Abstract
Elongation factor P (EF-P) is a conserved ribosome-binding protein that structurally mimics tRNA to enable the synthesis of peptides containing motifs that otherwise would induce translational stalling, including polyproline. In many bacteria, EF-P function requires post-translational modification with (R)-β-lysine by the lysyl-tRNA synthetase paralog PoxA. To investigate how recognition of EF-P by PoxA evolved from tRNA recognition by aminoacyl-tRNA synthetases, we compared the roles of EF-P/PoxA polar contacts with analogous interactions in a closely related tRNA/synthetase complex. PoxA was found to recognize EF-P solely via identity elements in the acceptor loop, the domain of the protein that interacts with the ribosome peptidyl transferase center and mimics the 3'-acceptor stem of tRNA. Although the EF-P acceptor loop residues required for PoxA recognition are highly conserved, their conservation was found to be independent of the phylogenetic distribution of PoxA. This suggests EF-P first evolved tRNA mimicry to optimize interactions with the ribosome, with PoxA-catalyzed aminoacylation evolving later as a secondary mechanism to further improve ribosome binding and translation control.
INTRODUCTION
Mimicry is the process by which one species or molecule evolves to be similar to another (1,2). At the molecular level (i.e. molecular mimicry), this is observed in several pathogens that have evolved molecular structures to resemble the host’s own structures to evade or trigger a response from it (2). An example of this is translation initiation factor 2A (eIF2A) that is mimicked by K3L from poxviruses. During viral infections, double-stranded RNA-dependent protein kinase (PKR) phosphorylates eIF2A preventing protein translation, which in turn decreases viral production. This cellular defense mechanism placed a selective pressure on the poxvirus, leading to the evolution of K3L (the mimic), an inhibitor that binds the active site of PKR, thereby preventing eIF2A phosphorylation (3).
In addition to the mimicry of molecules between interacting species, the term ‘molecular mimicry’ is also used to describe the convergent evolution of molecules that coexist within the same species. Some of the most intriguing examples of molecular mimicry are within the transcription and translation apparatus where several proteins mimic nucleic acids to control these processes (4–6). For instance, several translation factors mimic tRNA, which presumably allows these proteins to fit properly within binding sites on the ribosome (6,7). Perhaps one of the most striking examples is EF-P, which not only resembles a complete tRNA molecule in structure (Figure 1), but also emulates the tRNA before its interaction with the ribosome.
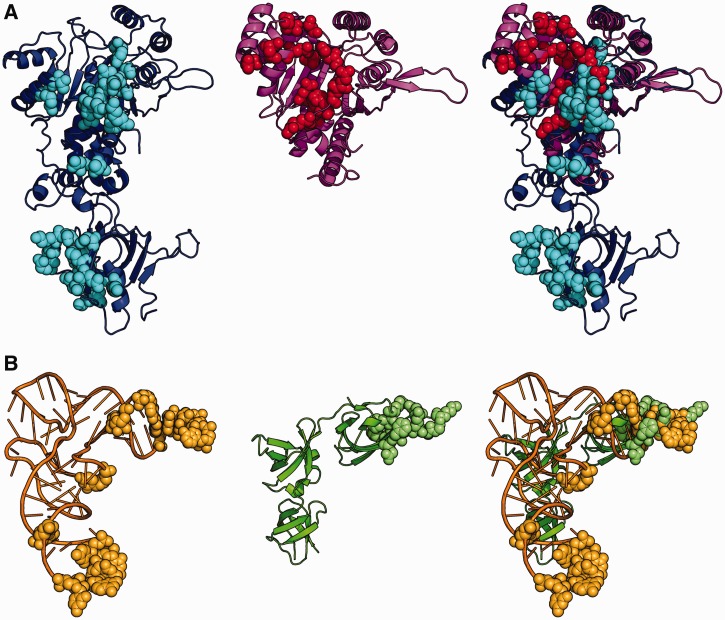
Figure 1.
Contact surface in EF-P/PoxA and tRNAAsp/AspRS complexes. The image shows a comparison of the polar contacts between EF-P and PoxA with polar contacts between tRNAAsp and AspRS. Image corresponds to a superposition of AspRS and PoxA from pdb files 1asy (chains A and R) and 3a5z (chains C and D). AspRS is in blue, tRNAAsp in orange, PoxA in magenta and EFP in green. Amino acids and nucleotides making polar contacts are marked in balls with the following colors: AspRS in light blue, tRNAAsp in light orange, EFP in light green and PoxA in red. (A) Enzymes. (B) Substrates.
EF-P is a non-essential elongation factor that has been shown to facilitate the translation of several proteins involved in pathogenesis and survival under other stressful conditions (8,9). Many EF-P-regulated proteins have a stretch of prolines or other sequence patterns that induce ribosome stalling during translation. EF-P is required for alleviating the stalling of these paused ribosomes, allowing for the translation of these genes to continue (10–12). To function efficiently in translation, EF-P is (R)-β-lysylated at Lys34 (Escherichia coli numbering), a position structurally equivalent to A76 in tRNA (10,13,14). The modification of EF-P is catalyzed by PoxA, a protein paralog of the catalytic domain of lysyl–tRNA synthetase (LysRS). All aminoacyl–tRNA synthetases (aaRS) including LysRS catalyze tRNA aminoacylation in a two-step reaction. First, the substrate amino acid is activated with ATP forming an aminoacyl adenylate (aa-AMP) that usually remains stably bound to the enzyme in the absence of tRNA. In a second step, the amino acid is transferred from AMP to the 3' or 2' OH of A76 at the 3'-end of tRNA:
Similar to all known aaRSs, PoxA also catalyzes the aminoacylation of EF-P in a two-step reaction, which in this case requires ATP and (R)-β-lysine ((R)-β-Lys), an analog of α-lysine that has the amino group in the beta instead of the alpha position (8,14):
(R)-β-Lys is transferred to Lys34 of EF-P forming an (R)-β-lysyl-lysine amino acid. This group is further hydroxylated to form (R)-β-lysyl-hydroxylysine in a reaction catalyzed by the product of the yfcM gene (15). Until now it has not been possible to find the physiological relevance of this hydroxylation, as it is not required for alleviating stalled ribosomes, and yfcM deletion strains do not present any obvious phenotype (10,16).
While PoxA modifies EF-P in a reaction that resembles those catalyzed by aaRSs, the extent of EF-P mimicry to tRNA is unknown. Therefore, we set out to investigate whether EF-P mimics tRNA, not only in structure, but also in its interaction with the LysRS paralog, PoxA. Our results show that PoxA recognizes EF-P mainly through interactions at positions mimicking the acceptor stem of tRNA. Thus, PoxA has retained and even strengthened interactions that were already relevant for recognition of tRNA by aaRSs. Most of the conserved positions in the EF-P sequence correspond to residues contacting the ribosome, with PoxA only recognizing a small subset of these conserved positions. This suggests EF-P first evolved tRNA mimicry to interact with the ribosome, whereas PoxA evolved afterward in a subset of bacterial species to recognize EF-P. The absence of PoxA in many bacteria and the presence of a different pathway for the modification of the EF-P paralog in archaea and eukaryotes (IF5a) further support the hypothesis that tRNA mimicry in this pathway originated to allow for interaction between EF-P and the ribosome, whereas PoxA recognition of the mimic is a more recent addition.
