At4g18590), two 32-kD middle-subunit genes (RPA2A/RPA2-1/ROR1, At2g24490; RPA2B, At3g02920) and five 70-kD large-subunit genes (RPA1A, At2g06510; RPA1B, At5g08020; RPA1C, At5g45400; RPA1D, At5g61000; RPA1E, At4g19130), consistent with previous studies in Arabidopsis (24,25,29). Since the large (70 kD) subunit family contained the most gene members and each appeared significantly distinct based on their amino acid sequences (all members display >19% non-identity), we hypothesized that individual members of this gene family likely represents more specialized functional subunits. Phylogenetic analysis of the large subunit protein sequences (Figure 1) shows two evolutionary distinct groups among plants, one group containing the Arabidopsis RPA1A, RPA1C and RPA1E (ACE group) and another group containing RPA1B and RPA1D (BD group). The branching pattern of the ACE group within the tree suggests that plant RPA1A is distinct from the RPA1C subgroup (that includes Arabidopsis RPA1E), the latter of which likely became more specialized sometime during the divergence of monocots and dicots. The BD group displays a similar pattern with Arabidopsis RPA1B and RPA1D diverging later in plant evolution from a common ancestor that also gave rise to rice RPA1B.
Figure 1.
Neighbor-joining distance tree of RPA1-like protein sequences. All known sequences for rice and Arabidopsis are shown. Numbers above each branch indicate bootstrap values (percentage) of 1000 replicates. Scale bar represents the expected number of amino acid substitutions per site.
Comparing differences in transcriptional regulation among the RPA1 family, we find that the Arabidopsis thaliana BD group has a higher basal level of transcription in young seedlings (Supplementary Figure S1) versus the ACE group. Interestingly, all members of the ACE group in Arabidopsis thaliana display strong transcriptional up-regulation in response to ionizing radiation in young seedlings, and this regulation is dependent upon functional ATM (33). In contrast, neither RPA1B nor RPA1D display significant changes in this analysis. These data along with the phylogenetic analysis described above suggest distinct functional divergence between these two groups.
Arabidopsis RPA1C and RPA1E have unique roles in response to DNA damage
To genetically define individual roles of the five RPA1 family members in DNA metabolism, we first identified homozygous T-DNA insertion lines of each respective subunit gene. Two of the homozygous lines used in this study, rpa1a (SALK_017580) and rpa1b (SALK_088429) have previously been characterized (25,28,37). For RPA1C, RPA1D and RPA1E we identified two T-DNA alleles for each gene (rpa1c, SALK_085556 and SALK_139567; rpa1d, SALK_140762 and SALK_149669; rpa1e, SALK_120368 and SALK_077939). Initial characterization of these alleles proved no differences during normal growth conditions or in response to DNA damaging agents (e.g. ionizing radiation, CPT and replication blocking agents). We chose individual lines SALK_085556, SALK_140762 and SALK_120368 for subsequent characterization and mutant combination construction since in each case the location of the T-DNA is within an exon (Supplementary Figure S2), designated herafter as rpa1c, rpa1d and rpa1e, respectively. Reverse transcription (RT)-PCR of these lines (rpa1c, rpa1d, rpa1e) revealed undetectable transcript downstream of their respective T-DNA insertion sites (Supplementary Figure S2), and in the case of rpa1e, there is also no detectible transcript upstream of the T-DNA. RT-PCR analysis of the rpa1a and rpa1b lines (SALK_017580 and SALK_088429) revealed results similar to rpa1c and rpa1d (upstream detected, downstream undetected), similar to previous studies (25,28). In addition, protein expression of RPA1B was absent in the rpa1b mutant employing a rice RPA1b antibody (25). We therefore suggest that these most likely represent null mutant lines, although we cannot unequivocally rule out the possibility of some functioning partial transcripts. Interestingly, all of the single mutant lines were viable and did not display obvious developmental deficiencies or alterations under standard growth conditions. One exception, however, is reduced fertility (seed set per silique) in the rpa1a mutant, as previously described (28).
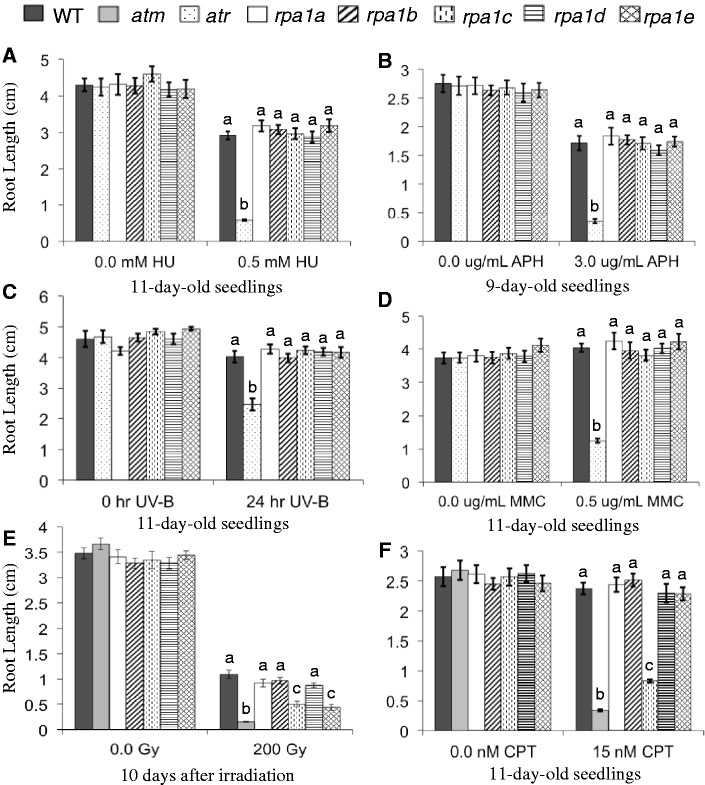
To test the role(s) of RPA1 family members in response to DNA damage resulting from agents that block replication, we employed a root-growth assay to measure hypersensitivity to various replication-blocking agents. In this assay, we simply measure primary root growth following germination on, or transfer to media containing the particular agent (an example experiment is shown in Supplementary Figure S3 and this particular experiment is further described below and in the next section). As shown in Figure 2A and B, none of the single rpa1 mutants (rap1a, rpa1b, rpa1c, rpa1d and rpa1e) displayed hypersensitivity (reduction in root growth compared to WT) to the replication-blocking agents HU or APH. HU blocks replication progression by inhibiting ribonucleotide reductase (RNR) leading to reduced dNTP pools, while APH directly inhibits replicative DNA polymerases (e.g. pol delta and epsilon). Since both of these agents do not directly damage DNA, we also tested MMC (an agent that produces inter-strand DNA crosslinks) and UV-B light (produces intra-strand DNA crosslinks between adjacent base pairs, such as cyclobutane pyrimidine dimers), which damage DNA directly and produce replication-blocking lesions. Similarly, none of the five single mutants displayed significant hypersensitivity to UV-B light or MMC (Figure 2C and D).
Figure 2.
