Abstract
The Y-family DNA polymerase Rev1 is required for successful replication of G-quadruplex DNA (G4 DNA) in higher eukaryotes. Here we show that human Rev1 (hRev1) disrupts G4 DNA structures and prevents refolding in vitro. Nucleotidyl transfer by hRev1 is not necessary for mechanical unfolding to occur. hRev1 binds G4 DNA substrates with Kd,DNA values that are 4–15-fold lower than those of non-G4 DNA substrates. The pre-steady-state rate constant of deoxycytidine monophosphate (dCMP) insertion opposite the first tetrad-guanine by hRev1 is ∼56% as fast as that observed for non-G4 DNA substrates. Thus, hRev1 can promote fork progression by either dislodging tetrad guanines to unfold the G4 DNA, which could assist in extension by other DNA polymerases, or hRev1 can prevent refolding of G4 DNA structures. The hRev1 mechanism of action against G-quadruplexes helps explain why replication progress is impeded at G4 DNA sites in Rev1-deficient cells and illustrates another unique feature of this enzyme with important implications for genome maintenance.
INTRODUCTION
Studies in several organisms indicate that G-quadruplex DNA (G4 DNA) forms in vivo, with important biological ramifications (1–7). G4 DNA sequences form highly stable structures (8), and motifs predicted to form G4 DNA structures are positioned non-randomly in functional regions of prokaryotic, archaeal and eukaryotic genomes (9). Effective G4 DNA maintenance depends on the action of several helicases such as Pif1, FANCJ and RecQ family members (10–18). G4 DNA sequences are converted to single-stranded (ss) intermediates during DNA replication. Kinetic analyses indicate that at least some intramolecular G4 DNA structures can refold on the millisecond timescale (19), and a recent study found that G-quadruplexes appear to be most abundant during S-phase (5). Thus, it is likely that intramolecular G-quadruplexes can spontaneously refold in cells and are encountered intermittently by DNA polymerases (pols) during replication. Refolding of the G-quadruplex is undoubtedly a complex event, but once accomplished, it would be expected to hinder fork progression. Studies in Saccharomyces cerevisiae have shown that ribosomal DNA (rDNA) repeats, which are enriched in G4 DNA motifs are efficient blocks to replication fork progression in unperturbed wild-type cells and that yeast requires Pif1-family helicases for Pol2 to move through these sites (16,20–23). Rigorous examination of G4 DNA replication by different pol families is lacking, as it would seem at face value to be primarily an issue left to the realm of single-stranded DNA (ssDNA) binding proteins and helicases. Importantly, recent reports have implicated so-called translesion DNA synthesis (TLS) pols in the biology of G4 DNA replication, as depletion of three of the four Y-family pol members has been shown to cause defects in G4 DNA maintenance (24–27). Of particular interest is Rev1, an enzyme with extraordinary structural features and functional properties that has been implicated in G4 DNA replication dynamics.
Rev1 is conserved from yeast to humans and plays an important role in both DNA repair and damage tolerance pathways (24–26). The Rev1 enzyme possesses an absolutely unique mechanism of nucleotide selection in which a protein side-chain from the so-called N-digit ejects the template base from the pol active site (27). In human Rev1 (hRev1), the side-chain of Leu358 displaces the template strand and the guanidinium moiety of Arg357 hydrogen bonds with the incoming deoxycytidine triphosphate (dCTP) (28). The displaced template base is positioned in a region formed by multiple domains of the enzyme called the ‘G-loop’ (27,28). The protein-template mechanism is also observed in yeast and is associated with nucleotide insertion across from DNA adducts (27,29). As such, Rev1 is primarily a deoxycytidyl transferase due to the favorable H-bonding between Arg357 and dCTP, but the enzyme can catalyze insertion of other 2′-deoxy-nucleoside triphosphates (dNTPs) under certain in vitro conditions (30). Recent studies have shown that DT40 cells require Rev1 for successful replication of G4 DNA sites, and that this process is coordinated to some degree with the action of the FANCJ helicase (31,32). The structural properties of Rev1 and the purported role of the enzyme in G4 DNA maintenance led us to hypothesize that hRev1 may actively aid in the unfolding of G4 DNA structures by sequestering tetrad-associated guanines in the G-loop, thereby disrupting the G4 tetrad.
MATERIALS AND METHODS
Materials
All chemicals were molecular biology grade or better. dNTPs were obtained from GE Healthcare Life Sciences Biosciences (Piscataway, NJ, USA). All oligonucleotides used in this work were synthesized by Integrated DNA Technologies (Coralville, IA, USA) and purified using high-performance liquid chromatography by the manufacturer, with analysis by matrix-assisted laser desorption time-of-flight MS. Dimethyl sulfate (DMS) was purchased from Sigma-Aldrich (St. Louis, MO, USA) and is a suspected carcinogen. Both the liquid and vapor forms of DMS are extremely severe irritants to the skin, eyes and mucous membranes. All DMS footprinting experiments were performed in a fume hood and extreme caution should be exercised when performing experiments with DMS.
DNA substrate preparation
All oligonucleotide stock solutions were resuspended in 50 mM HEPES (pH 7.5) buffer. Primer–template substrates were prepared in 50 mM HEPES (pH 7.5) buffer containing KCl (either 40 mM or 100 mM) by adding the primer complement (1:2, primer:template molar ratio) where indicated, heating the sample to 95°C for 5 min and then slow cooling to room temperature. Where indicated, KCl was omitted for ‘no salt’ experiments. DNA substrates were not frozen after re-annealing because we consistently observed a reduction in fluorescent changes for the stopped-flow assays if frozen aliquots were simply thawed and then used in the experiment. Single-strand DNA substrates were heated and slow cooled in an identical manner to the primer–template DNA.
Protein expression and purification
The core polymerase domain of hRev1 (residues 330–833) was expressed in bacteria. The His-GST-hRev1330−833 fusion protein was expressed in Escherichia coli BL21 (DE3) Gold cells (Agilent Technologies, Santa Clara, CA, USA). Cells were grown at 37°C and 250 rpm for 3 h (OD600 = 0.5–0.6), followed by induction for 3 h (37°C and 250 rpm) by addition of isopropyl β-d-1-thiogalactopyranoside (1 mM) and finally harvested by centrifugation. Buffer containing 50 mM Tris-HCl (pH 7.4), 0.5 M NaCl, 10% glycerol (v/v), 5 mM β-mercaptoethanol (β-ME), lysozyme (1 mg/ml) and a protease inhibitor cocktail (Roche, Basel, Switzerland) was added to the harvested pellet. The suspension was sonicated and supernatant recovered from an ultracentrifugation step (35 000 g, 1 h, 4°C). The protein was purified by two affinity steps using Ni Sepharose (GE Healthcare Life Sciences) followed by Gluthatione Sepharose 4B beads (GE Healthcare Life Sciences). Briefly, the protein was bound to a nickel-chelating column in 50 mM Tris-HCl (pH 7.4) buffer containing 0.5 M NaCl, 10% glycerol (v/v) and 5 mM β-ME. The column was washed with 40–60 mM imidazole and the protein eluted as a single peak in 400 mM imidazole. Following dialysis to remove imidazole and lower the concentration of NaCl to 0.15 M, the protein was added to the GST column in 50 mM Tris-HCl (pH 7.4) buffer containing 0.15 M NaCl, 10% glycerol (v/v) and 5 mM β-ME. After washing, the protein was then cleaved from the His-GST tag by treatment with PreScission protease (GE Healthcare Life Sciences) on the GST column, according to the methods suggested by the manufacturer. The final hRev1330−833 stock solution was >95% pure. The highly pure hRev1330−833 core protein was stored at −80°C in 50 mM HEPES buffer (pH 7.5) containing 0.1 M NaCl, 5 mM β-ME and 30% glycerol, at a final concentration of near 100 µM.