MATERIALS AND METHODS
Strains and vectors, cloning and mutagenesis
EF-P and PoxA were expressed in BL21 XJB (DE3) strains from plasmids EFP pTYB11 and PoxA pTYB11 (14). To add a phosphorylation site for the catalytic subunit of the cAMP-dependent protein kinase A (PKA) at the carboxy terminus of EFP in the pTYB11 plasmid, efp was amplified by polymerase chain reaction using the primers: EFP-fd (GCAACGTACTATAGCAACGAT) and EFP-PKA-XmaI-Rv (GTCACCCGGGCTCCGATTAAACAGATGCACGACGCTTCACGCGAGAGACG). The amplified DNA was digested and inserted into the EFP pTYB11 plasmid at XmaI restriction site and the efp internal EcoRI site. efp and poxA mutations were performed by polymerase chain reaction amplification and DpnI digestion using the primers listed in Supplementary Table S1 (PoxA) and Supplementary Table S2 (EFP) together with PoxA pTYB11 or pWN403 [pBAD18-poxA; (8)] and EFP pTYB11 as templates, respectively. All cloning and mutagenesis were confirmed through sequencing at the Plant Microbe Genomics Facility at The Ohio State University and then transformed in E. coli BL21 XJB (DE3) cells for overexpression and purification.
Sensitivity assays
Complementation assays were performed in Salmonella typhimurium 14028s using plasmid pWN403 (pBAD18-poxA) and mutation variants as previously described (8,9). Briefly, hypo-osmolarity was assessed by streaking Salmonella on antibiotic medium 2 agar (AB2; Difco, Detroit, MI, USA). Plates were incubated at 37°C for 16 h followed by visual assessment of colony size. For gentamicin and lauryl sulfobetaine sensitivity, overnight cultures of Salmonella strains were diluted 1:1000 into fresh Luria-Bertani medium containing either gentamicin (3.125 µg/ml) or lauryl sulfobetaine (6.25 mM). Growth was assessed by turbidity following 16 h of incubation at 37°C with shaking. In this condition, all strains grow up to 1–1.15 OD600 units in the absence of gentamicin and sulfobetaine.
Purification of PoxA, EF-P and variant proteins
All proteins were expressed using autoinduction media and then purified as previously reported (14). Briefly, cells were resuspended in buffer A (50 mM Tris HCl, pH 8.0, 500 mM NaCl and 10% glycerol) plus 0.05% triton X-100 and lysed by sonication. Cell debris was eliminated by centrifugation and the supernatant loaded to a chitin column. Column was washed with five volumes of buffer A plus 0.05% triton X-100, and then with 25 volumes of buffer A. Finally, the column was washed with 2 volumes of buffer A plus 100 mM dithiothreitol and incubated overnight at 4°C. After this, the protein was eluted with buffer A and concentrated using a filter centrifugation unit with a MWCO of 3 KDa. Then the protein was dialyzed against dialysis buffer (25 mM Tris HCl, pH 8.0, 150 mM NaCl and 4 mM 2-mercaptoethanol) plus 50% glycerol and stored at −20°C. EF-P used for kcat/ KM determinations was further purified through anion exchange using a Resource Q column. Protein was loaded on the column and washed with 10 volumes of buffer 1 (25 mM Tris HCl, pH 8, 50 mM NaCl and 2 mM 2-mercaptoethanol), then with additional 6 volumes of 10% buffer 2 (25 mM Tris HCl, pH 8, 1 M NaCl and 2 mM 2-mercaptoethanol) in buffer 1. Finally, the protein was eluted in a linear gradient to 14% buffer 2 in buffer 1, concentrated and dialyzed against dialysis buffer plus 20% glycerol. The protein was then stored in aliquots at −80°C. All proteins were quantified in 6 M guanidinium HCl based on absorbance at 280 nm and a molar extinction coefficient of 24750 M-1 cm-1 (EF-P) or 29870 M−1 cm−1 (PoxA) that were calculated using EMBOSS (17).
Phosphorylation of EF-P-PKA
EF-P-PKA was phosphorylated for 1 h at 30°C in a reaction mixture that contained 60 µM EF-P-PKA, 50 mM Tris HCl, pH 7.5, 10 mM MgCl2, 1.4 µM (∼9µCi/µl) 32P-γATP, 200 µM ATP and 30 U/ml PKA (NEB). Then the 32P-EF-P-PKA was purified using a DEAE sepharose column at room temperature. The protein was loaded and the column was cleaned with 50 volumes of buffer 1, then with additional 15 volumes of 5% buffer 2 in buffer 1 and eluted with 10% buffer 2 in buffer 1. The purified protein was dialyzed against dialysis buffer with 50% glycerol and stored in aliquots at −80°C.
Active site titration of PoxA
PoxA active site titration was made in a reaction mix containing 0.1 M glycine, pH 9.0, 30 mM KCl, 10 mM MgCl2, 30 µM ATP, 0.0065 µM (∼0.02 µCi/µl) 32P-αATP, 2.5 mM 2-mercaptoethanol, 4.2 U/ml pyrophosphatase and 4 mM (R)-β-Lys. Reactions were started by adding enzyme and performed at room temperature. At specific time points, aliquots were taken and quenched by mixing with 2.5 volumes of 0.2 M Na acetate, pH 5.0. Aliquots (0.5 µl) of each sample were separated on PEI-F cellulose plates using 0.1 M ammonium acetate plus 5% acetic acid as mobile phase. Plates were analyzed using phosphorimaging, and PoxA active sites were quantified from the plateau of the (R)-β-lysyl-AMP intermediate formation.
Aminoacylation of EF-P with 14C-α-lysine
EF-P aminoacylation with 14C-α-Lysine was performed at 37°C in a mixture containing 100 mM glycine, pH 9.0, 30 mM KCl, 10 mM MgCl2, 8 mM ATP, 3 mM 2-mercaptoethanol, 8.4 U/ml pyrophosphotase and 0.75 µM EFP. At defined time points, 6 µl of reaction mixture was precipitated on 5% trichloroacetic acid saturated 3MM Whatman paper. The papers were washed three times with 5% trichloroacetic acid, once with ethanol and dried at 85°C. Then the 14C aminoacylated EF-P was quantified using scintillation counting. Additionally, aliquots of the reaction were separated in a sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and 14C α-Lysyl-EF-P was detected by phosphorimaging to confirm aminoacylation of EF-P.
Aminoacylation of EF-P with (R)-β-Lys
Aminoacylation kinetics of EF-P with (R)-β-Lys were assayed at 37°C in a mixture containing 100 mM glycine, pH 9.0, 30 mM KCl, 10 mM MgCl2, 2.5 mM 2-mercaptoethanol, 0.1 mg/ml bovine serum albumin, saturating concentrations of ATP (2–8 mM depending on the PoxA mutant), 6–8 mM (R)-β-Lys, 4 nM 32P-EF-P-PKA as a probe for EF-P aminoacylation and variable concentrations of EF-P (0.5–2.5 µM). Reaction aliquots were taken at defined time points and quenched in an equal volume of stop solution (2% triton X-100, 1% 2-mercaptoethanol, 750 mM Na acetate, pH5.0). Aminoacylated and non-aminoacylated forms of 32P-EF-P-PKA were separated through isoelectric focusing using a 4.5–5.4 pH gradient (Pharmalyte 4.5–5.4 GE-Healthcare) and subsequently quantified using phosphorimaging.