Hypersensitivity analysis of rpa1 single mutants. Root-length measurements of (A) plants grown in the absence or presence of 0.5 mM hydroxyurea (HU); (B) plants grown in the absence or presence of 3.0 μg/mL APH (USBiological, Swampscott, MA, USA); (C) plants grown for 5 days, treated with 0.4 J/s/m2 UV-B for 24 h or left untreated and grown for an additional 5 days; (D) plants grown in the absence or presence of 0.5 μg/mL MMC; (E) seeds gamma-irradiated (0 or 200 Gy) and grown for 10 days; (F) plants grown in the absence or presence of 15 nM CPT. Data are mean ± SE (n > 30). To analyze statistical difference with in each treatment group F-test (ANOVA) and LSD were carried out at P ≤ 0.05. Bars with different letters indicate significant differences.
However, in response to ionizing radiation (γ-radiation), both the rpa1c and rpa1e single mutants display a hypersensitivity response. As shown in Figure 2E, rpa1c and rpa1e root growth is reduced ∼60% in comparison to the WT control group at 10 days post irradiation of seeds, while rpa1a, rpa1b and rpa1d show no significant difference in comparison to WT. For comparison, we included in this analysis the extremely hypersensitive mutant atm (33,38).
Since γ-radiation induces a variety of DNA-damage lesions that primarily include double-strand breaks, but also DNA base damage, we further tested the double-strand break inducing agent CPT. CPT blocks the re-ligation step of DNA Topoisomerase I, and ultimately produces a double-strand break in the presence of DNA replication. As is shown in Figure 2F and Supplementary Figure S3A, both the atm mutant and rpa1c mutant display strong hypersensitivity to CPT. Unlike γ-radiation however, the rpa1e mutant did not display significant hypersensitivity. Taken together, these results suggest that RPA1C plays a leading role (among other RPA1 family members) in the repair of double-strand breaks, and that RPA1E may play a role in repair of auxiliary damage specific to ionizing radiation, such as base damage or recruitment of DNA-end-processing complexes, for example. Because CPT specifically produces DSBs in the presence of ongoing DNA replication, it is possible that RPA1E is specific for DSB repair in cell-cycle stages outside of S-phase (e.g. G1 phase).
RPA1C, RPA1E and ATR act in parallel in response to double-strand breaks
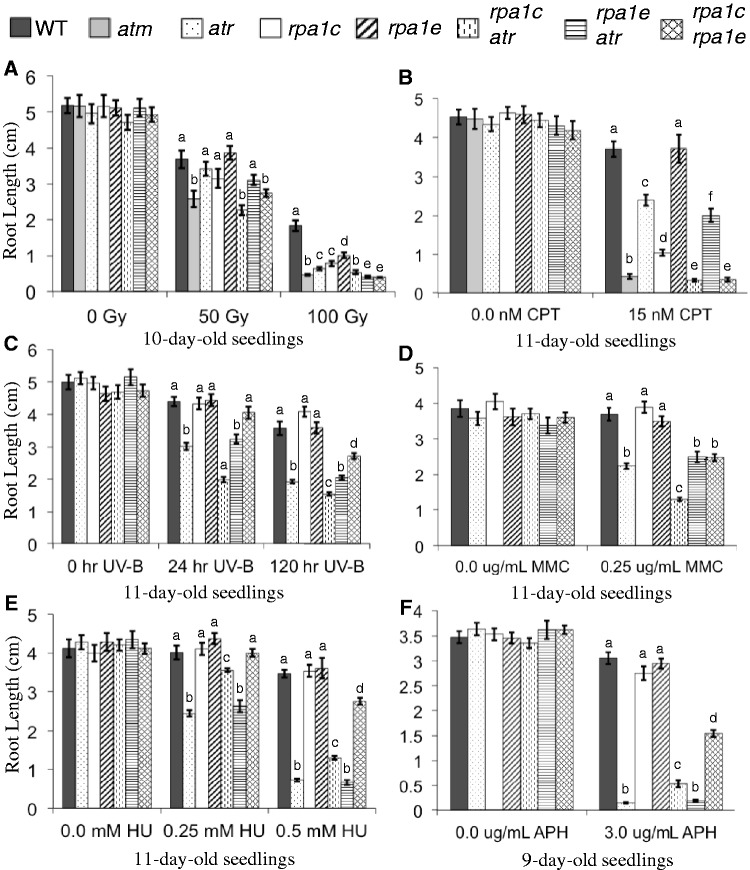
Since the rpa1c and rpa1e single mutants display hypersensitivity to DNA damage, we first wanted to test the genetic interactions of RPA1C and RPA1E with ATR, a central player in the DNA-damage response and that is (at least in part) activated by RPA in animal models. To this end, we constructed rpa1c atr and rpa1e atr double mutant lines. Under standard growth conditions, neither of these double mutants displayed obvious abnormal development of the shoot (including flower development and fertility) or root length (Figure 3, untreated all panels). However, in response to γ-radiation or CPT, the rpa1c atr double mutant displays an additive hypersensitivity phenotype (reduction of root growth) over either of the single mutants (Figure 3A and B and Supplementary Figure S3B). This suggests that RPA1C and ATR act in parallel during the response to double-strand breaks, since a consistent trend is seen in response to both CPT and ionizing radiation.
Figure 3.
Hypersensitivity analysis of rpa1c, rpa1e and atr mutant combinations. Root-length measurements of (A) 5-day-old seedlings were gamma-irradiated (0, 50 or 100 Gy) and grown for 5 days; (B) plants grown in the absence or presence of 15 nM CPT; (C) plants grown for 5 days, treated with 0.4 J/s/m2 UV-B for 24 h, 120 h or left untreated, and grown for an additional 5 days; (D) plants grown in the absence or presence of 0.25μg/mL MMC; (E) plants grown in the absence or presence of 0.25 mM, 0.5 mM hydroxyurea (HU); (F) plants grown in the absence or presence of 3.0 μg/mL APH (A.G. Scientific Inc., San Diego, CA, USA). Data are mean ± SE (n > 30). To analyze statistical difference with in each treatment group F-test (ANOVA) and LSD were carried out at P ≤ 0.05. Bars with different letters indicate significant differences.
The rpa1e atr double mutant however displayed hypersensitivity to CPT and γ-radiation similar to the atr single mutant (Figure 3A and B). At lower doses of CPT and γ-radiation we see comparable root lengths of both atr and rpa1e atr and these differences are not significant. Only at higher doses do we see a significant difference between the atr and rpa1e atr mutant lines, but this difference is minimal in response to CPT. Since the double mutant is no more hypersensitive than the atr mutant, this initially suggests that either RPA1E functions within an ATR-dependent pathway, or functions within an ATR-independent pathway while exhibiting genetic redundancy with RPA1C. If the latter is the case, we might expect to see a ‘supra-additive’ effect whereby elimination of both RPA1C and RPA1E creates a phenotype that is above and beyond addition of phenotypes.
To test this, we constructed an rpa1c rpa1e double mutant line and determined the hypersensitivity responses to CPT and γ-radiation. As shown in Figure 3A, the rpa1c rpa1e double mutant does in fact display a supra-additive hypersensitivity phenotype in response to CPT, but we did not observe any obvious developmental defects in the absence of these agents. In response to γ-radiation, we see a similar supra-additive pattern at the lower (50 Gy) dose, and a more additive hypersensitivity pattern at the higher dose (100 Gy), which is likely due to the fact that the roots simply do not grow less than ∼1 mm, regardless of the hypersensitivity (Figure 3B). Nonetheless, this supports a model that includes redundancy between RPA1C and RPA1E in the absence of either protein.