DMS footprinting assays
Template oligonucleotides with a 6-carboxyfluorescein (FAM) label at the 5′-end were used to monitor G4 DNA formation by DMS footprinting. The 42-mer Myc2345 G1423T sequence was 5′-/FAM/TGAGGGTGGGTAGGGTGGGTGCGTCTGCGGCTGGCTCGAGGC-3′. The 23-mer primer sequence was 5′-GCCTCGAGCCAGCCGCAGACGCA-3′. The oligonucleotides were resuspended in 50 mM HEPES (pH 7.5) buffer containing the indicated concentrations of KCl (either no, 40 mM or 100 mM). The primer:template molar ratio was 1:2. As before, the substrates (both ss-42-mer and primer-template) were heated to 95°C for 5 min and then slow cooled to room temperature. The DNA substrates (5 pmoles) were incubated in 50 mM HEPES (pH 7.5) buffer containing KCl (where indicated), 0.1 µg salmon sperm DNA and DMS (1 µl of DMS:ethanol, 4:1, v/v). The final volume of the reaction was 72 µl. The reaction was allowed to proceed for 10 min at room temperature. The reaction was quenched with 18 µl of stop buffer (1.5 M sodium acetate, pH 7.0, 1 M βME). Following ethanol precipitation, the samples were centrifuged under vacuum to dryness, and the DNA pellet was resuspended in 90 µl of 1 M piperidine. The reaction was incubated at 95°C for 30 min and then placed on ice for 2 min. The ethanol precipitation step was repeated and the resulting pellet resuspended in 100 µl of dH2O. The samples were centrifuged under vacuum to dryness and then resuspended in 10 µl of loading dye [95% formamide (v/v)/20 mM ethylenediaminetetraacetic acid (EDTA)/0.01% (w/v) bromophenol blue]. The samples were heated at 95°C for 5 min, and 4 µl (∼2 pmoles) was separated by electrophoresis on a 12% (w/v) polyacrylamide/7 M urea denaturing gel. To optimize band separation, the samples were run overnight at constant power (2W). All gel imaging was performed with a Typhoon imaging system (GE Life Sciences). The relative reactivity at each nucleotide on the DNA substrate was calculated by quantifying the intensity of each band in the lane, then dividing that value by the total lane intensity and multiplying by 100 to yield a percentage.
Measurement of DNA binding affinity by fluorescence polarization
Four DNA substrates were prepared for DNA binding assays. The sequence of the 28-mer G4 DNA template was 5′-/FAM/TGGGTGGGTAGGGTGGGTAGGGGAGGAT-3′ and the 28-mer non-G4 DNA template was 5′-/FAM/TATGTGACTATGCTGGGTAGGGGAGGAT-3′. The sequence of the unlabeled primer was 5′-ATCCTCCCCTA-3′. Substrates were prepared as described above. DNA binding affinity was measured by incubating DNA substrates (1 nM) labeled at the 5′-terminus with FAM with varying concentrations of hRev1330−833 (0–4 µM). Fluorescence polarization was measured in a Biotek SynergyH4 plate reader using the appropriate filter sets (λex = 485 ± 20 nm and λem = 525 ± 20 nm). All titrations were performed at 25°C in 50 mM HEPES buffer (pH 7.5) containing 10 mM KOAc, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM β-ME and 0.1 mg/ml bovine serum albumin (BSA). Polarization was determined using Equation 1:
 |
(1) |
where F|| equals fluorescence intensity parallel to the excitation plane and F⊥ equals the fluorescence intensity perpendicular to the excitation plane. The resulting change in polarization units was plotted against protein concentration and fit to a quadratic equation (equation 2).
 |
(2) |
where P = the measured change in fluorescence polarization, P0 = minimum polarization value measured, Pmax = maximum polarization value measured, Etotal = the concentration of enzyme, DNAtotal = the concentration of FAM-labeled DNA and Kd is the measured equilibrium dissociation constant for enzyme binding to DNA. The binding curves were fit such that DNAtotal was fixed to be constant (1 nM).
Fluorescence quenching assay to monitor G4 DNA folding and unfolding
The sequence of the 42-mer Myc2345 G1423T oligonucleotide used to monitor G4 DNA dynamics was 5′-/FAM/TGAGGGTGGGTAGGGTGGGTGCGTCTGCGGCTGGCTCGAGGC/Dab/-3′. An 18/42-mer primer-template DNA substrate was used for some G4 DNA unfolding experiments. The primer sequence was 5′-GCCTCGAGCCAGCCGCAG-3′. Stopped-flow experiments were performed on an SX.18MV instrument (Applied Photophysics) maintained at 25°C by a circulating water bath. Changes in FAM fluorescence over time were measured after a 515-nm-cutoff filter (Newport Optical Filter #51294) with excitation at 495 nm through 1-mm slits. G4 DNA folding/unfolding experiments were performed at 25°C in 10 mM Tris (pH 7.4 at 25°C) buffer containing KCl at the indicated concentrations. For the G4 DNA-folding reactions, both KCl and hRev1 were incubated in a separate syringe from the DNA substrate. For the G4 DNA unfolding reactions, KCl was included in both syringes. Where indicated, hRev1330−833, MgCl2 and dCTP were incubated in a separate syringe from the DNA. The final concentrations of MgCl2 and dCTP were 2 mM and 50 µM, respectively. The final concentration of DNA in the reaction was 20 nM, and hRev1330−833 concentration was varied as indicated. The resulting curves represent the average of two to four experiments. The G4 DNA-folding reactions were fit to Equation 3, which describes a single-exponential decay:
| (3) |
The fluorescence changes observed for G4 DNA unfolding were fit to Equation 4:
 |
(4) |
where A = amplitude of the change in fluorescence signal, n = number of steps to form product, kobs = observed rate of product formation and k2 = linear phase. For data sets exhibiting a lag in product formation, a two-step exponential equation (n = 2) was used.
Steady-state kinetic analysis of dCTP insertion by hRev1330−833
Single nucleotide insertion by hRev1330−833 was measured over a range of dCTP concentrations. Three primer–template DNA substrates were used for each 42-mer template strand for six substrates: (i) 19/42-mer non-G4 DNA, (ii) 21/42-mer non-G4 DNA, (iii) 23/42-mer non-G4 DNA, (iv) 19/42-mer G4 DNA, (v) 21/42-mer G4 DNA and (vi) 23/42-mer G4 DNA. The 42-mer non-G4 DNA sequence was 5′-GTGAGATGTTGACCATGGGTGCGTCTGCGGCTGGCTCGAGGC-3′. The sequence of the 42-mer G4 DNA sequence was 5′-TGAGGGTGGGTAGGGTGGGTGCGTCTGCGGCTGGCTCGAGGC-3′. The sequence of the 19-mer primer was 5′-/FAM/-TTTGCCTCGAGCCAGCCGCAGA-3′. The sequence of the 21-mer primer was 5′-/FAM/-TTTGCCTCGAGCCAGCCGCAGACG-3′. The sequence of the 23-mer primer was 5′-/FAM/-TTTGCCTCGAGCCAGCCGCAGACGCA-3′. The FAM-labeled primer was annealed to the template in a 1:2 ratio.
To obtain estimates for Michaelis–Menten kinetic parameters, the DNA substrate (200 nM) was pre-incubated with hRev1330−833 (10 nM), and the reactions were initiated by addition of dCTP (0–250 µM) and MgCl2 (5 mM). All the enzymatic reactions were carried out at 37°C in 40 mM HEPES buffer (pH 7.5) containing 100 mM KCl, 5 mM dithiothreitol, 0.1 mg ml−1 BSA and 5% (v/v) glycerol. At the desired time points, 15 μl of reaction was quenched with 85 μl of 95% formamide (v/v), 20 mM EDTA and 0.1% bromophenol blue (w/v) solution and were separated by electrophoresis on a 12% polyacrylamide (w/v)/7 M urea gel. The products were then visualized using a Typhoon imager (GE Healthcare Life Sciences) and quantified using ImageJ software (33). The rate was plotted as a function of dCTP concentration and fit to the Michaelis–Menten equation in Prism (GraphPad, San Diego, CA, USA). The turnover number (kcat) and Michaelis constant (KM,dNTP) are reported. To allow hRev1330−833 to perform multiple insertions opposite the first three template guanines, the DNA substrate (200 nM) was pre-incubated with hRev1330−833 (5 nM), and the reactions were initiated by addition of dCTP (250 µM) and MgCl2 (5 mM). Time points were taken out to 1 h, processed and visualized as described above. In this way, we were able to observe multiple insertion events by hRev1330−833 on both non-G4 and G4 DNA.
