Abstract
CpG methylation in mammalian DNA is known to interfere with gene expression by inhibiting the binding of transactivators to their cognate sequence motifs or recruiting proteins involved in gene repression. An Epstein–Barr virus-encoded transcription factor, Zta, was the first example of a sequence-specific transcription factor that preferentially recognizes and selectively binds DNA sequence motifs with methylated CpG residues, reverses epigenetic silencing and activates gene transcription. The DNA binding domain of Zta is homologous to c-Fos, a member of the cellular AP-1 (activator protein 1) transcription factor family, which regulates cell proliferation and survival, apoptosis, transformation and oncogenesis. We have identified a novel AP-1 binding site termed meAP-1, which contains a CpG dinucleotide. If methylated, meAP-1 sites are preferentially bound by the AP-1 heterodimer c-Jun/c-Fos in vitro and in cellular chromatin in vivo. In activated human primary B cells, c-Jun/c-Fos locates to these methylated elements in promoter regions of transcriptionally activated genes. Reminiscent of the viral Zta protein, c-Jun/c-Fos is the first identified cellular member of the AP-1 family of transactivators that can induce expression of genes with methylated, hence repressed promoters, reversing epigenetic silencing.
INTRODUCTION
AP-1 (activator protein 1) is a family of transcription factors involved in cell proliferation and survival, apoptosis, transformation and oncogenesis. AP-1 proteins contain a basic leucine zipper (bZIP) domain and belong to three different subfamilies: Jun (c-Jun, JunB and JunD), Fos (c-Fos, FosB, Fra-1 and Fra-2) and ATF (activating transcription factor: ATFa, ATF2, ATF3, ATF4 and B-ATF). To bind to DNA, members of the different subfamilies need to form dimers, which show different affinities for two classes of DNA sequences. They are 12-O-Tetradecanoylphorbol-12-Acetate (TPA) response elements (TRE) and cyclic adenosine monophosphate (cAMP) response elements (CRE), which encompass the consensus sequences 5′-TGAC/GTCA-3′ and 5′-TGACGTCA-3′, respectively. In general, Jun/Jun and Jun/Fos dimers show a higher affinity for TRE sites, whereas Jun/ATF and Fos/ATF dimers preferentially bind to CRE sites (1).
Different cellular signaling pathways, referred to as mitogen-activated protein kinase (MAPK) cascades, regulate both the expression and post-translational modifications of AP-1 proteins. The prototypic AP-1 member is the c-Jun/c-Fos heterodimer, and both proteins are regulated by MAPK pathways (2). Many different extracellular stimuli, such as growth factors and cytokines, trigger the MAPK cascades consisting of a hierarchy of sequential kinases that finally activate the three main MAP kinases called JNK, ERK and p38. These proteins can phosphorylate different transcription factors, which in turn activate the expression of c-jun and c-fos genes. Once expressed, the c-Jun and c-Fos proteins form dimers and bind to their target DNA sequence motifs, but they also need to be phosphorylated, a modification that can be introduced by MAPKs as well, to activate transcription of target genes with AP-1 binding sites in their promoters (1).
In eukaryotic cells, transcriptional activation depends on chromatin structure, nucleosomal occupancy, histone modifications and DNA methylation, which are diverse processes that play important roles in transcriptional regulation. Transcriptionally repressed heterochromatin is characterized by the frequent occurrence of cytosine nucleotides with a methyl group at the 5′ position of the cytosine pyrimidine ring followed by guanine nucleotides, termed CpG. DNA methylation affects the transcription of genes in two ways. First, methylation of DNA itself may physically impede the binding of transcriptional proteins to promoters and, second, methylated DNA may be bound by proteins known as methyl-CpG-binding domain proteins (MBDs). MBD proteins then recruit additional proteins to the locus, such as histone deacetylases and other chromatin remodeling proteins that modify histones and promote the formation of compact, inactive and silenced chromatin, leading to gene repression (3).
CpG methylation is indicative of transcriptional repression, but we and others have recently found that a viral transcription (Zta) factor preferentially binds to CpG-methylated DNA sequence motifs activating epigenetically repressed viral promoters (4,5). The Zta protein (also termed BZLF1, ZEBRA, Z or EB1) is the major immediate early transactivator of a human herpes virus, Epstein–Barr virus (EBV). Zta’s bZIP domain is highly homologous to cellular AP-1 members, especially c-Fos (6). In a systematic approach, we mapped all Zta response elements (ZREs) within promoters of viral genes and identified two classes of ZREs, which are characterized by two distinct binding motifs. One class encompasses nearly all ZREs, which bind to an AP-1-related DNA motif. The second class encompasses ZREs with a CpG motif. Zta binds to the majority of these ZREs only when the cytosine in the CG dinucleotide carries a 5′-methyl group. Consequently, these sites were termed meZREs. Zta binds to meZRE motifs and dramatically induces viral gene expression but in a methylation-dependent fashion (5). This exceptional feature is essential for this herpes virus to escape from its latent phase of infection, which is governed by transcriptional silencing of the majority of viral genes including extensive and widespread CpG methylation of viral DNA (7).