To determine functional redundancy between RPA1A, RPA1C and RPA1E in response to double-strand breaks, we also generated the double mutant combinations rpa1a rpa1c and rpa1a rpa1e. In response to CPT treatment, rpa1a rpa1c showed similar hypersensitivity as the rpa1c single mutant, while the rpa1a rpa1e double mutant showed no additional sensitivity over the rpa1a, rpa1e single mutants and wild type (Supplementary Figure S4). This suggests no functional redundancy between RPA1A and either RPA1C or RPA1E, and argues against a role for RPA1A in response to CPT. However, further hypersensitivity analyses of the triple mutant rpa1a rpa1c rpa1e in response to a range of damaging agents will be needed to fully characterize the genetic interactions of the ACE group.
If RPA1E were acting within an ATR-pathway, we would expect that an rpa1c rpa1e atr triple mutant would display a hypersensitivity phenotype similar to the rpa1c atr double mutant. To test this, we constructed the rpa1c rpa1e atr triple mutant and find that this combination results in severe seedling defects that is, in most cases, lethal. Although this result will require additional analysis to understand why some triple mutant seedlings survive, this nevertheless suggests the triple mutant is hypersensitive to the relatively low levels of endogenous DNA damage, and therefore suggests a supra-additive interaction. Overall, these data suggest a model in which RPA1C, RPA1E and ATR are acting largely in independent pathways in response to double-strand breaks, and that genetic redundancy plays a role in the severity of individual rpa1c or rpa1e hypersensitivity phenotypes.
RPA1C and RPA1E act in parallel in response to UV-B and MMC
We further tested the rpa1c atr, rpa1e atr and rpa1c rpa1e double mutants to agents that create replication-blocking bulky lesions, UV-B light and MMC. In response to a chronic UV-B exposure over a course of 120 h (Figure 3C), the rpa1c atr double mutant displayed less root growth than the (non-hypersensitive) rpa1c and (hypersensitive) atr single mutants resulting in a supra-additive response. In contrast the rpa1e atr double mutant did not display additional hypersensitivity over the atr single mutant. Combination of rpa1c and rpa1e however resulted in supra-additive hypersensitivity. This was surprising since neither single mutant displayed significant hypersensitivity. These general trends in response to UV-B were also observed in response to chronic exposure to MMC (Figure 3D). Overall, the additive phenotypes of rpa1c seen here with rpa1e and atr are consistent with the notion that RPA1C plays a largely unique and separate role, in this case in response to lesions that block DNA replication. One example could be that defective repair responses in atr or rpa1e leads to double-strand breaks that require RPA1C for repair.
The rpa1c mutation partially suppresses atr hypersensitivity to the replication-blocking agents HU and APH
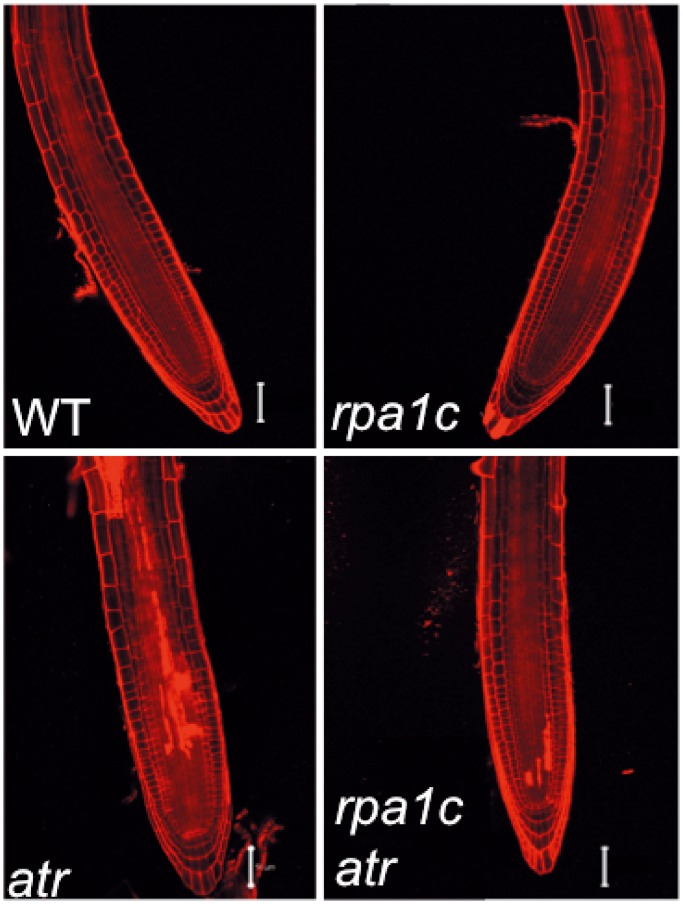
As discussed above, none of our single rpa1 mutants displayed significant hypersensitivity to HU or aphidicolin (APH), while the atr mutant displays a strong hypersensitivity response to both agents (30) (Figure 3E and F and Supplementary Figure S5). Surprisingly, combination of rpa1c with atr results in less hypersensitivity to HU and APH (increased root growth) over the single atr mutant (Figure 3E and F). This suppression of hypersensitivity was most pronounced at a lower concentration of HU (0.25 mM), but was still significant at the higher dose (0.5 mM; Figure 3E). Moreover, we see reduced cell death in response to replication blocks (HU) in the rpa1c atr double mutant versus the atr single mutant (Figure 4), employing PI viability staining. In response to APH, the root growth trend is similar, albeit less pronounced in comparison (Figure 3F and Supplementary Figure S5), but still statistically significant. In contrast, the rpa1e atr double mutant did not display a similar suppression phenotype to either HU or APH, suggesting the hypersensitivity suppression of atr is rpa1c-specific. Similar to CPT, UV-B and MMC above, the rpa1c rpa1e double mutant displays supra-additive hypersensitivity to both HU (0.5 mM) and APH in comparison to either single rpa1 mutants. Again, this suggests RPA1C and RPA1E act in unique pathways, but each may partially compensate for the absence of either protein in each respective single mutant.
Figure 4.
PI viability staining of HU-treated root tips. WT, rpa1c, atr and rpa1c atr mutant lines were grown for 4 days, treated with 0.25 mM HU for 24 h, then transferred to media without HU for 24 h. Roots were stained with 5 mg/mL PI immediately prior to laser-scanning confocal microscopy. PI-filled cells represent dead cells. Bars = 50 µm.
In light of the suppression phenotype observed in the rpa1c atr mutant in response to HU and APH, increased dNTPs might be one way to circumvent slower DNA polymerases. Since the suppression phenotype is more pronounced in response to HU, one possibility is that RPA1C negatively regulates the transcription of RNR, the rate-limiting and highly regulated enzyme in the production of dNTPs (39,40). However, no significant (and/or relevant) transcriptional differences were observed for RNR1, RNR2A, RNR2B or TSO1/RNR2C comparing WT, the rpa1c and atr single mutants, and the rpa1c atr double mutant (Supplementary Figure S6). While this suggests that RNR transcriptional regulation is not a factor in the suppression phenotype for rpa1c atr, additional analyses will need to be employed to determine if post-transcriptional effects of RNR regulation are affected by the presence of RPA1C.