Sequence and structure analysis
Structure interfaces and polar contacts were determined using PDBe PISA (18). Additional polar contacts previously described were also included (13). Figures were made with The PyMOL Molecular Graphics System (version 1.2.0.3 Schrödinger, LLC). Alignments of 112 sequences of EF-P from organisms of diverse genera were performed with PROMALS3D (19) using EF-P structures from pdb 3a5z-D, 1ueb-A, 3oyy-A to guide the alignment. WebLogo representation of this alignment, as well as from a subset of the sequences with or without a Lys residue in the modification position (Supplementary Tables S3 and S4), was performed using the WebLogo online server (20). Variability of each position on the sequence alignments was calculated using the Shannon entropy analysis of the protein variability server (21).
RESULTS
Equivalent contacts between PoxA/EF-P and aspartyl-tRNA synthetase/tRNA are within the acceptor stem
To investigate how PoxA evolved from LysRS to acquire its substrate specificity for a protein rather than RNA, it would be ideal to first compare the structure of EF-P bound to PoxA with that of tRNALys bound to LysRS. tRNALys is not well resolved in the corresponding crystal structure (22), so instead we compared the EF-P/PoxA complex structure with that of tRNAAsp bound to aspartyl-tRNA synthetase (AspRS) (pdb 1asy). AspRS is closely related to LysRS in structure and, together with AsnRS, they form the class IIb group of aaRSs (23,24), which share common tRNA identity elements, including the discriminator base (25–27). In addition, the effects of several alanine substitution mutations on the contact surface of the tRNAAsp/AspRS complex have been studied (27), providing a useful reference for comparisons with EF-P/PoxA complex.
AspRS, in common with all class IIb aaRSs, has two domains: a catalytic domain that interacts with the acceptor stem of tRNA and an anticodon-binding domain that binds to the anticodon stem loop (Figure 1). Most of the binding energy comes from interactions with the anticodon-binding domain (27) that is absent from PoxA, which contains only the catalytic domain. Focusing on the polar contacts (Figure 1 and Table 1), we observe within the catalytic domain of both enzymes some interactions that have been lost from the EF-P/PoxA complex, others that are conserved (Glu102, Glu103, Arg106 and His108 from PoxA) and a group representing novel interactions not present in the AspRS complex (His52, Asp177, Asn180, Glu185, Gln193, Ser218, Arg235, Glu244 and Arg303 from PoxA). Most of the common contacts correspond to AspRS interactions with nucleotides 73 (discriminator base) and 74 of tRNAAsp. The equivalent positions are located in domain 1 (N-terminal) of EF-P along with all of the novel contacts. Non-polar and polar interactions between EF-P and PoxA have similar distributions, with the only notable differences being in a group of contacts formed between EF-P and the second monomer in the PoxA dimer (Pro63, Gly64, His65, Ser66 and Gln67 from PoxA) (Supplementary Figure S1). Overall, this distribution indicates that most of the EF-P surface does not contact PoxA and, furthermore, the protein is tilted ∼60° compared with tRNAAsp binding to AspRS, with domain III of EF-P displaced from where the anticodon binding domain would have been in a class IIb aaRS (Supplementary Figures S1 and S2) (13). To determine the roles of the retained and novel interactions, we constructed alanine replacement variants for all PoxA residues involved in polar interactions with EF-P and further analyzed their effect on EF-P aminoacylation.
Table 1.
Polar contacts between EF-P and PoxAa
| PoxA |
EFP |
||||
|---|---|---|---|---|---|
| Amino acid | Position | Atom | Amino acid | Position | Atom |
| His | 52 | O | Lys | 31 | NZ |
| His | 52 | O | Lys | 31 | CE |
| His | 52 | ND1 | Lys | 34 | O |
| His | 52 | ND1 | Gly | 35 | O |
| His | 52 | ND1 | Gln | 36 | N |
| Glu | 102 | O | Lys | 31 | NZ by H2O-mediated hydrogen bond |
| Glu | 103 | OE1 | Lys | 31 | NZ |
| Glu | 103 | OE2 | Lys | 31 | NZ |
| Arg | 106 | NH1 | Glu | 28 | OE1 |
| Arg | 106 | NE | Glu | 28 | OE2 |
| Arg | 106 | NH1 | Glu | 28 | OE2 |
| His | 108 | NE2 | Lys | 31 | O |
| Asp | 177 | OD1 | Lys | 54 | NZ |
| Asp | 177 | OD2 | Lys | 54 | NZ |
| Asn | 180 | ND2 | Thr | 55 | O |
| Glu | 185 | OE1 | Lys | 57 | NZ |
| Gln | 193 | NE2 | Gly | 33 | O |
| Gln | 193 | NE2 | Lys | 34 | O |
| Ser | 218 | O | Lys | 34 | NZ |
| Arg | 235 | NH2 | Gly | 33 | O |
| Glu | 244 | OE1 | Gly | 33 | N |
| Glu | 244 | OE2 | Gly | 33 | N |
| Arg | 303 | NH1 | Lys | 31 | O by H2O-mediated hydrogen bond |
| Arg | 303 | NH2 | Lys | 31 | O by H2O-mediated hydrogen bond |
aContacts determined using PDBE Pisa. Additional contacts previously reported are also included (13).
In vitro characterization of PoxA catalyzed amino acid activation and aminoacylation
The catalytic activities of aaRSs are commonly monitored using radiolabeled amino acids. As radioactive (R)-β-Lys is not commercially available, we developed alternative strategies to characterize PoxA activity in vitro. Active site titration was performed using α-32P-ATP and non-radioactive (R)-β-Lys, which allowed monitoring of ATP degradation as well as the formation of 32P-AMP and 32P-(R)-β-lysyl adenylate by TLC and phosphorimaging quantification. Some aaRS show half-of-the-sites reactivity, meaning that only one of the monomers in the dimer at a time can perform amino acid activation (28). To determine whether this is true for PoxA, we calculated the concentration of functional PoxA active sites, which were considered to be equivalent to the concentration of 32P-(R)-β-lysyl adenylate at the plateau of the reaction, a method commonly used with aaRSs. Doubling the enzyme concentration resulted in a 2-fold increase of the plateau level, confirming that the substrate concentration was saturating for the reaction. Enzyme concentrations calculated for purified WT PoxA were usually within 90–95% of concentrations calculated from spectrophotometric determinations (data not shown). The fact that active site titration of PoxA correlated reasonably well with spectrophotometric quantifications suggests that both PoxA monomers are active in the dimer at the same time. During active site titration, the concentration of 32P-(R)-β-lysyl adenylate plateaued after the first time point (15 s) for WT and most of the PoxA variants. Some PoxA variants were slower than WT (R106A, R235A and especially R303A), whereas others were almost completely inactive (E244A; Table 2). Owing to the low activity observed during (R)-β-lysyl adenylate formation, PoxA E244A was not analyzed further.