Pre-steady-state kinetic analysis of hRev1-catalyzed insertion of dCTP
Pre-steady-state experiments were performed using a KinTek RQF-3 model chemical quench-flow apparatus (KinTek Corp., Austin, TX, USA) maintained at 37°C by a circulating water bath. Assays were carried out in 40 mM HEPES buffer (pH 7.5) containing 100 mM KCl, 5 mM dithiothreitol, 0.1 mg ml−1 BSA and 5% (v/v) glycerol. hRev1330−833 (400 nM) was pre-incubated with 23/42-mer FAM-labeled primer-template DNA (100 nM). The reaction was initiated by rapid mixing of the enzyme·DNA solution with a solution containing MgCl2 (5 mM) and dCTP (500 µM). Polymerase catalysis was stopped by the addition of 200 mM EDTA (pH 8.5) over a range of time points (0.005–8 s). Product formation was analyzed as described previously for steady-state assays and the resulting curves were fit to Equation 3.
RESULTS
Binding of hRev1 exposes tetrad-guanines
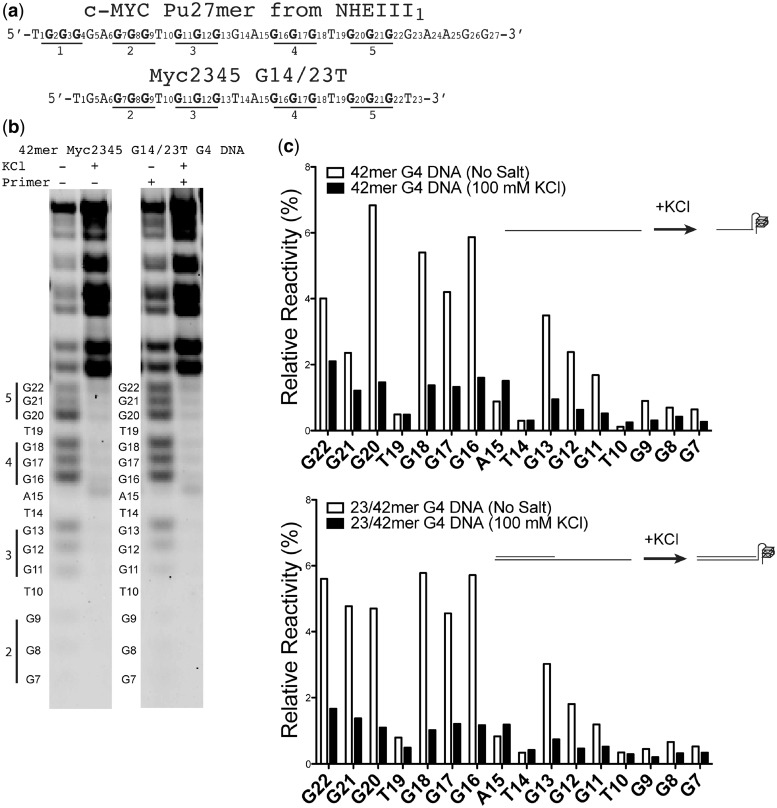
We chose the G4 DNA-forming sequence from the promoter of the c-MYC proto-oncogene as our in vitro model for understanding fundamental biochemical properties of G4 DNA replication by hRev1 (Figure 1 and Supplementary Table S1). A major advantage of using the c-MYC promoter to study G4 DNA replication is the abundant literature critically examining structural properties of this G-quadruplex-forming sequence (34–37), which permits the study of hRev1 catalysis against a substrate adopting a single G4 DNA fold, as opposed to a conformationally heterogeneous substrate. The 27-nt purine-rich (Pu27mer) NHEIII element in the c-MYC promoter contains five tracts of guanines that can adopt several G-quadruplex loop isomers (Figure 1a). The major G-quadruplex loop isomer formed by the c-MYC Pu27mer in K+ solution involves the second, third, fourth and fifth runs of guanines (Myc2345). The Myc2345 sequence forms an intramolecular G-quadruplex comprising three stacked G4 tetrads that are separated by three loops of 1–2 nt. Strategic mutations in Myc2345 can isolate at least four discrete loop isomers, all of which form parallel-stranded G4 DNA structures. We chose to study the most stable loop isomer identified so far: Myc2345 G14/23T (Tm = 74°C) (35), where the numbers 14 and 23 correspond to the position of the bases that are mutated from the wild-type Pu27mer sequence (e.g. G14 and G23 are both mutated to T).
Figure 1.
The c-MYC promoter sequence forms G4 DNA in vitro. (a) The c-MYC promoter contains a 27-nt sequence (Pu27mer) with five runs of tandem guanines (1–5) that can form several G4 DNA structures. Tetrad-associated guanines are marked in bold, and the Pu27mer numbering scheme is in accordance with previous reports in the literature. A mutant of the wild-type Pu27mer, the Myc2345 G14/23T loop isomer, was chosen for our study of hRev1 activity on G4 DNA substrates. (b) DMS footprinting was performed with Myc2345 G14/23T 42-mer DNA in the presence or absence of an annealed 23-mer primer. Tetrad-associated guanines are protected from reaction with DMS on addition of potassium, indicative of G4 DNA folding. The addition of a primer does not alter the protection afforded to the G4 DNA-forming sequences. The nucleotide numbering scheme depicted to the left of each gel follows that of the Pu27mer (a). (c) Quantification of template base reactivity to DMS for experiments performed with each substrate either with KCl (100 mM, black bars) or without KCl (white bars) reveals a strong protection of guanines predicted to participate in the G4 DNA structure, indicating that both G4 DNA substrates (i.e. with or without 23-mer primer) behave as expected. A cartoon illustration of the DNA structures that depicts our interpretation of the experimental results is shown for clarity.
Before performing experiments with hRev1, we confirmed the stable formation of the G-quadruplex structures in our substrates by DMS footprinting assays and circular dichroism (CD) spectroscopy. G-quadruplexes display characteristic spectral properties when analyzed by CD (38,39). DMS is most reactive with the N7 atom of guanine (40) and is often used to monitor formation of the G-quadruplex structures, as tetrad formation protects guanine from this reaction. As expected, we observe a clear salt-dependent protection from DMS reactivity for the guanines predicted to participate in the G4 DNA structures (Figure 1b), and CD reveals spectral properties characteristic of G4 DNA formation (Supplementary Figures S1 and S2). The protection from DMS reactivity is observed for both ssDNA and primer–template substrates. There does appear to be a reduction in the amount of DNA recovered in the DMS experiments performed without salt. On average, we find that the total amount of DNA recovered from DMS reactions without salt is ∼55–67% of that obtained with salt (either 40 mM or 100 mM K+) when the experiments are performed side by side. However, the conclusions drawn from the DMS footprinting experiments do not depend on the total amount of DNA loaded on the gel, as the relative reactivity for each band is calculated as a percentage of the total signal in the lane. Quantification of the gels reveals that G22-G20 and G18-G16 are ∼2–3-fold less reactive in the presence of K+ (Figure 1c). The other tetrad guanines (G13–G11 and G9–G7) are also protected, but the amount of product formed in the DMS reaction is lower, which makes quantitative comparisons more tenuous. Still, it is clear that the addition of K+ affords significant protective effects to the guanines predicted to participate in G4 DNA formation.