Elimination of RPA1C and RPA1E is not sufficient to block activation of ATR-dependent cell-cycle arrest
RPA activates ATR-dependent responses to genomic insults through the ATR interacting protein ATRIP (scDdc2, spRad26), based on current models in animals and yeasts (41,42). In plants, an ATRIP ortholog has been identified and mutants in this gene display nearly identical hypersensitivity responses and cell-cycle defects as the atr mutant (12,13). This suggests that plants employ a similar RPA-dependent mechanism to activate downstream responses of ATR in response to DNA damage. Since RPA1C and RPA1E likely play important roles in response to DNA damage, we wanted to further test if either is involved in activating cell-cycle arrest, a key aspect of the DNA damage response. Previous studies suggest that plant ATR and ATRIP play key roles in the G2/M transition (checkpoint) in response to replication blocks and double-strand breaks (13,30,33). These studies employed a PcyclinB1;1:GUS promoter/reporter fusion construct (34) to monitor accumulation of G2 phase cells in root meristems (see Materials and methods section for more information). Briefly, the construct is transcriptionally expressed during S-phase to a maximum in G2. Inclusion of the CyclinB1;1 mitotic destruction box upstream of the GUS reading frame results in degradation of GUS reporter in M-phase.
To test whether RPA1C or RPA1E regulates the G2/M transition similar to ATR, we constructed our various single and double rpa1c, rpa1e and atr mutant combinations with the PcyclinB1;1:GUS construct. This was accomplished by crossing the original parental lines (atr, rpa1c, rpa1e) with an established individual PcyclinB1;1:GUS wild-type (Col-0) line. As shown in Supplementary Figure S7, all WT and mutant lines show a similar basal level of GUS expression (a few GUS positive cells within the meristematic region) grown under standard conditions without γ-radiation (Supplementary Figure S7A). Upon treatment with γ-radiation, we see an accumulation of GUS-positive meristematic cells, consistent with G2-phase arrest, in WT, rpa1c, rpa1e and the rpa1c rpa1e double mutant. However, this accumulation is largely absent in the atr, rpa1c atr and rpa1e atr lines and both rpa1/atr double mutants are similar to the atr single mutant. This suggests that RPA1C and RPA1E are not exclusive regulators of ATR-dependent cell-cycle arrest in response to DNA damage and genetic redundancy from other RPA1 subunits may play a role in activating cell-cycle responses to DNA damage.
RPA1C functions synergistically with RPA1A during meiosis
A recent study demonstrated that RPA1A is required for class I crossover formation, playing a role in second-end capture of homologous recombination (HR) during crossing over (28). However, these authors found no evidence of meiotic chromosomal fragmentation during metaphase I or subsequent stages in the rpa1a mutant, suggesting RPA1A is not essential for meiotic DSB repair. The authors also produced antibodies to Arabidopsis RPA1A to show meiotic localization consistent with its function at later stages of prophase I. Based on the requirement of RPA in meiotic DSB repair in yeast, the authors further argue that additional RPA1 paralogs likely play a role early on in the recombination process. In comparison to the partially fertile rpa1a mutant, we find here that the rpa1a rpa1c double mutant is infertile producing no viable seeds (Supplementary Figure S8). In addition, the infertility of the rpa1a rpa1c double mutant is unique among the other double mutant combinations constructed during the course of this study: the rpa1c rpa1e is fertile and develop normal siliques, rpa1a rpa1e is partially fertile similar to rpa1a and rpa1b rpa1d is partially fertile, but may be due to developmental or pre-meiotic replication defects (see below). This suggests a key role for RPA1C in the processing of meiotic DSBs. Since the rpa1c single mutant displays no obvious meiotic defects, this may suggest that RPA1A can fulfill the role of RPA1C in its absence.
To determine the role of RPA1C during meiosis, we prepared chromosomal spreads of pollen-mother cells from WT, rpa1a, rpa1c and the double mutant rpa1a rpa1c. As shown in Figure 5, rpa1a displays visually normal chromosome synapsis at pachytene, but displayed univalents during metaphase and resulted in missegregation of chromosomes at the first and second meiotic divisions as previously described (28). The rpa1c single mutant displayed no obvious meiotic abnormalities in all stages of meiosis I and II (not all stages are represented in Figure 5). However, in the rpa1a rpa1c double mutant we are unable to identify any fully synapsed pachytene stages (>40 zygotene stages were observed) or stages that resembled WT metaphase. Instead, we observe highly fragmented chromosomes during anaphase I followed by similarly defective meiosis II stages. By comparison, the rpa1a rpa1c fragmentation resembles chromosome fragmentation seen previously in rad51 and atr atm mutants (43). This suggests that both RPA1C, and RPA1A (perhaps only in the absence of RPA1C), play primary roles in the initiation of HR events during meiosis.
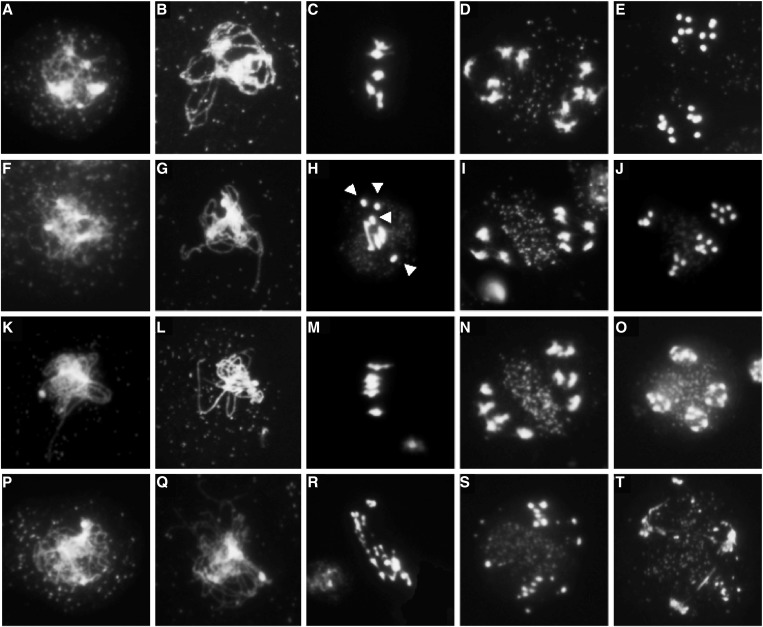
Figure 5.
Meiotic stages of pollen-mother cells from WT and rpa1 mutants. (A–E) Wild-type, (F–J) rpa1a, (K–O) rpa1c, (P–T) rpa1a rpa1c. Stages included are zygotene (A, F, K, P), pachytene (B, G, L), a zygotene stage representing the absence of a fully synapsed pachytene stage in the rpa1a rpa1c double mutant (Q), metaphase I (C, H, M, R), anaphase I (D, I, N, S) and anaphase II (E, J, O, T). In wild-type and rpa1c, homologs are segregated in equal number at anaphase I (D, N), followed by the separation and segregation of sister chromatids at anaphase II (E, O). In rpa1a, zygotene (F) and pachytene (G) stages are similar to WT and rpa1c. However, univalents (arrows) are present at the metaphase plate (H) leading to unequal segregation of homologous chromosomes at anaphase I (I), and anaphase II (J). In rpa1a rpa1c, multiple fragmented chromosomes were present at metaphase plate (R) and at anaphase I (S) leading to abnormal anaphase II (T).