Table 2.
Effect of alanine substitutions on PoxA activity
| PoxA variant (AspRS equivalent) | Area of tRNA contacted in AspRSa | Effect on amino acid activation | kcat/KM (µM−1 s−1)b | Relative kcat/KM (Rc) | ΔΔGbd(kcal/mol) |
|---|---|---|---|---|---|
| Wild type | 0.05 ± 0.005 | 1 | – | ||
| H52A (gap) | Gap | Active | 0.018 ± 0.007 | 0.37 | 0.62 |
| E102A (N328) | Acceptor arm (G73) | Active | 0.049 ± 0.022 | 0.98 | 0.011 |
| E103A (S329) | Acceptor arm (G73 +C74) | Active | Inactive | – | – |
| R106A (T331) | Acceptor arm (G73) | Reduced | 0.055 ± 0.005 | 1.1 | −0.057 |
| H108A (H334) | Acceptor arm (C74) | Active | Inactive | – | – |
| D177A (gap) | Gap | Active | 0.027 ± 0.003 | 0.53 | 0.39 |
| N180A (gap) | Gap | Active | 0.037 ± 0.008 | 0.73 | 0.19 |
| E185A (gap) | Gap | Active | 0.013 ± 0.002 | 0.27 | 0.81 |
| Q193A (D421) | No contact | Active | 0.055 ± 0.009 | 1.1 | −0.057 |
| S218A (E451) | No contact | Active | 0.047 ± 0.009 | 0.93 | 0.044 |
| R235A (S469) | No contact | Reduced | Inactive | – | – |
| E244A (E478) | No contact | Inactive | Inactive | – | – |
| R303A (R531) | No contact | Reduced | 0.044 ± 0.004 | 0.87 | 0.086 |
aData from Eriani and Gangloff.27
bkcat/KM was estimated using sub-saturating EF-P concentrations from the slope of the equation, V = kcat [E][S]/KM.
cCompared with wild-type.
dΔΔGb represents differences in the transition state binding ΔG and was estimated to be RTln[(kcat/KM)mut/(kcat/KM)WT] (29).
PoxA recognizes EF-P through residues that mimic the tRNA 3'-end
To compare the role of the contacts that are specific to the EF-P/PoxA complex with those that are common to other class IIb aaRSs, we measured the effect of the corresponding alanine substituted variants on PoxA’s ability to aminoacylate EF-P with (R)-β-Lys. Modification of EF-P with (R)-β-Lys adds a positive charge that allows for the separation of the modified protein from the non-aminoacylated form by isoelectric focusing. To allow quantification of both modified and unmodified EF-P, we added a phosphorylation site at the carboxy terminus of EF-P far from the interaction surface with PoxA. Phosphorylation of this site using the catalytic subunit of the cAMP-dependent protein kinase (PKA) and γ-32P ATP allowed us to quantify the fraction of lysylated EF-P by analyzing phosphor images of the samples separated by isoelectric focusing (Supplementary Figure S3). Based on their aminoacylation activity, the PoxA variants can be divided in three main groups (Table 2). The majority of amino acid replacements have little or no effect on PoxA activity. These include PoxA E102A, R106A, N180A, Q193A, S218A and R303A, which have a kcat/KM of at least 70% of the WT protein. A second group composed of H52A, D177A and E185A showed a moderate loss of activity, with a kcat/KM between 20 and 50% of WT PoxA. The third group composed of E103A, H108A and R235A retains no detectable activity for aminoacylation of EF-P with (R)-β-Lys.
All inactivating replacements map to PoxA residues that interact with a loop region of EF-P domain 1 where Lys34, the modification site, is located (Figure 2). One of these residues is Glu103 that hydrogen bonds to Lys31 of EF-P (2.9 Å). The equivalent interaction in the tRNAAsp/AspRS complex corresponds to Ser329 (Glu264 in E. coli LysRS) that is at hydrogen bonding distance from the discriminator base (G73) (2.5 Å) and is also in close contact with C74 (3.7 Å) of tRNAAsp. Replacement of Ser329 as well as any of the two other amino acids of AspRS that contact the discriminator base (Asn328 and Thr331) had only a mild effect on efficiency of tRNAAsp aminoacylation (27). Replacement of the PoxA residues that correspond to Asn328 and Thr331 (Glu102 and Arg106) did not affect EF-P aminoacylation. In contrast, replacement of Glu103 in PoxA completely inactivated the enzyme, indicating a much more important role for this position than observed in the corresponding aaRSs.
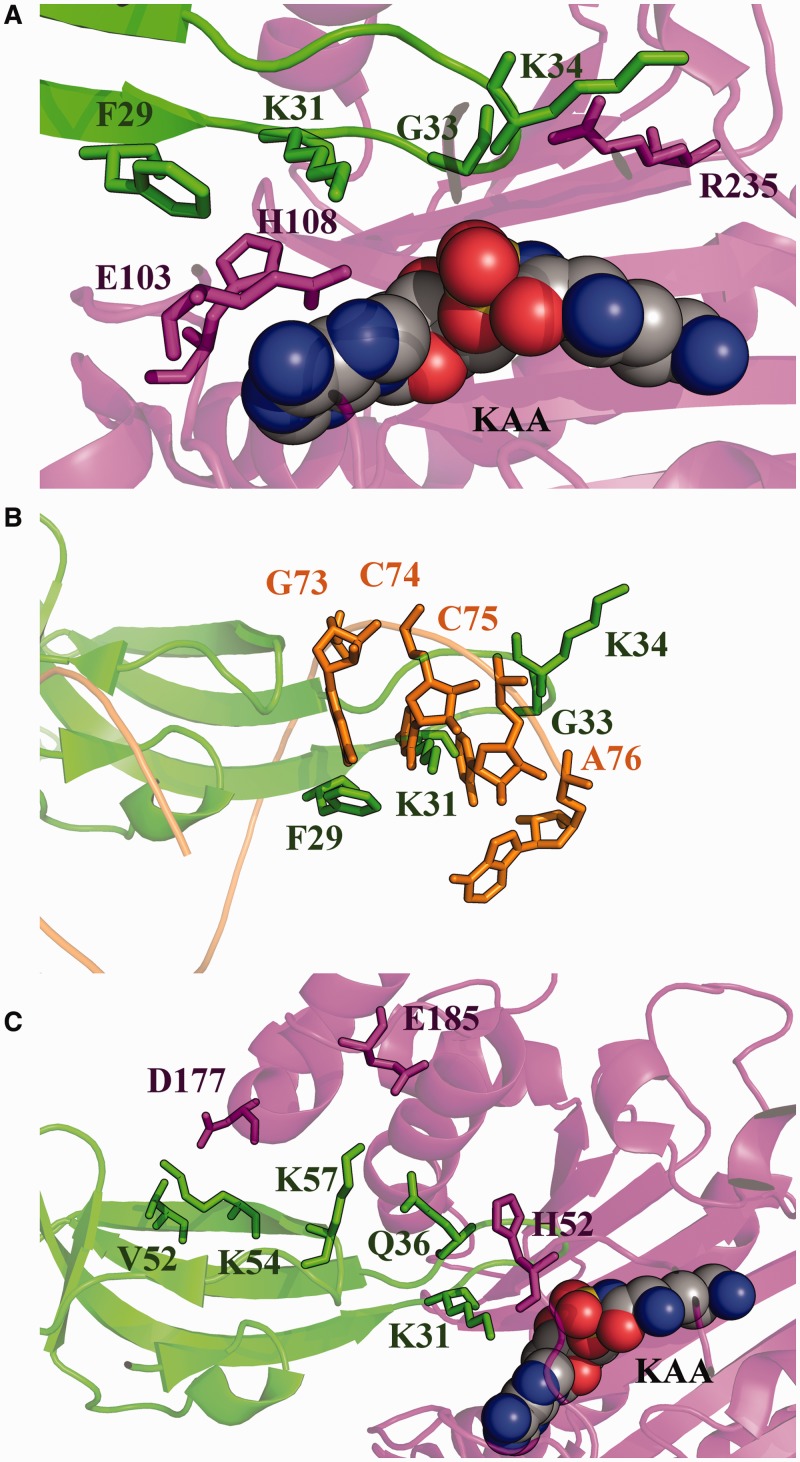
Figure 2.