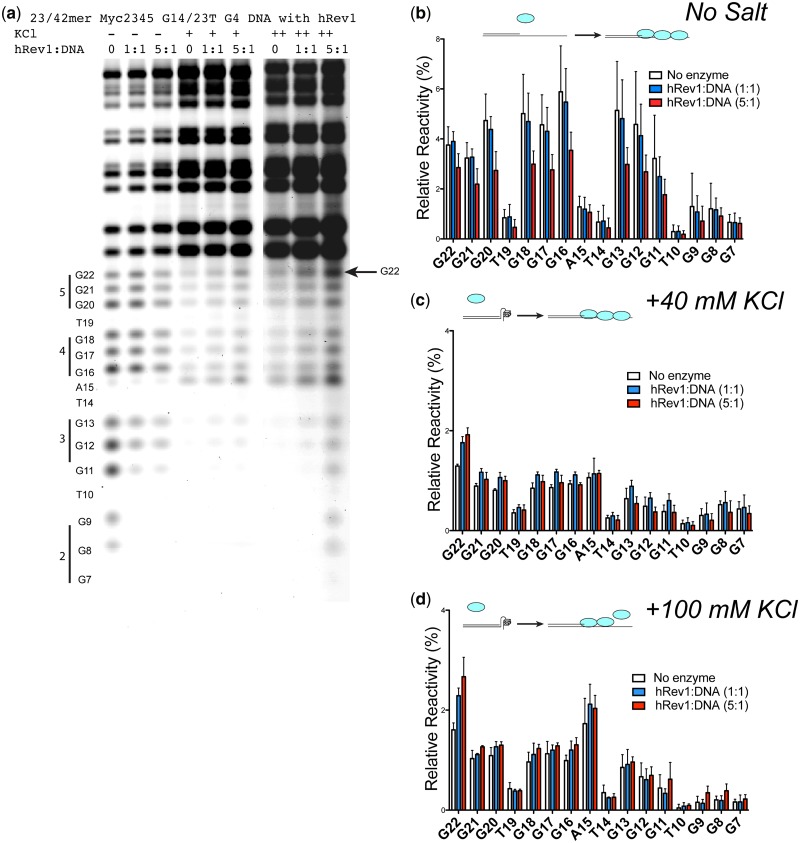
We then tested whether addition of hRev1330−833 altered the cleavage pattern for the Myc2345 G14/23T substrate under conditions that do not promote G4 DNA formation. When hRev1330−833 is incubated with 23/42-mer G4 DNA in the absence of K+ (i.e. no G4 DNA formed), there is a modest protective effect all along the template strand (Figure 2a and quantified in Figure 2b). There is a slight increase in reactivity for G22 and G21, near the primer–template junction, on addition of a 1:1 hRev1330−833:DNA molar ratio. However, this difference is not statistically significant (P = 0.67). At a 5:1 hRev1330−833:DNA molar ratio, both G22 and G21 are protected, and when compared with the no enzyme control, the difference is statistically significant (P = 0.037 and 0.015 for G22 and G21, respectively). The protective effect of hRev1 is even more pronounced for guanines G20–G7 (e.g. P = 0.0037 when comparing the results for the no-enzyme control and 5:1 hRev1330−833:DNA experiments at position G20). We conclude from the no-salt DMS foot-printing experiments that hRev1 can simultaneously bind and protect guanines at both the primer–template junction and on the ssDNA portion of the substrate.
Figure 2.
hRev1330-833 alters the pattern of DMS reactivity for G4 DNA-forming sequences. (a) Primer–template G4 DNA substrate (23/42mer, 5 pmoles) was incubated with hRev1330-833 (5 and 25 pmoles) either without salt (−) or in the presence of KCl (40 mM, +; 100 mM, ++), and DMS footprinting was performed. (b) Quantification of template base reactivity for experiments performed without salt (a) either in the absence of enzyme (‘white’ bars) or in the presence of hRev1330–833 (5 pmoles, ‘blue’ bars; 25 pmoles, ‘red’ bars) reveals that hRev1330–833 binds to unstructured ssDNA and attenuates reactivity with DMS. The mean ± SD is shown (n = 6). (c) Quantification of template base reactivity for experiments performed with KCl (40 mM, a) either in the absence of enzyme (‘white’ bars) or in the presence of hRev1330–833 (5 pmoles, ‘blue’ bars; 25 pmoles, ‘red’ bars) reveals that the tetrad-associated guanine (G22) at the extreme 3'′-terminus is deprotected on addition of hRev1330–833. The mean (± SD is shown (n = 3). (d) Quantification of template base reactivity for experiments performed with KCl (100 mM, a) either in the absence of enzyme (‘white’ bars) or in the presence of hRev1330–833 (5 pmoles, ‘blue’ bars; 25 pmoles, ‘red’ bars) reveals that the tetrad-associated guanines in the template strand are deprotected on addition of hRev1330–833. The mean ± SD is shown (n = 3). A cartoon illustration of the DNA structures that depicts our interpretation of the effect of hRev1330–833 binding is shown for clarity.
We then examined the effect of hRev1 binding on the stability of G4 DNA structures by performing DMS footprinting experiments at two concentrations of K+ (40 and 100 mM). These experiments were performed in the absence of metal ions and dNTPs. Incubating the 23/42-mer G4 DNA substrate with hRev1330−833 in the presence of 40 mM K+ leads to a reproducible and pronounced increase in the reactivity of G22, the tetrad-associated guanine at the extreme 3′-end of the G4 DNA-forming sequence, consistent with disruption of the G4 tetrad on binding of hRev1 (Figure 2a and c). A similar effect is observed at 100 mM K+ (Figure 2a and d). The change in G22 reactivity attributable to hRev1 is statistically significant at both concentrations of K+ (e.g. P = 0.0033 and 0.0045 when comparing the reactivity of G22 for no enzyme control with the 1:1 hRev1330−833:DNA experiment at 40 mM and 100 mM K+, respectively). These results suggest that binding of hRev1330−833 is sufficient to remove G22 from the first tetrad of the c-MYC G4 DNA structure. The crystal structure of hRev1 in complex with template dG predicts that interactions with the G-loop maintain the base in an ‘extrahelical path’ outside of the enzyme active site (28), which may account for the most pronounced change in guanine reactivity being observed at G22.
Comparing the reactivity of the other tetrad-associated guanines at either 40 or 100 mM K+ reveals a more complex pattern. For the 1:1 hRev1330−833:DNA experiments, there is a general trend for increased reactivity at both salt concentrations. However, at the lower salt concentration (40 mM), adding more protein attenuates the reactivity for guanines G21–G7 (Figure 2c), indicative of a protective effect on the ssDNA template similar to what we observe for the no-salt experiments (Figure 2b). Increasing the K+ to 100 mM results in a trend toward increased reactivity of guanines at both 1:1 and 5:1 hRev1330−833:DNA, especially for G11–G7 (Figure 2d). Thus, at higher salt concentrations, the unfolded tetrad-guanines are not as protected by the ability of hRev1 to bind ssDNA. We were intrigued by these findings and decided to examine hRev1330−833 interactions with G4 DNA substrates in more detail.
hRev1 preferentially binds G4 DNA
Next, we used fluorescence polarization to measure the affinity of hRev1330−833 for G4 DNA and non-G4 DNA substrates. hRev1330−833 was titrated into a solution containing one of four fluorescently-labeled DNA substrates: (i) ss-non-G4 DNA, (ii) primer–template non-G4 DNA, (iii) ss-G4 DNA or (iv) primer–template G4 DNA. Rev1 is known to bind ssDNA, a property that does not appear to be shared with other Y-family members, such as hpol η (41). Analysis of fluorescence polarization assays revealed that hRev1330−833 binds ss-G4 DNA-containing substrates with ∼4-fold greater affinity than non-G4 DNA control ssDNA (Figure 3a). The effect is even more pronounced for primer–template substrates, where hRev1330−833 binds G4 DNA substrates ∼15-fold tighter than the non-G4 DNA control (Figure 3b). These results indicate that hRev1330−833 preferentially forms binary complexes with G4 DNA substrates.