RPA1B and RPA1D are required for normal DNA replication during root and shoot development
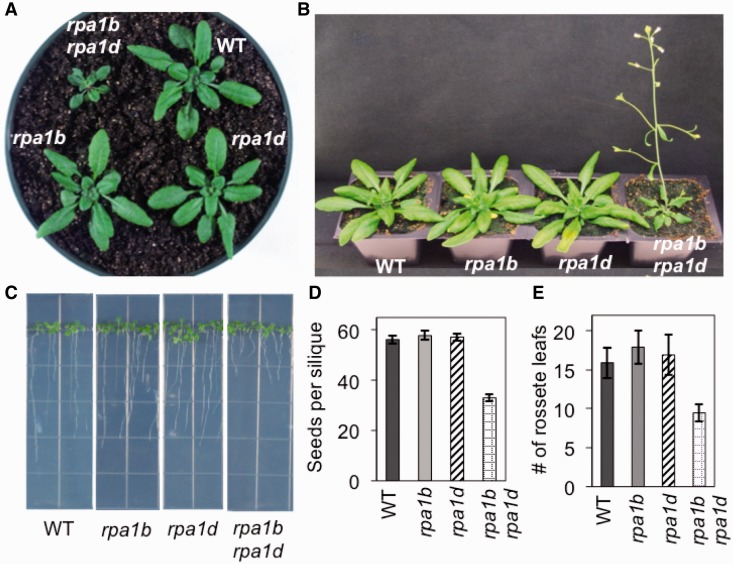
Since individual mutations in the closely related RPA1B and RPA1D genes revealed no obvious phenotypes throughout development, we suspected functional redundancy may also play a role in the BD group similar to the ACE group described above. To test this, we constructed an rpa1b rpa1d double mutant line. As is shown in Figure 6, both root and shoot growth of the double mutant is severely reduced under normal growth conditions versus WT and either single mutant line. Early development of the rosette is delayed (Figure 6A) and fewer leaves are produced (Figure 6E). Interestingly, the resulting plants flower early (Figure 6B), similar to other factors involved in chromatin remodeling [FAS1, FAS2 (44); BRU1 (45); TEB (46) for example], or DNA replication [DNA polymerase subunits (47,48); for review see (49)]. The plants also produce smaller siliques and fewer seeds (Supplementary Figure S9 and Figure 6D). Preliminary analysis of meiotic chromosomal spreads shows no obvious visual abnormalities during meiosis I and II in either rpa1b or rpa1d single mutants, nor the rpa1b rpa1d double mutant, possibly suggesting defects during pre-meiosis replication. However, a more complete analysis will be needed to determine the cause of reduced fertility in the rpa1b rpa1d double mutant. The double mutant also displays >50% reduction in root growth versus WT and the single mutants (Figure 6C).
Figure 6.
Phenotypes of the rpa1b rpa1d double mutant line. (A) Twenty-six-day-old plants grown on soil. (B) Thirty-one-day-old plants grown on soil. (C) Eleven-day-old wild-type and mutant plants grown on MS phytoagar media. (D) Measurements of seeds produced by individual siliques of wild-type and mutant plants. (E) Flowering time measurements as counted by the number of rosette leaves when the inflorescence is 1-cm long.
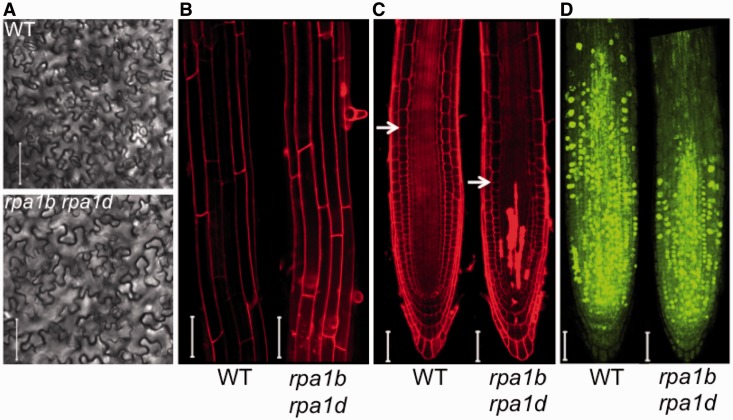
Since the most prominent defect in the double mutant is reduced growth, this would suggest developmental defects in cell elongation, cell division or both. If cell elongation were defective in the double mutant, we would expect to see differences in cell sizes of differentiated tissues. As shown in Figure 7A and B, and quantified in Supplementary Figure S10, we find no clear differences in cell size of both pavement cells of leaves, and differentiated root cells in the elongation zone, suggesting that cell elongation is normal in the rpa1b rpa1d double mutant compared to WT. In addition, the meristematic region is reduced and the double mutant displays cell death in stem cells and their descendants (Figure 7C). To determine if DNA replication is defective in the root meristem, we employed an EdU (nucleotide analog) incorporation assay to measure cellular DNA replication levels. Interestingly, as shown in Figure 7D we find that the number of actively replicating cells in root meristems is reduced ∼50% overall. EdU positive cells are located primarily within the meristematic region and unlike WT, incorporation largely does not extend into the elongation region. Overall, these data suggest that cell division is defective in the rpa1b rpa1d double mutant, and the resulting cell death accounts for its reduced growth.
Figure 7.
Leaf and root cell phenotypes of the rpa1b rpa1d double mutant. (A) Outlines of leaf epidermal cells in wild-type (upper) and mutant (lower) plants. Calculated average cell areas for WT and rpa1b rpa1d were (2137 ± 1403) mm2 and (2055 ± 1300) mm2, respectively. (B) Laser-scanning confocal microscope images of PI stained 9-day-old wild-type and rpa1b rpa1d double mutant root tips. Calculated average cell length for WT and rpa1b rpa1d was 148 ± 44 mm and 162 ± 51 mm, respectively. (C) Laser-scanning confocal microscope images of PI stained 9-day-old wild-type and rpa1b rpa1d double mutant root tips. Arrows indicate starting point of the cell elongation zone. (D) Laser-scanning confocal microscope images of EdU stained 9-day-old wild-type and rpa1b rpa1d double mutant root tips. Fluorescent nuclei represent actively incorporating (replicating) nuclei. Bars = 50 µm.
Because RPA1B and RPA1D show overlapping functions that promote normal DNA replication, it is possible they may also have overlapping functions in response to DNA damage. However, the reduced root growth seen in the double mutant in the absence of DNA damaging agents complicates analysis of hypersensitivity through simple comparison of root length. To test the rpa1b rpa1d double mutant for hypersensitivity to DNA damage, we employed relative root growth rates (percentage of root growth reduction) as a measure of hypersensitivity (see Materials and methods section for description of assay). In response to UV-B and CPT we see no significant hypersensitivity (Supplementary Figure S11), and a small but significant, suppression effect (less growth reduction) in response to MMC. It is possible the latter suppression effect is a result of reduced DNA replication rates, perhaps allowing more time for repair of this type of lesion. Nevertheless, this suggests RPA1B and RPA1D play only minor roles, if any, in DNA damage repair. Again, redundancy with ACE group subunits could play a role here but overall these data argue against primary DNA-repair roles for RPA1B and RPA1D.