EF-P identity elements. (A) Active site of PoxA. PoxA residues required for aminoacylation of EF-P and the corresponding EF-P contact residues are highlighted. The α-lysyl-adenylate analog 5′-O-[(l-lysylamino) sulfonyl] adenosine (KAA) is also represented. (B) tRNA structural mimicry by EF-P identity elements. EF-P identity elements and the relevant tRNA nucleotides are shown as localized after superposition of PoxA and AspRS. (C) Active site of PoxA. PoxA amino acids from the medium effect group and the EF-P amino acids contacted by them are highlighted. Colors used in this figure correspond to the color scheme of Figure 1.
His108 of PoxA also forms a hydrogen bond with Lys31 through interactions to the backbone oxygen (3.3 Å), and additionally is in close contact with Phe29 (3.4 Å). PoxA His108 corresponds to one of the most conserved residues in structural motif 2 of AspRS, His334 (His270 in E. coli LysRS), which contacts C74 of tRNAAsp. As with His108 of PoxA, replacement of His334 in AspRS severely reduced substrate binding affinity and aminoacylation (27). In AspRS, substitution of this His with Ala also reduced the rate of amino acid activation (27). In contrast, replacement of His108 in PoxA did not produce any significant effect on the amino acid activation reaction (Table 2), indicating that the role of this His residue has partially diverged between the two enzymes.
The third replacement that inactivates PoxA corresponds to R235A. Arg235 contacts the backbone oxygen of Gly33 (3.6 Å) and Lys34 (4.0 Å) of EF-P. This arginine corresponds to Ser469 of AspRS, which does not contact tRNAAsp. E. coli LysRS has an arginine (Arg412), which structurally co-localizes with Arg235 of PoxA. Although tRNALys is not well resolved in the LysRS crystal structure, superposing LysRS to AspRS reveals that one of the A76 phosphate oxygens from tRNAAsp is a short distance from Arg412 of LysRS (5.2 Å), suggesting a possible direct contact with tRNA. Independently of this, we consider Lys34 of EF-P to be functionally analogous to A76 of tRNA, as both are sites of aminoacylation. Alternatively, Lys34 may be viewed as a structural mimic of C75 of tRNAAsp, as both co-localize in the structural alignment between AspRS and PoxA (Figure 2B). Structural alignment of the EF-P/PoxA complex with tRNAAsp/AspRS shows that Phe29 of EF-P localizes in a position similar to G73 in tRNAAsp and Lys31 is near C74. These different EF-P residues are grouped together in what we refer to as the acceptor loop, which structurally and functionally mimics the 3'-end of the acceptor stem of tRNA (Figure 2A and B).
Replacement of most amino acids involved in the polar contacts that are unique to the PoxA/EF-P complex (i.e. absent from the tRNAAsp/AspRS complex) have a minimal effect on aminoacylation activity. The only exceptions were R235A (discussed earlier) along with three other variants with modest effects on activity, H52A, D177A and E185A (Figure 2C). His52 contacts the acceptor loop of EF-P (with contacts to Lys31, Lys34, Gly35 and Gln36), but the other two amino acids from this group interact with amino acids outside the acceptor loop in a region that is not used by AspRS to contact tRNAAsp.
EF-P identity elements
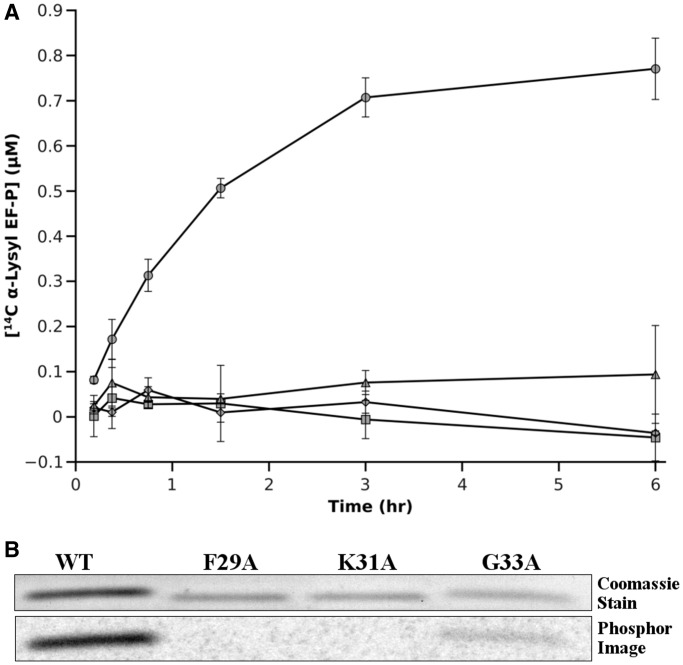
Replacement of most of the amino acids in PoxA involved in polar contacts with EF-P did not lead to any significant effect on the aminoacylation kinetics, with the exception of three positions that contact Phe29, Lys31, Gly33 and Lys34 in the acceptor loop. To confirm the relevance of these contacts, we replaced each of these amino acids with Ala in the EF-P acceptor loop and analyzed their effect on aminoacylation with 14C-α-lysine (Figure 3). Previous experiments have shown that substituting Lys34 with either Arg or Ala prevents aminoacylation of EF-P (13,14). Replacements of Phe29 and Lys31 with Ala also completely abolished PoxA aminoacylation, confirming their role as identity elements (Figure 3). Substitution of Gly33 with Ala also greatly diminished PoxA’s ability to modify EF-P, although some aminoacylation activity was retained. Taken together, our data indicate that residues Phe29, Lys31, Gly33 and Lys34 are the identity elements of EF-P for PoxA aminoacylation and that these amino acids are clustered in the acceptor loop. These contacts mimic those of the tRNA acceptor stem with the aaRS, while novel contacts elsewhere have at most a secondary role in this interaction. Although EF-P and tRNAs are composed of different monomers and bind their respective enzymes with different geometries (Supplementary Figure S2), our data indicate that the EF-P/PoxA complex evolved a molecular recognition strategy that is analogous to the one used by class IIb aaRSs, the evolutionary ancestors of PoxA.