Figure 3.
hRev1330–833 preferentially binds to G4 DNA-containing substrates. (a) hRev1330–833 was titrated into a solution containing either FAM-labeled ss-G4 DNA (square) or ss-non-G4 DNA (circle) oligonucleotides (1 nM). Changes in fluorescence polarization were measured, and the resulting data fit to a quadratic equation to yield the following equilibrium dissociation constants: ss-G4 DNA, Kd,DNA = 15.7 ± 8.7 nM; ss-non-G4 DNA, Kd,DNA = 65.7 ± 8.8 nM. (b) hRev1330–833 was titrated into a solution containing either FAM-labeled primer–template G4 DNA (square) or primer–template non-G4 DNA (circle) oligonucleotides (1 nM). Changes in fluorescence polarization were measured, and the resulting data fit to a quadratic equation to yield the following equilibrium dissociation constants: primer–template G4 DNA, Kd,DNA = 8.4 ± 2.5 nM; primer–template non-G4 DNA, Kd,DNA = 129 ± 20 nM. The reported values represent the mean ± SD (n = 3).
hRev1 disrupts G4 DNA structures
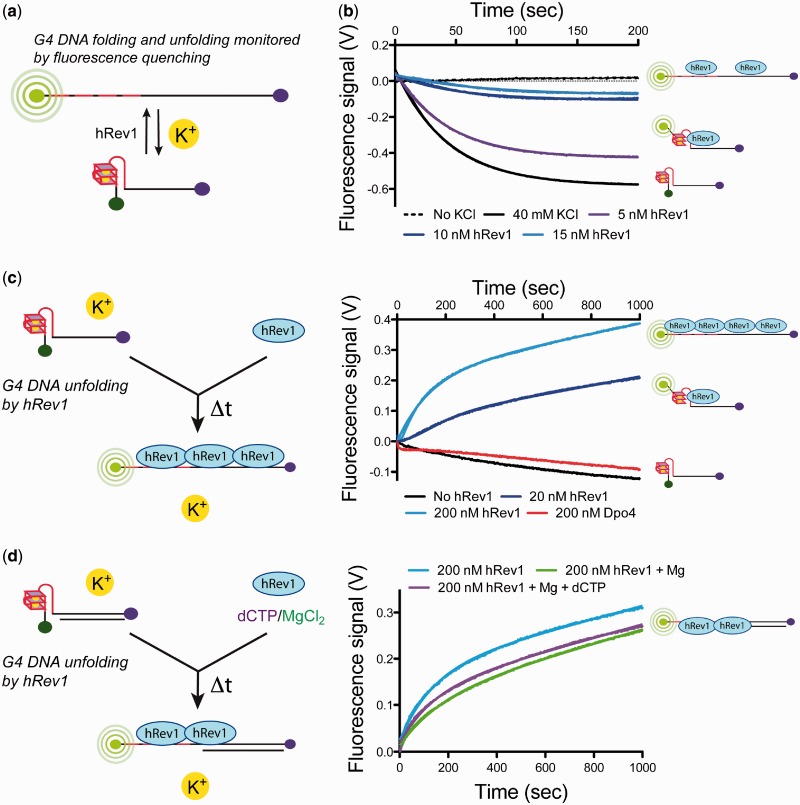
Although the DMS footprinting assays were suggestive of G4 DNA disruption by hRev1330−833 on binding, we sought a complementary approach for monitoring G4 DNA dynamics to further assess the ability of hRev1330−833 to unfold these structures. Toward this end, an oligonucleotide with an FAM label at the 5′-terminus and a dabcyl (Dab) quencher at the 3′-terminus were used to monitor G4 DNA folding/re-folding (42). Quenching of FAM signal by Dab occurs when the molecules are brought in proximity to one another (i.e. when the G4 DNA folds). Conversely, the fluorescence signal increases as the G4 DNA structure unfolds (Figure 4a).
Figure 4.
hRev1330–833 can disrupt G4 DNA structures. (a) Schematic illustration of the fluorescence-quenching assay used to monitor G4 DNA folding and unfolding. FAM fluorescence is quenched by dabcyl (Dab) moiety when G4 DNA folds and increases when G4 DNA unfolds. (b) Fluorescence quenching for FAM/Dab-42-mer ss-G4 DNA (20 nM) was monitored by stopped-flow following mixing with buffer without KCl (dashed line), 40 mM KCl (black trace), 40 mM KCl with either 5 nM (‘purple’ trace), 10 nM (‘dark blue’ trace) or 15 nM (‘light blue’ trace) hRev1330–833. The resulting curves were fit to Equation 3 to yield the following kinetic parameters: 40 mM KCl, no hRev1330–833: A = −58.2 ± 0.1 × 10−2 V, kobs = 22.9 ± 0.1 × 10−3 s−1; 40 mM KCl, 5 nM hRev1330–833: A = −42.8 ± 0.1 × 10−2 V, kobs = 23.1 ± 0.1 × 10−3 s−1; 40 mM KCl, 10 nM hRev1330–833: A = −10.6 ± 0.1 × 10−2 V, kobs = 20.0 ± 0.5 × 10−3 s−1; 40 mM KCl, 15 nM hRev1330–833: A = −7.73 ± 0.05 × 10−2 V, kobs = 14.7 ± 0.6 × 10−3 s−1. The fit of the data to Equation 3 is shown. (c) G4 DNA unfolding was monitored for a pre-formed FAM/Dab-42-mer ss-G4 DNA (20 nM) following addition of buffer (‘black’ trace) and either 20 nM (‘dark blue’ trace), 200 nM (‘light blue’ trace) hRev1330–833 or 200 nM Dpo4 (‘red’ trace). The data for experiments with hRev1330–833 were fit to Equation 4 to yield the following kinetic parameters: 20 nM hRev1330–833: n = 2, A = 6.6 ± 0.4 × 10−2 V, kobs = 153.4 ± 0.1 × 10−4 s−1, k2 = 14.7 ± 0.1 × 10−5 V s−1; 200 nM hRev1330–833: n = 1, A = 23.1 ± 0.1 × 10−2 V, kobs = 87.1 ± 0.4 × 10−4 s−1, k2 = 17.9 ± 0.6 × 10−5 V s−1. The fit of the data to Equation 4 is shown. (d) G4 DNA unfolding was monitored for FAM/Dab-18/42-mer G4 DNA (20 nM) following addition of either 200 nM hRev1330–833 alone (‘light blue’ trace), 200 nM hRev1330–833 with 2 mM MgCl2 (‘green’ trace), or 200 nM hRev1330–833 with 50 µM dCTP and 2 mM MgCl2 (‘purple’ trace). All experiments were performed in the presence of KCl (40 mM). The data for all three experiments were fit to Equation 4 to yield the following kinetic parameters: hRev1330–833 alone: n = 1, A = 15.2 ± 0.7 × 10−2 V, kobs = 72.0 ± 0.7 × 10−4 s−1, k2 = 13.9 ± 0.1 × 10−5 V s−1; hRev1330–833 with MgCl2: n = 1, A = 9.9 ± 0.6 × 10−2 V, kobs = 58.4 ± 0.8 × 10−4 s−1, k2 = 15.2 ± 0.1 × 10−5 V s−1; hRev1330–833 with dCTP/MgCl2: n = 1, A = 11.1 ± 0.6 × 10−2 V, kobs = 71.4 ± 0.9 × 10−4 s−1, k2 = 14.6 ± 0.1 × 10−5 V s−1. The fit of the data to Equation 4 is shown. The data represent the average of two to four experiments, and the error represents the standard error of the fit. A cartoon schematic is shown to the left of each reaction trace to better illustrate the experimental design and our interpretation of the results.
Stopped-flow analysis was used to monitor temporal aspects of c-MYC G4 DNA dynamics. Time-resolved fluorescence quenching showed that the c-MYC G4 DNA used in our study folds quickly on mixing with a solution containing potassium (Figure 4b, ‘black’ trace and Supplementary Table S2). The data were fit to an equation describing a one-phase exponential decay (Equation 3, A = −58.2 ± 0.1 × 10−2 V, kobs = 22.9 ± 0.1 × 10−3 s−1). The rate of folding is similar to that previously reported for intramolecular c-MYC G4 DNA-forming sequences (43). Simultaneous addition of hRev1330−833 and potassium prevents fluorescence quenching, indicating that hRev1 can bind to the unfolded ss-G4 DNA and hinder G4 DNA formation (Figure 4b, ‘purple’, ‘dark blue’ and ‘light blue’ traces and Supplementary Table S2). At a 1:4 hRev1330−833:G4 DNA molar ratio (5 nM hRev1330−833), the data again follow a single-exponential decay (A = −42.8 ± 0.1 × 10−2 V, kobs = 23.1 ± 0.1 × 10−3 s−1). Although the observed rate of G4 DNA folding is essentially unchanged under conditions where the DNA is in excess of the enzyme, the amplitude of the change is reduced, which indicates that fewer DNA molecules have folded into G-quadruplex structures. Further increasing the amount of hRev1330−833 in the reaction dramatically reduces the amplitude of fluorescence change, which we interpret as strong evidence for the prevention of G4 DNA formation by hRev1330−833 (Figure 4b, ‘dark blue’ and ‘light blue’ traces). The observed rate of folding at 1:2 hRev1330−833:G4 DNA was kobs = 20.0 ± 0.5 × 10−3 s−1, but there was an ∼5.5-fold reduction in amplitude (A = −10.6 ± 0.1 × 10−2 V) compared with the control G4 DNA-folding reaction with no enzyme (Figure 4b, compare the ‘black’ trace with the ‘light’ and ‘dark blue’ traces). The observed rate of folding at nearly equimolar hRev1330−833:G4 DNA was kobs = 14.7 ± 0.6 × 10−3 s−1, with a 7.5-fold reduction in amplitude (A = −7.73 ± 0.05 × 10−2 V) compared with the no enzyme control. Thus, the amplitude of the fluorescence change, which directly monitors the amount of G4 DNA present in the reaction, is severely attenuated on addition of the enzyme, which indicates that hRev1330−833 can prevent G4 DNA folding.