DISCUSSION
In this study, we find that Arabidopsis encodes five RPA1 subunits (A–E), two RPA2 subunits (A; At2g24490, B; At3g02920) and two RPA3 subunits (At3g52630; At4g18590) consistent with previous accounts of RPA genes in Arabidopsis (24,25,28). An initial examination of Arabidopsis RPA1A and RPA1B T-DNA insertion mutant and RNAi knockdown lines suggests that elimination of RPA1A is lethal, while elimination of RPA1B results in hypersensitivity to DNA damaging agents (25). However, two subsequent studies (not including this study) isolated viable rpa1a homozygous mutants from this particular T-DNA line (SALK_017580). In one, rpa1a mutants displayed hypersensitivity to a variety of genotoxic agents, including hydroxyurea, bleomycin and MMS, as well as defective telomere-length homeostasis (37). In the other, the rpa1a mutant displays reduced fertility, manifested by defects in the later stages of meiotic recombination required for the formation of class I crossovers (28). To perform a more comprehensive comparison of the RPA1 family, we have isolated and confirmed T-DNA insertion lines of all five Arabidopsis RPA1 genes, including those published previously. With the exception of a mild fertility defect in rpa1a, all of these individual lines display normal development under standard conditions. Since RPA is required for viability in other (non-plant) systems, this suggests overlapping functions within the RPA1 family.
The RPA1 gene family encodes two main functional groups
To obtain the broadest picture of RPA function in Arabidopsis, we have focused here on the largest RPA subunit gene family, RPA1. We suggest that the RPA1 family is roughly divided into two main functional groups, the ACE group comprised of RPA1A, RPA1C and RPA1E, and the BD group that is comprised of RPA1B and RPA1D. Further, we hypothesize that the primary function of the ACE group is related to DNA repair/recombination activities, while the BD group promotes genomic DNA replication activities. This is supported by phylogenetic analysis of RPA1 protein sequences, analysis of rpa1 mutant phenotypes and expression analysis of RPA1 transcription under normal conditions and in response to DNA damage, as discussed below.
First, our phylogenetic analysis of RPA1 protein sequences shows that RPA1A, RPA1C and RPA1E fall into a clade with strong bootstrap support (100%/1000 replicates) that includes rice RPA1A and RPA1C. An additional separate clade, also with strong bootstrap support (100%/1000 replicates), includes Arabidopsis RPA1B and RPA1D and rice RPA1B. This analysis is not robust enough to clearly determine where these clades fall with respect to non-plant organisms, and where the ancestral duplication that gave rise to both the ACE and BD group occurred. Additional plant, algal, fungal and animal sequences from completed and well-annotated genomes will be required (due to questions about orthologous groups) to address this and related questions about the evolution of the RPA1 family in plants. However, it is clear that additional specialization occurred in plants, likely after the separation of monocots and dicots. It is also clear from our analysis that the divergence of RPA1C and RPA1E, and RPA1B and RPA1D in their respective groups is not likely a result of recent gene duplications in Arabidopsis thaliana.
Second, genetic analysis of single and double mutants of RPA1 gene members also suggests at least two distinct functional groups. While none of the rpa1 single mutants display obvious hypersensitivity to replication blocking agents such as HU or APH, or to the DNA damaging agents UV-B and MMC, rpa1c and rpa1e display hypersensitivity to ionizing radiation, and rpa1c displays additional hypersensitivity to CPT. Moreover, the rpa1c rpa1e double mutant displays additive or supra-additive hypersensitivity to all of the agents tested. As stated above, previous genetic analyses of rpa1a (37) and rpa1b (25) described hypersensitivity phenotypes of each to a variety of damaging and replication-blocking agents, including HU and UV-B. However, under our experimental conditions, we were unable to replicate these hypersensitivity phenotypes. It is possible that the hypersensitivity of rpa1a and rpa1b is relatively mild, and undetectable under the conditions we employed. Nonetheless, based on our analysis of hypersensitivity responses we suggest that RPA1C and RPA1E play leading roles in the repair responses to DNA damage among the RPA1 gene family members.
Obviously, RPA1 is known in yeasts and animals as a component of the single-stranded binding protein required for ‘normal’ genome replication. In contrast to the ACE group, we present here evidence that suggest the primary role of the BD group is to promote normal genome replication throughout development of the plant. Neither the rpa1b and rpa1d single mutants nor the rpa1b rpa1d double mutant display obvious DNA damage hypersensitivity. The pleiotropic-like developmental defects observed in the rpa1b rpa1d double mutant in the absence of exogenous DNA damage suggest a fundamental cellular role of both genes throughout development. Essentially, the plants appear normal but are slower to develop (shorter roots, smaller leaves, reduced silique production, etc.), perhaps suggesting either a reduction in cell size (cell elongation), or a reduction in cell division. However, we do not detect any obvious differences in cell size or morphology in both root and shoot tissues. In contrast, we find root meristematic regions that are significantly smaller than either WT or the single rpa1b or rpa1d mutants. These findings are similar to a mutant defective in RPA2A (ROR1) (29), a member of the middle subunit (RPA32/RPA2) family. In this study, ror1 was identified as a suppressor of ros1 (‘repressor of silencing 1’), a repressor of transcriptional gene silencing. Single mutant (ror1) plants also display reduced vegetative growth similar to the rpa1b rpa1d double mutant, while defects in ROR1 in the ros1 background are not affected in final cell sizes (cell elongation) compared to WT or ros1 single mutants. Based on this, the authors concluded that mutations in RPA2A/ROR1 affect cell division rather than cell size. Here, we find discernably less DNA replication occurring in root meristems in the rpa1b rpa1d double mutant versus WT and single mutants, and accumulation of dead cells throughout the meristem. Therefore, these data suggest RPA1B, RPA1D and RPA2A (perhaps by interacting in heterotrimeric complexes) cooperate during cell division to promote faithful DNA replication of normal developing cells in the absence of exogenous DNA damage. Since the rpa1b rpa1d is not lethal this further suggests partial overlap of the ACE group to complete genome replication, albeit inefficient.
Third, comparison of genomic expression data in young seedlings reveals that RPA1B and RPA1D display higher basal levels of transcription than RPA1A, RPA1C or RPA1E. However, in response to DNA damage (ionizing radiation) this is reversed, as RPA1A, RPA1C and RPA1E are all strongly up-regulated shortly after irradiation (33). This further supports the notion that the BD group plays important roles during normal development, such as normal DNA replication, while the ACE group is specialized for repair responses.