Figure 3.
Replacement of F29, K31 or G33 with alanine prevents EF-P recognition by PoxA. WT and mutant EF-P were aminoacylated with PoxA using 14C-α-lysine. (A) Plot shows aminoacylation of WT (circle), F29A (square), K31A (rhombus) and G33A (triangle) EF-P with 14C-α-Lys. (B) After 6 h, aminoacylation samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. Top: coomassie stained gel; bottom: phosphor image of the gel.
Conservation of EF-P binding sites to PoxA and the ribosome
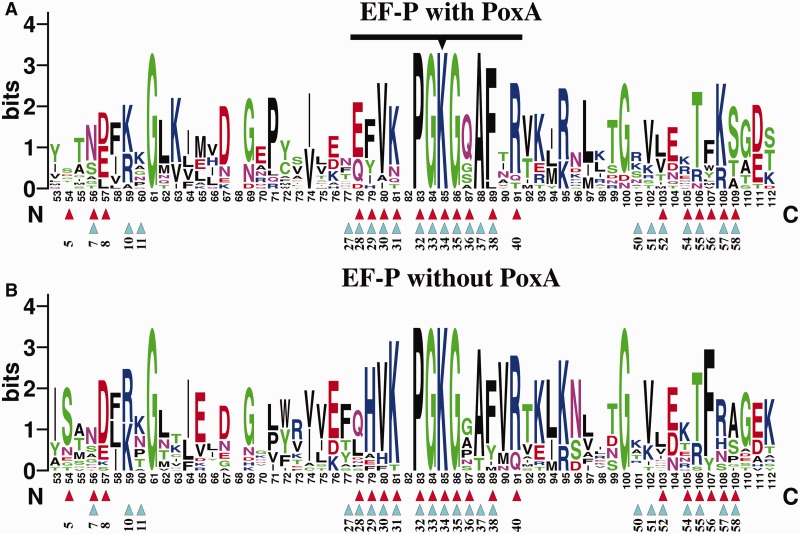
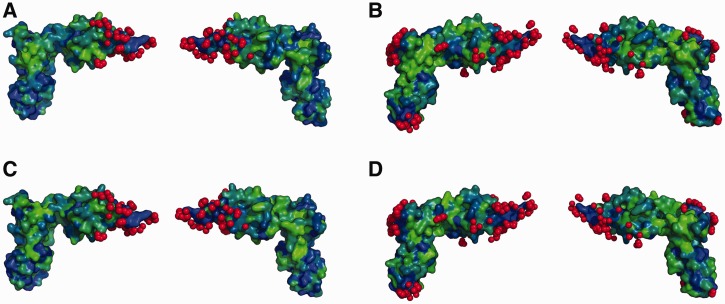
It is well established that positions in a protein that interact with other molecules tend to be more conserved, as their variability during evolution is constrained by the need to recognize their molecular partners, which imposes a strong selective pressure (21,30). Thus, one would expect in organisms where PoxA is present that the contacts made to EF-P would be well conserved, especially in the case of the four identity elements that are essential for EF-P aminoacylation. Similarly, it would be predicted that this would not be the case for EF-P from organisms that lack the poxA gene, as these positions are not under the same selective constrains. The acceptor loop and other PoxA contacting positions are well conserved in EF-P sequences from organisms that encode a poxA gene (Table 3 and Figures 4 and 5 and Supplementary Figures S4 and S5). The most conserved residues include the modification site (Lys34 following E. coli EF-P numbering) and some surrounding positions that are completely invariant (32, 33 and 35). Most contacts outside of the acceptor loop are also well conserved, with some minor differences observed for organisms that lack the poxA gene but retain a Lys at position 34. Position 29, one of the four EF-P identity elements, is usually Phe or Tyr in organisms that have the poxA gene, but it is frequently His in organisms that lack poxA. Other positions interacting with PoxA such as residues 28 and 36 of the acceptor loop or 57 and 58 distal from it show a similar pattern of conservation. Overall, most positions in EF-P contacting PoxA are strictly conserved, regardless of whether or not the corresponding organism has a poxA gene. This includes all the other identity elements (EF-P residues 31, 33 and 34), as well as several other contacting positions (e.g. residues 8, 30, 32, 38, 40 and 55). The strong conservation of these residues in EF-P, regardless of the presence of the poxA gene, suggests that there are other constrains on their variability beyond the need to bind PoxA. The most likely selective pressure that constrains variability of EF-P’s surface amino acids is the need to maintain functional interactions with the ribosome to prevent translational stalling. Most of the EF-P residues that contact PoxA, including all of the identity elements, also correspond to positions that interact with the ribosome or tRNA (Figures 4 and 5, Supplementary Figures S4 and S5), consistent with their high degree of conservation even in the absence of PoxA.
Table 3.
Conservation of the amino acid sequence of the acceptor loop from EF-P
| Positiona | Organisms with PoxA |
Organisms without PoxA |
||
|---|---|---|---|---|
| Variabilityc | Amino acid identityd | Variabilityc | Amino acid identityd | |
| Glu28 | 1.198 | Usually acid (71%) | 1.993 | Rarely acid (6%) |
| Phe29b | 1.611 | Usually Phe or Tyr (79%) | 0.952 | Usually His (82%) |
| Val30 | 0.735 | Usually Val (86%) | 0.834 | Usually Val (82%) |
| Lys31b | 1.292 | Usually Lys (71%) | 0.323 | Usually Lys (94%) |
| Pro32 | 0.000 | Pro (100%) | 0.000 | Pro (100%) |
| Gly33b | 0.000 | Gly (100%) | 0.000 | Gly (100%) |
| Lys34b | 0.000 | Lys (100%) | 0.000 | Lys (100%) |
| Gly35 | 0.000 | Gly (100%) | 0.000 | Gly (100%) |
| Gln36 | 1.484 | Usually Gln (71%) | 2.190 | Never Gln (0%) |
| Ala37 | 0.000 | Ala (100%) | 0.523 | Usually Ala (88%) |
| Phe38 | 0.371 | Usually Phe (93%) | 1.277 | Usually Phe or Tyr (88%) |
| Ala39 | 2.414 | Variable | 0.834 | Usually Val (82%) |
| Arg40 | 0.735 | Usually Arg (86%) | 0.672 | Usually Arg (82%) |
aPositions are indicated according to E. coli EF-P numbering. Positions that contact PoxA are highlighted in bold letters.
bIdentity elements for PoxA aminoacylation.
cShanon entropy was calculated as an index of variability using Protein Variability Server. A value of 0 indicates no variability at all, whereas 4.322 indicates maximum variability (equal probability to find any of the 20 amino acids) (21).
dPercentage of occurrence of the amino acids in each position is in parentheses.
Figure 4.
Conservation of EF-P residues involved in PoxA contacts. WebLogo representation of a fragment of the alignment of diverse EF-Ps that contain a lysine in the equivalent to the modification position. The top WebLogo (A) corresponds to an alignment of 14 EF-P sequences from organisms that have a poxA gene encoded in their genome. The bottom WebLogo (B) corresponds to an alignment of 17 EF-P sequences from organisms that do not have a poxA gene. The acceptor loop is marked with a black line, with the modification position highlighted with a triangle. Contacts to PoxA (based on pdb 3a5z) are marked with red triangles and contacts to the ribosome (pdb 3huw and 3hux) are indicated with cyan triangles. Numbering of the WebLogo positions corresponds to the full alignment, and corresponding positions on EF-P from E. coli are indicated below the triangles (for full alignments see Supplementary Figure S4).