We then monitored hRev1330−833-mediated unfolding of a pre-formed G4 DNA structure. The addition of hRev1330−833 to a solution of ss-G4 DNA produces an increase in fluorescence signal, indicative of G4 DNA disruption (Figure 4c and Supplementary Table S3). At a 1:1 hRev1330−833:G4 DNA molar ratio, there is an apparent lag in the fluorescence change indicative of a multiple-step reaction (Figure 4c, ‘dark blue’ trace). The curve was fit to Equation 3 (n = 2, A = 6.6 ± 0.4 × 10−2 V, kobs = 153.4 ± 0.1 × 10−4 s−1, k2 = 14.7 ± 0.1 × 10−5 V s−1). Increasing the concentration of hRev1330−833 in the reaction mixture 10-fold eliminates the lag phase and a larger absolute change in fluorescence is observed (Figure 4c, ‘light blue’ trace). The resulting curve was fit to Equation 3 (n = 1, A = 23.1 ± 0.1 × 10−2 V, kobs = 87.1 ± 0.4 × 10−4 s−1, k2 = 17.9 ± 0.6 × 10−5 V s−1). The rate of G4 DNA-unfolding in the presence of hRev1330−833 is ∼2–4-fold faster than the apparent first-order rate of c-MYC G4 DNA unfolding stimulated by hybridization to a 4000-fold excess of the complement strand, as measured by single-molecule experiments (44). Also, the amplitude of the fluorescence change increases sharply with higher amounts of enzyme, indicative of more complete G4 DNA disruption occurring as a function of hRev1330−833 concentration. Another Y-family pol, Dpo4 from the crenarchaeote Sulfolobus solfataricus fails to induce a change in fluorescence signal for the G4 DNA substrate (Figure4c, ‘red’ trace), suggesting that G4 DNA disruption is unique to hRev1.
Finally, we tested whether hRev1330−833 could induce a fluorescence change on a G4 DNA substrate possessing a primer strand (Figure 4d and Supplementary Table S4). Similar to results with ss-G4 DNA, hRev1330−833 can promote G4 DNA dissolution with the 18/42-mer primer–template G4 DNA substrate (Figure 4d, ‘light blue’ trace). Adding either Mg2+ or Mg2+:dCTP to the reaction mixture does not alter the rate or the amplitude of the increase in fluorescence that accompanies G4 DNA unwinding (Figure 4d, ‘green and purple’ traces, respectively). Collectively, these results support the idea that hRev1330−833 can disrupt G4 DNA structures without performing nucleotidyl transfer.
hRev1 inserts dCMP on G4 DNA substrates
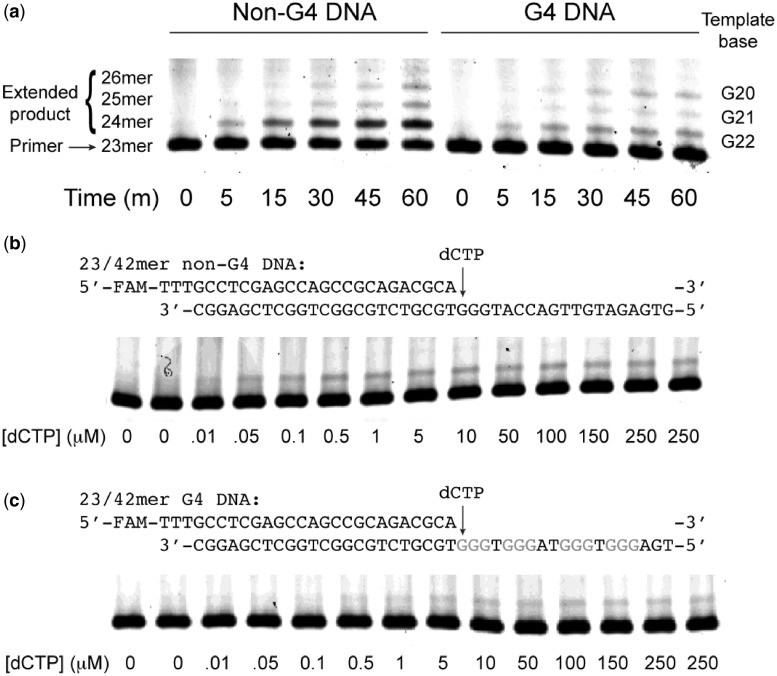
Next, we wanted to examine the effect of G4 DNA on hRev1-catalyzed insertion of deoxycytidine monophosphate (dCMP). Initially, we compared hRev1-catalyzed dCMP insertion over an extended time frame. We find that, even though it is an extremely non-processive enzyme, hRev1330−833 is able to extend the primer across the first run of three guanines by inserting dCMP opposite G22–G20 on both non-G4 and G4 DNA substrates (Figure 5a). The amount of product formed over time is diminished in the presence of G4 DNA. The addition of calcium inhibits hRev1 catalysis on both substrates (Supplementary Figure S3). To systematically quantify the impact of G4 DNA on hRev1 catalysis, we performed steady-state kinetic analysis of dCMP insertion using three different primer–templates that position the hRev1 active site 4 (19/42 mer), 2 (21/42 mer) and 0 (23/42 mer) nt away from the first tetrad-guanine (G22), respectively. Three non-G4 DNA primer-template substrates were prepared as controls, for a total of six substrates. In this way, we systematically evaluated the relative effect on nucleotidyl transfer when hRev1 is positioned at different distances from the G4 DNA structure. It should be noted that for all of the kinetic measurements, we were careful to use conditions that only allowed incorporation of the first dCMP on the templates with three guanines adjacent to one another (Figure 5b and c).
Figure 5.
Representative dCMP insertion results for 23/42-mer DNA substrates. (a) Results of a time-course experiment measuring hRev1330–833 (5 nM) insertion of dCTP (1 mM) on 23/42-mer non-G4 DNA or G4 DNA substrates (200 nM). hRev1330–833 can insert dCMP opposite the first three tetrad-guanines (G22–G20) but the rate of insertion is less than that observed for the non-G4 DNA substrate. Representative gels for steady-state kinetic analysis of hRev1 (10 nM) insertion of dCTP at the concentrations indicated on either (b) 23/42-mer non-G4 DNA or (c) 23/42-mer G4 DNA (200 nM) substrates. The reaction time for non-G4 DNA substrates was 5 min and that for G4 DNA substrates was 10 min.
We find that the specificity constant (kcat/KM,dNTP) for hRev1330−833-catalyzed dCTP insertion on G4 DNA substrates is ∼70% of the non-G4 DNA control when the polymerase is positioned 4 nt away from the G-quadruplex-forming sequence (Table 1). Moving hRev1 to within 2 nt of the G4 DNA structure diminishes the specificity constant to ∼16% of the non-G4 DNA control, indicating some impairment of hRev1 catalysis as it nears the G-quadruplex. The change in activity on the 21/42-mer G4 DNA substrate is driven by an increase in the Michaelis constant (KM,dNTP), as the turnover number (kcat) is essentially identical for both G4 DNA and non-G4 DNA 21/42-mer substrates (Table 1). At the insertion point opposite the first tetrad-associated guanine (G22), hRev1 steady-state activity is perturbed to <10% of its normal activity by an apparent reduction in the kcat and an increased KM,dNTP for dCMP insertion opposite G22. The specificity constant for the non-G4 DNA 23/42-mer control is substantially higher than that observed for either the 19/42mer or the 21/42-mer non-G4 substrates (Table 1). This increase may be suggestive of a sequence-dependent effect observed for hRev1 on templates with more than one guanine located adjacent to the primer–template junction. However, any sequence-dependent increase in activity for the control does not appear to be carried over to the G4 DNA substrate. The kinetic results suggest that dCMP insertion opposite tetrad-guanines (i.e. on the 23/42-mer G4 DNA substrate) remains impaired for hRev1330−833, despite favorable binding interactions and the disruption of G4 DNA.