Genetic interactions of RPA1C, RPA1E and ATR
In an effort to better understand the role of RPA in DNA repair induction through ATR, we focused on RPA1C and RPA1E since (i) their respective mutants displayed the most obvious hypersensitivity, (ii) RPA1C and RPA1E show the highest transcriptional induction within the ACE group and (iii) the rpa1a mutant did not display obvious hypersensitivity either as a single mutant, or additively in combination with rpa1c or rpa1e. Overall, we find that combination of rpa1c, rpa1e and atr mutants create additive (in some cases supra-additive) hypersensitivity phenotypes in response to DNA-damaging agents (ionizing radiation, CPT, UV-B and MMC). This is best exemplified by hypersensitivity responses to double-strand breaks created by CPT; the rpa1c atr double mutant displays increased levels of hypersensitivity over either single (hypersensitive) mutant (additive), while the rpa1c rpa1e double mutant displays supra-additive hypersensitivity since the rpa1e mutant is no more sensitive than WT. Based on the supra-additive effects we see, and the fact that the rpa1c rpa1e double mutant is hypersensitive to all DNA damaging and replication-blocking (HU, APH) agents tested, it seems likely that genetic redundancy (synergy) between RPA1C and RPA1E is playing a role to compensate for the loss of one subunit. Therefore, our current working model is that the primary functions of RPA1C, RPA1E and in some cases ATR (for double-strand breaks), are required for separate repair pathways.
In yeasts and mammals, ATR is activated in part through interaction of ssDNA-bound RPA molecules and the ATR-associated ATRIP (scDdc2, spRad26) subunit. This interaction has been shown to be important in variety of cellular responses to DNA damage, including regulation of the cell cycle (41). We have previously shown that Arabidopsis mutants in ATR and ATRIP (HUS2) display defects in cell-cycle regulation in response to DNA damage and replication blocking agents (12,13,30). Employing a similar strategy, we find here that neither RPA1C nor RPA1E appear to directly regulate ATR-dependent (and by extension ATRIP-dependent) cell-cycle responses to genomic insults. This may suggest that (i) functional redundancy compensates for loss of RPA1C, RPA1E or both by other active RPA1 paralogs or (ii) that RPA complexes involved in normal genomic replication (e.g. RPA1B, RPA1D) are primarily involved in activating ATR-dependent cell-cycle responses to genomic insults. Careful analysis of cell-cycle responses in both the rpa1a rpa1c rpa1e triple mutant and the rpa1b rpa1d double mutant will be needed to better determine if/how RPA regulates these ATR-dependent responses.
In response to DNA damage, root meristem cells undergo programmed cell death located primarily at root initials and their daughter cells (50–52). Elimination of either ATR or ATM effectively reverses this cell death in roots treated with modest amounts (i.e. 40 Gy of gamma radiation) of damage (52). These latter results led the study’s authors to conclude that ATR and ATM promote programmed cell death in the presence of modest amounts of DNA damage within the root. In response to replication blocking agents (HU) however, we show that WT roots do not accumulate dead cells, while loss of ATR results in cell death throughout the root meristem (primarily the initials and their daughter cells) grown in the presence of modest levels of replication blocking agents (e.g. 0.25 mM HU causes <25% root growth inhibition in WT). Obviously, the resulting cell death most likely causes the severe hypersensitivity root phenotype. In the case of replication blocks from polymerase inhibition (HU and APH), it is likely that loss of ATR prevents the ability of the cell to restart replication following DNA polymerase stalling, although this has not specifically been shown in plants to date.
Interestingly, we find combination of the atr mutant with the rpa1c mutant resulted in less sensitivity to replication blocking agents, and less overall cell death (suppression of atr). One way to overcome a lack dNTP production (as is the case of cells treated with HU) would be to simply increase RNR activity. RNR is transcriptionally regulated in plants (40), although little is known about post-transcriptional regulation. Employing real-time PCR of each RNR catalytic and regulatory subunit genes, we do not find any evidence to suggest increased transcriptional regulation of any RNR subunit in rpa1c mutant lines. In addition, we find that the rpa1c mutant also suppresses APH hypersensitivity (albeit to a lesser, but statistically significant amount), a direct inhibitor of DNA polymerase that has no direct effect on cellular dNTP levels. Taken together, this suggests that rpa1c suppression of atr hypersensitivity to replication blocking agents is not a result of increases in RNR activity in the rpa1c mutant.
Mutations in chromatin modifying factors (SET1 and CHD1) have been shown to suppress the HU hypersensitivity of mec1 mutants (an ATR ortholog) in Saccharomyces cerevisiae (53). SET1 encodes a histone H3 (K36) methyltransferase and CHD1 encodes a chromatin remodeling factor. Both genes appear to play roles in modification of transcribed regions that can affect RNA polymerase action (54–60). However, suppression of mec1 HU hypersensitivity by set2 and/or chd1 suggests a role for SET1 and CHD1 in negatively regulating DNA replication (53). As described above, defects in RPA2A/RPA2-1 (ror1/rpa2-1 mutants) in Arabidopsis results in altered transcriptional gene silencing (29,61,62). In addition, ror1 plants display similar defects in development (reduced growth of root and shoot) and cell division as we find in our rpa1b rpa1d double mutant (29), suggesting a role for RPA2A in genomic DNA replication. While RPA is not known to directly modify chromatin per se, it is possible that RPA could play a role in maintenance of chromatin structure that could affect progression of polymerases on DNA. By extension, it is possible RPA1C could play a role in negatively controlling DNA polymerase progression (perhaps through chromatin modification) whereby, in the absence of ATR, it could increase sensitivity to HU by further inhibiting DNA replication.
Another possibility is that RPA1C promotes aspects of replication fork restart, such as HR, and is regulated by ATR. Stalled replication forks must be restarted in order to complete replication. Checkpoint related kinases, such as Mec1 and Rad53 (Chk2 ortholog) in yeast, play important roles in regulating restart by activating appropriate pathways to resolve fork blockage (63,64). One important step in this process is to reduce the frequency of HR that could lead to destabilization of replication forks and generate toxic HR intermediates (63,64). In the case of rpa1c suppression of atr-dependent hypersensitivity to HU and APH, we propose a possible model: in WT cells, ATR regulates efficient restart by promoting fork regression and reloading of the replication machinery (polymerase, etc.), while preventing unnecessary HR. In the case of depletion of dNTPs (HU) or polymerase inhibition (APH), HR-mediated repair pathways would generally not be required since in most cases of fork stalling, there is no lesion to be removed. In the absence of ATR (atr mutant), cells lose regulation of the restart mechanism and HR (promoted through RPA1C) is unchecked leading to lethal recombination intermediates. Thus, elimination of RPA1C reduces atr-induced hypersensitivity (atr rpa1c double mutant) by reducing the number of lethal HR intermediates. In an analogous example, unrestrained HR in yeast defective in the Srs2 helicase leads to increased cell death (65), and can be suppressed by elimination of Rad51-dependent HR pathways (66–68). This suggests that promotion of HR processing in the absence of required (SRS2 helicase) factors might lead to toxic HR intermediates. Nonetheless, considering the hypersensitivity of rpa1c mutants to double-strand breaks (γ-radiation and CPT), the requirement of both RPA1C and ATR in response to DNA lesions (UV-B and MMC) and contributing defects in meiosis suggest a critical and unique role for RPA1C in HR-dependent repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Abigail Boduch, Stacy Wong and William Wyman for assistance with mutant construction, hypersensitivity assays and EdU staining protocols.