Figure 5.
EF-P contacts with PoxA and the ribosome. Figure shows EF-P structures from pdb 3a5a-D (A and C) or 3huw-V (B and D). Atoms from PoxA (C and E) or the ribosome and tRNA (D and F) that contact EF-P are highlighted in red. Variable positions of the alignments (Figure 4 and Supplementary Figure S4) are highlighted on the EF-P surface for organisms with (C and D) or without (E and F) poxA. Variable positions are in green and non-variable are in blue.
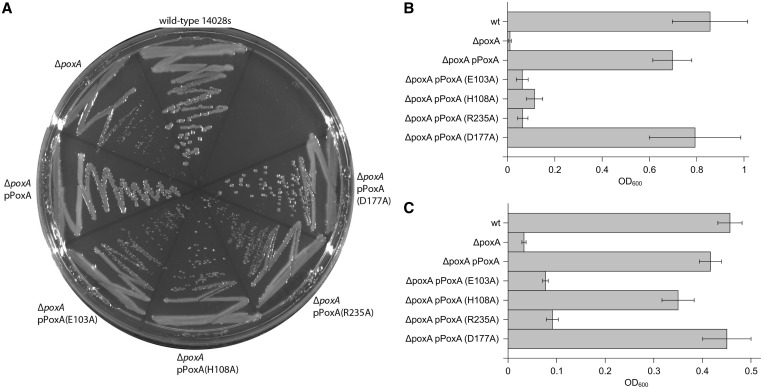
The ability of EF-P to functionally interact with both PoxA and the ribosome poses questions as to the origins of tRNA mimicry in this system. Amino acids contacting PoxA are concentrated in domain 1, and the results described above indicate that EF-P mimics only the tip of the tRNA acceptor stem in its interaction with PoxA. Conversely, amino acids contacting the ribosome are distributed throughout the three domains of EF-P (Figure 5 and Supplementary Figures S4 and S5), suggesting that the overall similarity to the tRNA structure was selected for optimal interaction with the ribosome. This hypothesis is supported by the fact that PoxA deletions have less severe phenotypes than EF-P deletions, indicating that unmodified EF-P retains some activity on the ribosome. For example, while both are clearly defective compared with WT, Δefp Salmonella strains grow more slowly and display a lower gentamicin MIC than isogenic ΔpoxA strains (9). Similarly, PoxA E103A and R235A that inactivate PoxA aminoacylation activity are unable to complement ΔpoxA strains, while transformation with a plasmid coding for PoxA D177A (that retains ∼50% of PoxA activity in vitro) complemented all the tested WT phenotypes. PoxA H108A restored only some of the phenotypes (growth on lauryl sulfobetaine) (Figure 6), indicating that it retains some activity, although we were unable to detect aminoacylation in vitro. These observations also support the hypothesis that emergence of PoxA aminoacylation activity in bacterial evolution could have gradually improved fitness. These results are in line with recent reports indicating that unmodified EF-P can stimulate translation of proteins with polyproline stretches, albeit with a much lower efficiency than the modified protein (10), further suggesting that EF-P may have originally functioned without post-translational modification.
Figure 6.
Complementation of Salmonella ΔpoxA with poxA variants. Figure shows complementation assays for phenotypes associated with poxA deletion mutants. Sensitivity to low osmolarity was assessed by the size of colonies grown on solid AB2 media (A), whereas sensitivity to gentamicin (3.125 µg/ml) (B) and lauryl sulfobetaine (6.25 mM) (C) was assessed as turbidity of cells grown on liquid media in presence of the corresponding compound.
DISCUSSION
Our results demonstrate that EF-P is recognized mainly through contacts with residues that mimic the 3′-end of the acceptor stem of tRNA. These amino acids are all localized in a loop of domain 1 at the amino terminus of EF-P that we have termed the acceptor loop, as it resembles the acceptor stem of tRNA in both structure and function. It is surprising that although EF-P closely resembles almost the complete structure of a tRNA, PoxA only relies on a small region of EF-P for its recognition (Figure 5 and Supplementary Figure S4). This is in contrast to the recognition of tRNA by a canonical aaRS where the contacts are spread throughout the entire tRNA (Figure 1) and identity elements are usually located at both the acceptor and anticodon stems (31). The only known exception is GluQRS, a paralog of the catalytic domain of GluRS that aminoacylates the queuosine-modified wobble base of tRNAAsp with glutamate rather than the acceptor stem of tRNAGlu (32,33). In this case, the tRNAAsp anticodon stem and loop mimic the acceptor stem of tRNAGlu and are recognized by GluQRS (34). Molecular docking experiments showed that GluQRS does not contact most of the tRNAs (34) and indicate that tRNAAsp nevertheless maintains a complete canonical tRNA shape due to the evolutionary constrains of its need to interact with AspRS, the translation machinery and several nucleotide-modifying enzymes. In a similar manner, the structure of EF-P is also constrained by the need to maintain its interactions not only with PoxA, but also with YfcM and the ribosome. Ribosome contacts are well conserved and distributed throughout the structure of EF-P (Figures 4 and 5 and Supplementary Figures S4 and S5), in contrast to contacts with PoxA that are confined to a subset of these residues (Figure 5 and Supplementary Figures S4 and S5), supporting the idea that functionality at the ribosome is the strongest determinant underlying the structural and functional mimicry of tRNA by EF-P. Furthermore, PoxA is mostly present in proteobacteria and scarcely found in other bacteria (35), suggesting that post-translation modification with (R)-β-Lys is a feature that appeared only recently in evolution to improve the affinity of EF-P for the ribosome. Deletion of poxA generates phenotypes less severe than an efp deletion, indicating that unmodified EF-P retains part of its activity on the ribosome. Additionally, recent reports show that the role of EF-P modification is mainly to enhance binding to the ribosome and that unmodified EF-P can still stimulate synthesis of proteins with polyproline stretches, although with a much lower efficiency than its aminoacylated version (10). The alternative hypothesis, where EF-P originally interacted with PoxA and interaction with the ribosome appeared later in evolution, is highly unlikely and unsupported by our data. Such a scenario would imply that independent of its binding to the ribosome, EF-P had an alternative unknown function that would have allowed for its retention during evolution. Additionally, the alternative hypothesis implies that all species except the proteobacteria lost poxA or that there was widespread lateral transfer of efp to all domains of life. Therefore, we favor the hypothesis that EF-P originally interacted with the ribosome, as it is the most parsimonious.
The modification of EF-P by PoxA is analogous to the modification of IF5a (the homolog of EF-P) with a hydroxylated polyamine by two enzymes, deoxyhypusine synthase and deoxyhypusine hydroxylase (36,37). The presence of two alternative pathways that produce a similar outcome highlights the relevance of EF-P modification, but at the same time raises the question of how EF-P functions in organisms that lack PoxA or deoxyhypusine synthase. In some cases, the lack of PoxA may be indicative of a lack of EF-P modification, which, in turn, may result from alternative mechanisms of ribosome binding or optimization of polyproline translation. In other instances, organisms may have instead evolved an alternative pathway for modifying EF-P to enhance binding to the ribosome, similar to the previously mentioned IF5a modification in archaea and eukaryotes.