Table 1.
Steady-state kinetic parameters for hRev1 catalysis on G4 DNA and non-G4 DNA substrates
 |
Data represent the mean ± SD calculated from three independent experiments.
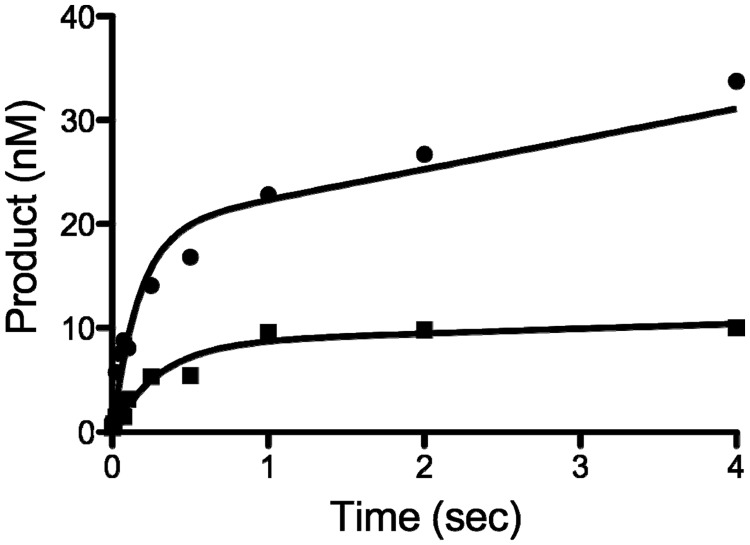
To better understand the effect of G4 DNA on nucleotidyl transfer, we then examined hRev1 insertion of dCMP opposite G22 using rapid chemical-quench method. Pre-steady-state kinetic experiments reveal that the presence of G4 DNA reduces the amplitude of product formation from 19.4 nM for non-G4 DNA to 8.5 nM when G4 DNA is present (Figure 6). A reduced burst-amplitude in pol assays is often attributed to non-productive ternary complex formation. Notably, the observed rate-constant for dCMP insertion opposite G4 DNA-associated G22 is only reduced to ∼56% of the control value in the pre-steady-state (Figure 6), suggesting that the actual rate-constant defining polymerization for productive complexes is not strongly inhibited by G4 DNA structures.
Figure 6.
Pre-steady-state kinetic analysis of hRev1330–833 activity on G4 DNA substrates. hRev1330–833 (400 nM) was pre-incubated with 23/42-mer DNA (100 nM), and the reaction initiated by addition of dCTP (250 µM) and MgCl2 (2 mM). Product formation was plotted as a function of time and the data fit to Equation 4 (n = 1), yielding the following kinetic parameters: non-G4 DNA (circle), A = 19.4 ± 1.6 nM, kobs = 5.9 ± 1.4 s−1, k2 = 2.9 ± 0.4 nM s−1; G4 DNA (square), A = 8.5 ± 0.8 nM, kobs = 3.3 ± 0.8 s−1 and k2 = 0.46 ± 0.17 nM s−1.
In summary, kinetic assays show that hRev1330−833 has substantial catalytic efficiency when positioned a few nucleotides back from the tetrad-associated guanines but that dCMP insertion opposite tetrad-associated guanine G22 is inhibited under steady-state conditions. The reduced product amplitude in the pre-steady-state may indicate non-productive complex formation when the enzyme attempts catalysis opposite G22, which seems reasonable considering the dynamic nature of G4 DNA substrates. The fact that the observed pre-steady-state rate constant for dCMP insertion opposite G22 is only reduced ∼1.8-fold in the pre-steady-state suggests that hRev1330−833 is able to effectively copy tetrad-associated guanines when productive complexes are formed. We do observe dCMP insertion opposite G22–G20 despite the fact that hRev1 is an extremely non-processive enzyme, indicating that the enzyme can copy the first three tetrad-associated guanines.
DISCUSSION
We have investigated the in vitro biochemical mechanism of G4 DNA replication by hRev1, a central regulator of translesion DNA synthesis in eukaryotes. The most important finding is that hRev1 preferentially binds to and can disrupt G4 DNA structures, consistent with its role in maintaining fork progression at these sites. Although previous work showed that Rev1 facilitates G4 DNA replication efficiency and the maintenance of chromatin marks near these sites (31,32), it was unclear whether the enzyme participated in the manipulation of nucleic acids or simply acted as a scaffold for other G4 DNA-interacting proteins (31,32). While our results certainly do not exclude the latter possibility, they are consistent with the notion that Rev1 can help maintain fork progress by assisting in the disruption of G4 DNA structures that are present during replication, most likely in coordination with DNA helicases, such as Pif1, FANCJ and RecQ enzymes.
Based on our findings, we propose that the mechanism of hRev1-mediated G4 DNA disruption can occur either before or after the G4 DNA structure has formed. To date, no other pol has been shown to be capable of either disrupting or preventing the formation of G4 DNA. The effect of hRev1 on G4 DNA stability can be attributed to at least two distinct properties of the enzyme. First, disruption of the G4 DNA structure by hRev1 is evident from the exposure of G22 near the primer-template junction in DMS footprinting assays, which is indicative of an effect mediated by the active site pocket of hRev1. Yet, disruption of the G-quadruplex does not require the binding of divalent metal cations or nucleotidyl transfer by hRev1, which leads us to hypothesize that G4 unfolding near the primer–template junction could involve interactions with Arg357 and Leu358, as these two residues are central to the protein template-directed mechanism for hRev1. Experiments are underway to test this possibility. A second property of hRev1 that contributes to the dissolution of the G4 DNA structure is its ability to bind to ssDNA. In this way, hRev1 is acting in a manner analogous to RPA (45–48). The added effect of hRev1 binding to ssDNA on G4 disruption is manifest in the DMS footprinting results, but it is also apparent in the stopped-flow fluorescence assays where more pronounced changes in the amplitude of the signal are observed when the amount of enzyme is increased. The stopped-flow fluorescence assay measures the distance between the FAM fluorophore and the Dabcyl quencher. At a 1:1 molar ratio of hRev1:DNA, we observe a lag in G4 unfolding, but an increase in fluorescence is observed nonetheless. At a 10:1 molar ratio of hRev1:DNA, there are 10 molecules of hRev1 for every 1 molecule of G4 DNA substrate. Adding more protein molecules to each G4 DNA substrate apparently results in a larger increase in the distance between FAM and Dabcyl; hence, the greater change in signal compared with the distance between FAM and Dabcyl is maintained by a single molecule of hRev1. The change observed at a 1:1 ratio of DNA to enzyme is a testament to the specificity of the binding/unwinding by hRev1. Additional structural studies will be required to more accurately assess the quaternary structure of hRev1 on DNA substrates. However, the fluorescence changes are consistent with the DMS footprinting results in that both suggest that multiple molecules of hRev1 can bind to ssDNA templates and disrupt G4 DNA structures.
In terms of catalysis on G-quadruplex substrates, hRev1-catalyzed insertion of dCMP on G4 DNA templates is fairly robust when the enzyme is positioned 2–4-nt away from tetrad-guanines. In pre-steady-state experiments, catalysis directly opposite tetrad-guanines is impaired, with a rate-constant that is about half that of the non-G4 DNA control and with a reduced burst amplitude, suggestive of ternary complex formation that is non-productive on G4 DNA substrates. The ability of Rev1 to insert a few bases near or opposite to tetrad-guanines may be important during G4 DNA replication in cells, based on experiments in avian cells revealing only ∼50% complementation with a catalytically inactive Rev1 mutant when replication of a G-quadruplex containing plasmid in rev1-deficient cells was assayed (32). Primer extension by hRev1, even if not directly opposite tetrad-guanines, may aid extension by other pols from the hRev1 C-terminal scaffold. Thus, understanding the role of Rev1 in facilitating extension by pols eta and kappa is an immediate priority, as deletion of the polymerase-interacting region diminishes G4 DNA replication efficiency almost to levels observed for rev1-deficient DT40 cells (32).