FUNDING
New Hampshire Agricultural Research Station (USDA) (Graduate Student Research Apprenticeship to B.B.A.); University of New Hampshire [Undergraduate Research Fellowships (UROP and SURF) to RS]; NSF [grant MCB-0818603]; New Hampshire Agricultural Research Station (HATCH) [grant NH00543]; NSF ADVANCE [grant UNH147493 to K.M.C.]. This is scientific contribution number 2541 from the New Hampshire Agricultural Experiment Station. Funding for open access charges: [NH00543].
Conflict of interest statement. None declared.
REFERENCES
- 1.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Ann. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 3.Abraham R. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 4.Hurley PJ, Bunz F. ATM and ATR: components of an integrated circuit. Cell cycle (Georgetown, Tex) 2007;6:414–417. doi: 10.4161/cc.6.4.3886. [DOI] [PubMed] [Google Scholar]
- 5.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 6.Byun TS, Pacek M, Yee M-C, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 10.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 11.Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto AN, Lan VTT, Puripunyavanich V, Hase Y, Yokota Y, Shikazono N, Nakagawa M, Narumi I, Tanaka A. A UVB-hypersensitive mutant in Arabidopsis thaliana is defective in the DNA damage response. Plant J cell Mol. Biol. 2009;60:509–517. doi: 10.1111/j.1365-313X.2009.03974.x. [DOI] [PubMed] [Google Scholar]
- 13.Sweeney PR, Britt AB, Culligan KM. The Arabidopsis ATRIP ortholog is required for a programmed response to replication inhibitors. Plant J. 2009;60:518–526. doi: 10.1111/j.1365-313X.2009.03975.x. [DOI] [PubMed] [Google Scholar]
- 14.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 16.Carty MP, Zernik-Kobak M, McGrath S, Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu JS, Kuo SR, McHugh MM, Beerman TA, Melendy T. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J. Biol. Chem. 2000;275:1391–1397. doi: 10.1074/jbc.275.2.1391. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhou XY, Wang H, Huq MS, Iliakis G. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J. Biol. Chem. 1999;274:22060–22064. doi: 10.1074/jbc.274.31.22060. [DOI] [PubMed] [Google Scholar]
- 19.Ariza RR, Keyse SM, Moggs JG, Wood RD. Reversible protein phosphorylation modulates nucleotide excision repair of damaged DNA by human cell extracts. Nucleic Acids Res. 1996;24:433–440. doi: 10.1093/nar/24.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 2010;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshav KF, Chen C, Dutta A. Rpa4, a homolog of the 34-kilodalton subunit of the replication protein A complex. Mol. Cell Biol. 1995;15:3119–3128. doi: 10.1128/mcb.15.6.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishibashi T, Kimura S, Sakaguchi K. A higher plant has three different types of RPA heterotrimeric complex. J. Biochem. (Tokyo) 2006;139:99–104. doi: 10.1093/jb/mvj014. [DOI] [PubMed] [Google Scholar]
- 24.Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF. Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 2007;144:1697–1714. doi: 10.1104/pp.107.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishibashi T, Koga A, Yamamoto T, Uchiyama Y, Mori Y, Hashimoto J, Kimura S, Sakaguchi K. Two types of replication protein A in seed plants. Febs J. 2005;272:3270–3281. doi: 10.1111/j.1742-4658.2005.04719.x. [DOI] [PubMed] [Google Scholar]
- 26.Singh DK, Islam MN, Choudhury NR, Karjee S, Mukherjee SK. The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res. 2007;35:755–770. doi: 10.1093/nar/gkl1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marwedel T, Ishibashi T, Lorbiecke R, Jacob S, Sakaguchi K, Sauter M. Plant-specific regulation of replication protein A2 (OsRPA2) from rice during the cell cycle and in response to ultraviolet light exposure. Planta. 2003;217:457–465. doi: 10.1007/s00425-003-1001-z. [DOI] [PubMed] [Google Scholar]
- 28.Osman K, Sanchez-Moran E, Mann SC, Jones GH, Franklin FC. Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 2009;28:394–404. doi: 10.1038/emboj.2008.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia R, Wang J, Liu C, Wang Y, Wang Y, Zhai J, Liu J, Hong X, Cao X, Zhu J-K, et al. ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell. 2006;18:85–103. doi: 10.1105/tpc.105.037507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Culligan KM, Tissier A, Britt AB. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell. 2004;16:1091–1104. doi: 10.1105/tpc.018903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso JEA. Genome-wide insertional mutagenesis of Arabidopsis. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 32.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 34.Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P. Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20:503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 35.Higgins JD, Armstrong SJ, Franklin FC, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 37.Takashi Y, Kobayashi Y, Tanaka K, Tamura K. Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis. Plant Cell Physiol. 2009;50:1965–1976. doi: 10.1093/pcp/pcp140. [DOI] [PubMed] [Google Scholar]
- 38.Garcia V, Bruchet H, Camescasse D, Fabienne G, Bouchez DL, Tissier A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15:119–132. doi: 10.1105/tpc.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elledge SJ. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 40.Roa H, Lang J, Culligan KM, Keller M, Holec S, Cognat V, Montane MH, Houlne G, Chaboute ME. Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol. 2009;151:461–471. doi: 10.1104/pp.109.140053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem. J. 2011;436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Culligan KM, Britt AB. Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 2008;55:629–638. doi: 10.1111/j.1365-313X.2008.03530.x. [DOI] [PubMed] [Google Scholar]
- 44.Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Mittelsten Scheid O, Shibahara K, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18:782–793. doi: 10.1101/gad.295404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inagaki S, Suzuki T, Ohto MA, Urawa H, Horiuchi T, Nakamura K, Morikami A. Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 2006;18:879–892. doi: 10.1105/tpc.105.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.del Olmo I, Lopez-Gonzalez L, Martin-Trillo MM, Martinez-Zapater JM, Pineiro M, Jarillo JA. EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. Cell Mol. Biol. 2010;61:623–636. doi: 10.1111/j.1365-313X.2009.04093.x. [DOI] [PubMed] [Google Scholar]
- 48.Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y. The catalytic subunit of Arabidopsis DNA polymerase alpha ensures stable maintenance of histone modification. Development. 2013;140:156–166. doi: 10.1242/dev.084624. [DOI] [PubMed] [Google Scholar]
- 49.He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Curtis MJ, Hays JB. Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair. 2011;10:1272–1281. doi: 10.1016/j.dnarep.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Curtis MJ, Hays JB. Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: requirements for DNA translesion polymerases eta and zeta. DNA Repair. 2007;6:1341–1358. doi: 10.1016/j.dnarep.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl Acad. Sci. USA. 2009;106:20984–20988. doi: 10.1073/pnas.0909218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biswas D, Takahata S, Xin H, Dutta-Biswas R, Yu Y, Formosa T, Stillman DJ. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics. 2008;178:649–659. doi: 10.1534/genetics.107.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 57.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PloS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biswas D, Dutta-Biswas R, Stillman DJ. Chd1 and yFACT act in opposition in regulating transcription. Mol. Cell. Biol. 2007;27:6279–6287. doi: 10.1128/MCB.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu J-K. Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr. Biol. CB. 2005;15:1912–1918. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 62.Elmayan T, Proux F, Vaucheret H. Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. CB. 2005;15:1919–1925. doi: 10.1016/j.cub.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 63.Branzei D, Foiani M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 64.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair. 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 65.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 66.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McVey M, Kaeberlein M, Tissenbaum HA, Guarente L. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 2001;157:1531–1542. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.