Our data suggest that tRNA mimicry originally allowed EF-P to bind the ribosome, thereby increasing the fitness of certain organisms by facilitating synthesis of polyproline containing proteins. PoxA evolution from LysRS likely occurred as a secondary process facilitated by the fact that EF-P already had several key features similar to that of a tRNA. The emergence of the PoxA modification pathway would then have provided additional advantages, as modified EF-P is far more efficient in stimulating translation of polyproline stretches than the unmodified form (10). It is critical that PoxA concurrently lost the ability to recognize the canonical substrates of LysRS to prevent synthesis of (R)-β-Lys-tRNA, which would not be a substrate for translation (38,39). Taken together, these and previous data illustrate the precise mechanism by which molecular mimicry can be used to enhance protein synthesis, and thus, the fitness of the cell.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM065183 to M.I.]; Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN) [386286-10 to W.W.N.]; Vanier Canada Graduate Scholarship from NSERC (to S.B.Z.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Medha Raina and Andrei Rajkovic for helpful discussions.
REFERENCES
- 1.Pasteur G. A classificatory review of mimicry systems. Annu. Rev. Ecol. Syst. 1982;13:169–199. [Google Scholar]
- 2.Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 2009;7:787–797. doi: 10.1038/nrmicro2222. [DOI] [PubMed] [Google Scholar]
- 3.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene JD. RNA surfaces as functional mimetics of proteins. Chem. Biol. 1996;3:505–513. doi: 10.1016/s1074-5521(96)90139-8. [DOI] [PubMed] [Google Scholar]
- 5.Nissen P, Kjeldgaard M, Nyborg J. Macromolecular mimicry. EMBO J. 2000;19:489–495. doi: 10.1093/emboj/19.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsonis PA, Dwivedi B. Molecular mimicry: structural camouflage of proteins and nucleic acids. Biochim. Biophys. Acta. 2008;1783:177–187. doi: 10.1016/j.bbamcr.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura Y, Ito K. tRNA mimicry in translation termination and beyond. Wiley Interdiscip. Rev. RNA. 2011;2:647–668. doi: 10.1002/wrna.81. [DOI] [PubMed] [Google Scholar]
- 8.Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou SB, Hersch SJ, Roy H, Wiggers JB, Leung AS, Buranyi S, Xie JL, Dare K, Ibba M, Navarre WW. Loss of elongation factor P disrupts bacterial outer membrane integrity. J. Bacteriol. 2012;194:413–425. doi: 10.1128/JB.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 11.Hersch SJ, Wang M, Zou SB, Moon K-M, Foster LJ, Ibba M, Navarre WW. Divergent protein motifs direct elongation factor p-mediated translational regulation in Salmonella enterica and escherichia coli. MBio. 2013;4:e00180–13. doi: 10.1128/mBio.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 13.Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat. Struct. Mol. Biol. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 14.Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat. Chem. Biol. 2011;7:667–669. doi: 10.1038/nchembio.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peil L, Starosta AL, Virumäe K, Atkinson GC, Tenson T, Remme J, Wilson DN. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat. Chem. Biol. 2012;8:695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- 16.Bullwinkle TJ, Zou SB, Rajkovic A, Hersch SJ, Elgamal S, Robinson N, Smil D, Bolshan Y, Navarre WW, Ibba M. (R)-β-lysine-modified elongation factor P functions in translation elongation. J. Biol. Chem. 2013;288:4416–4423. doi: 10.1074/jbc.M112.438879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice P, Longden I, Bleasby A. EMBOSS: the european molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 2008;36:W35–W41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cusack S, Yaremchuk A, Tukalo M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996;15:6321–6334. [PMC free article] [PubMed] [Google Scholar]
- 23.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullwinkle TJ, Ibba M. Emergence and evolution. Top. Curr. Chem. 2013 doi: 10.1007/128_2013_423. March 12 (doi:10.1007/128_2013_423; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. In vitro study of E.coli tRNA(Arg) and tRNA(Lys) identity elements. Nucleic Acids Res. 1992;20:2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Commans S, Lazard M, Delort F, Blanquet S, Plateau P. tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases. J. Mol. Biol. 1998;278:801–813. doi: 10.1006/jmbi.1998.1711. [DOI] [PubMed] [Google Scholar]
- 27.Eriani G, Gangloff J. Yeast aspartyl-tRNA synthetase residues interacting with tRNA(Asp) identity bases connectively contribute to tRNA(Asp) binding in the ground and transition-state complex and discriminate against non-cognate tRNAs. J. Mol. Biol. 1999;291:761–773. doi: 10.1006/jmbi.1999.3012. [DOI] [PubMed] [Google Scholar]
- 28.Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. USA: Landes Bioscience/: Eurekah.com. Georgetown, TX; 2005. [Google Scholar]
- 29.Wilkinson AJ, Fersht AR, Blow DM, Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983;22:3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- 30.Lovell SC, Robertson DL. An integrated view of molecular coevolution in protein-protein interactions. Mol. Biol. Evol. 2010;27:2567–2575. doi: 10.1093/molbev/msq144. [DOI] [PubMed] [Google Scholar]
- 31.Saks ME, Sampson JR, Abelson JN. The transfer RNA identity problem: a search for rules. Science. 1994;263:191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- 32.Dubois DY, Blaise M, Becker HD, Campanacci V, Keith G, Giegé R, Cambillau C, Lapointe J, Kern D. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp. Proc. Natl Acad. Sci. USA. 2004;101:7530–7535. doi: 10.1073/pnas.0401634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salazar JC, Ambrogelly A, Crain PF, McCloskey JA, Söll D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc. Natl Acad. Sci. USA. 2004;101:7536–7541. doi: 10.1073/pnas.0401982101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaise M, Becker HD, Keith G, Cambillau C, Lapointe J, Giegé R, Kern D. A minimalist glutamyl-tRNA synthetase dedicated to aminoacylation of the tRNAAsp QUC anticodon. Nucleic Acids Res. 2004;32:2768–2775. doi: 10.1093/nar/gkh608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailly M, de Crécy-Lagard V. Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol. Direct. 2010;5:3. doi: 10.1186/1745-6150-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolff EC, Kang KR, Kim YS, Park MH. Posttranslational synthesis of hypusine: evolutionary progression and specificity of the hypusine modification. Amino Acids. 2007;33:341–350. doi: 10.1007/s00726-007-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. eIF5A promotes translation of polyproline motifs. Mol. Cell. 51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartman MCT, Josephson K, Lin CW, Szostak JW. An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS One. 2007;2:e972. doi: 10.1371/journal.pone.0000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilreath MS, Roy H, Bullwinkle TJ, Katz A, Navarre WW, Ibba M. β-Lysine discrimination by lysyl-tRNA synthetase. FEBS Lett. 2011;585:3284–3288. doi: 10.1016/j.febslet.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.