The ability of hRev1 to bind tightly to and disrupt G4 DNA contrasts somewhat with the impaired catalysis observed for dCMP insertion opposite G22. This paradox can be reconciled by the fact that G4 DNA disruption does not appear to require the nucleotidyl transfer activity of hRev1. Using both DMS footprinting and stopped-flow fluorescence experiments, we show that binding of hRev1 can disrupt G4 DNA in the absence of nucleotidyl transfer and that this effect is especially pronounced as the enzyme concentration increases. In terms of catalytic properties, we show that hRev1 can insert dCMP across the first three tetrad-guanines under conditions where the G4 DNA substrate is in 20–40-fold excess of enzyme (Figure 5a). Moreover, it is efficient at nucleotidyl transfer 2–4 nt away from the first tetrad-guanine, G22. However, the efficiency of dCMP insertion opposite G22 is impaired based on the specificity constant (kcat/KM,dNTP) and the reduced burst-amplitude in the pre-steady-state. For the steady-state results where DNA is in large excess of enzyme, it is easy to envision a partially unfolded G4 DNA structure impairing catalysis. For the pre-steady-state results, the enzyme is in excess of DNA substrate, and based on our DMS footprinting/stopped-flow data, hRev1 should coat the ssDNA template under these conditions. So, either binding of additional hRev1 molecules is inhibitory to nucleotidyl transfer or the G4 motif is intrinsically difficult for hRev1 to copy because it spontaneously adopts non-productive template structures, even when coated with excess hRev1. This second scenario would be analogous to the enzyme attempting to wrestle a coiled spring into a smooth structure (or like grappling with a snake that keeps coiling around your arm). Although there are features of the hRev1 mechanism of action on G4 DNA that remain unresolved, the results presented here are internally consistent if DNA binding and catalytic features of hRev1 activity are considered separately.
Because G4 DNA structures are generally considered globular in nature, it is at first conceptually difficult to rationalize how a polymerase, such as Rev1, could interact with G4 DNA, much less disrupt the tetrads. Some insight into this idea may be found in a recent single-molecule study that probed mechanical features related to telomeric G4 DNA folding/unfolding (49). A major conclusion from the study was that the primary energetic barrier to G4 DNA unfolding involves the disruption of one or a few tetrad-associated guanines. The hRev1 mechanism of action would seem to present an ideal solution to such an energetic landscape. By pulling even a single tetrad-associated guanine free and sequestering the base away from other template guanines, hRev1 can destabilize the G4 DNA structure, although complete unfolding may require additional proteins/enzymes. This model could also prove applicable to non-processive DNA helicases, such as Pif1, which are known to unwind only a few base pairs before dissociation from the DNA substrate (15,16,50). Thus, nature may have retained enzymes for G4 DNA replication with short-range functions, analogous to general properties observed for translesion synthesis past DNA adducts. In conclusion, hRev1 can accomplish two important tasks necessary for replication of G4 DNA (i) hRev1 can disrupt G4 DNA structures that might otherwise block fork progression and (ii) it can prevent refolding of G4 DNA when the template is single-stranded. Based on our kinetic analysis dCMP insertion, a role for hRev1 catalysis in fork progression past G4 DNA would likely be limited to nucleotide insertion across from template bases near the G4 motif but insertion opposite tetrad-guanines may be performed by other pols, such as hpol η or κ. Experiments to identify the active-site features promoting G4 DNA disruption by hRev1 and the role of hRev1 as a scaffold for extension across G4 DNA sequences by other Y-family pols are ongoing. While it remains uncertain whether Y-family pols encounter fully folded G4 DNA structures in cells, an ability to actively disassemble G4 DNA would explain why loss of Rev1 impedes replication fork progression and subsequent epigenetic stability at sites predicted to form these secondary structures in cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
FUNDING
U.S.P.H. service [R00 GM084460 to R.L.E.]; NRF [2012R1A1A2042391 to J-Y.C.] from MEST Korea and by the Arkansas Breast Cancer Research Program (to R.L.E.), with additional support from the University of Arkansas for Medical Sciences Translational Research Institute (CTSA Award [UL1TR000039]). Funding for open access charge: The UAMS, College of Medicine.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank K.D. Raney for critically reading their manuscript and for many helpful suggestions related to the study.
REFERENCES
- 1.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson E, Hardin CC, Walk SK, Tinoco I, Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 3.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 4.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 7.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 11.Damerla RR, Knickelbein KE, Strutt S, Liu FJ, Wang H, Opresko PL. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012;11:3036–3044. doi: 10.4161/cc.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson JE, Cao K, Ryvkin P, Wang LS, Johnson FB. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2009;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA.J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang AY, Balasubramanian S. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J. Am. Chem. Soc. 2012;134:19297–19308. doi: 10.1021/ja309851t. [DOI] [PubMed] [Google Scholar]
- 20.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brewer BJ, Fangman WL. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 22.Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 23.Linskens MH, Huberman JA. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem. 2002;277:15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 25.Prakash L. Effect of Genes controlling radiation sensitivity on chemically induced mutations in Saccharomyces Cerevisiae. Genetics. 1976;83:285–301. doi: 10.1093/genetics/83.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 28.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human Rev1-DNA-dNTP ternary complex. J. Mol. Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure. 2008;16:239–245. doi: 10.1016/j.str.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 31.Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2011;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol. Cell. 2011;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 35.Hatzakis E, Okamoto K, Yang D. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry. 2010;49:9152–9160. doi: 10.1021/bi100946g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathad RI, Hatzakis E, Dai J, Yang D. c-MYC promoter G-quadruplex formed at the 5'-end of NHE III1 element: insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011;39:9023–9033. doi: 10.1093/nar/gkr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Antonio M, Rodriguez R, Balasubramanian S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods. 2012;57:84–92. doi: 10.1016/j.ymeth.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vorlickova M, Kejnovska I, Sagi J, Renciuk D, Bednarova K, Motlova J, Kypr J. Circular dichroism and guanine quadruplexes. Methods. 2012;57:64–75. doi: 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masuda Y, Kamiya K. Role of single-stranded DNA in targeting REV1 to primer termini. J. Biol. Chem. 2006;281:24314–24321. doi: 10.1074/jbc.M602967200. [DOI] [PubMed] [Google Scholar]
- 42.Rachwal PA, Fox KR. Quadruplex melting. Methods. 2007;43:291–301. doi: 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Halder K, Chowdhury S. Kinetic resolution of bimolecular hybridization versus intramolecular folding in nucleic acids by surface plasmon resonance: application to G-quadruplex/duplex competition in human c-myc promoter. Nucleic Acids Res. 2005;33:4466–4474. doi: 10.1093/nar/gki750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirude PS, Ying L, Balasubramanian S. Single molecule conformational analysis of the biologically relevant DNA G-quadruplex in the promoter of the proto-oncogene c-MYC. Chem. Commun. (Camb.) 2008;7:2007–2009. doi: 10.1039/b801465e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prakash A, Kieken F, Marky LA, Borgstahl GE. Stabilization of a G-quadruplex from unfolding by replication protein a using potassium and the porphyrin TMPyP4. J. Nucleic Acids. 2011;2011:529828. doi: 10.4061/2011/529828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prakash A, Natarajan A, Marky LA, Ouellette MM, Borgstahl GE. Identification of the DNA-binding domains of human replication protein A that recognize G-Quadruplex DNA. J. Nucleic Acids. 2011;2011:896947. doi: 10.4061/2011/896947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qureshi MH, Ray S, Sewell AL, Basu S, Balci H. Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J. Phys. Chem. B. 2012;116:5588–5594. doi: 10.1021/jp300546u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long X, Parks JW, Bagshaw CR, Stone MD. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013;41:2746–2755. doi: 10.1093/nar/gks1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramanagoudr-Bhojappa R, Chib S, Byrd AK, Aarattuthodiyil S, Pandey M, Patel SS, Raney KD. Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J. Biol. Chem. 2013;288:16185–16195. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.