To our knowledge, only two reports indicate that cellular transcription factors also can target methylated DNA sequence motifs supporting gene transcription in metazoan cells in vivo. Sp1/Sp3 binding is not commonly impaired by CpG methylation (8) but rather enhanced in the context of a tissue-specific promoter (9). When methylated, cAMP response elements (CRE) or composite CRE and C/EBP sequence elements generate novel binding sites for C/EBPα activating certain promoters (10), which is an unexpected finding (11).
Herpes viruses acquire cellular genes through recombination with host cell DNA. Cellular genes adopted and further modified by this class of viruses regulate apoptosis, control the cell cycle of infected cells and block host immune responses (12). Very likely, the viral BZLF1 gene encoding Zta is derived from a cellular member of the AP-1 family (6), has evolved to act as the critical immediate-early gene of EBV to allow exit from latently infected cells (13) and activates resting primary human B cells, the target of EBV infection (14). As described earlier in the text, the bZIP domain of Zta is highly conserved when compared with bZIP domains of AP-1 members and mediates binding to CpG-methylated DNA sequence motifs. Zta might have inherited this peculiar feature from cellular AP-1 members that could target and activate repressed cellular promoters encompassing CpG-methylated DNA as well. This hypothesis would substantiate a previously unknown type of gene regulation and constitute a new example of a cellular transcription factor capable of activating gene expression from epigenetically repressed promoters through direct binding to methylated DNA.
MATERIALS AND METHODS
Cells
HEK293, Raji and K562 cells were maintained in RPMI 1640 medium with 10% fetal calf serum (FCS), 1% penicillin–streptomycin and 1% sodium pyruvate at 37°C and 5% CO2. Wi-38 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) medium with 10% FCS, 1% penicillin–streptomycin and 1% non-essential amino acids at 37°C and 5% CO2. Adenoid B cells were maintained in RPMI 1640 medium supplemented with 10% FCS, 100 µg/ml streptomycin, 100 U/ml penicillin, 100 nM sodium selenite and 1 µg/ml cyclosporin A at 37°C and 5% CO2. For cellular activation, B cells were cultured on irradiated CD40L feeder cells in the presence of 2 ng/ml IL-4 for 5 days as described (15).
Plasmids
The wild-type maxi-EBV plasmid (p2089) contains the complete genome of the EBV prototype B95.8 strain and has been described (16). The DNA binding and dimerization domains of human c-Jun and c-Fos (amino acid residues 123–331 and 1–206, respectively), separated by a flexible linker (SG4)5 (17), were cloned downstream of the coding region of enhanced green fluorescence protein (eGFP) in pEGFP-C1 (Clontech), to generate the expression plasmid p4271, or downstream of the tandem StrepII/FLAG-tag (18) to yield the plasmid p4548. The plasmid expressing VP16:c-jun/c-fos (p4316) was generated by replacing the gfp gene by the VP16 transactivation domain (TAD) of the VP16 protein from HSV-1 (amino acid residues 404–490). Luciferase plasmids were constructed by inserting pentamers of single meAP-1 sites into a basic luciferase reporter plasmid with a minimal EF1a promoter. The plasmid backbone of this reporter plasmid is free of CpGs (19).
DNA transfection
Transfection of DNA into HEK293 cells was performed using polyethylenimine (Sigma-Aldrich) as previously described (14). For protein extracts, 1 × 107 cells per 130-mm dish were seeded the day before transfection. Each plate was transfected with 30 µg of plasmid DNA. For reporter assays, 5 × 105 HEK293 cells were seeded into 6-well plates the day before transfection. Each well was cotransfected with 0.5 µg of reporter plasmid together with 1 µg of transactivator and 0.01 µg of DNA of a renilla-expressing plasmid as an internal control for data normalization.
In vitro DNA methylation
CpG methylation in vitro was performed with the de novo methyltransferase M.SssI and S-adenosyl methionine as described (5,20).
In vitro immunoprecipitation assays with GFP:c-Jun/c-Fos
For the in vitro pull-down assays, the plasmid p4271 encoding gfp:c-jun/c-fos was transiently transfected into HEK293 cells. Nuclear extracts containing the GFP:c-Jun/c-Fos protein were used in in vitro immunoprecipitation assays with Escherichia coli-derived genomic EBV DNA as described (5).
Recombinant adenovirus generation and cell infection
The adenoviral expression vector was created using the Gateway Recombination Cloning Technology (Invitrogen). Shortly, the gfp:c-jun/c-fos coding region was amplified by polymerase chain reaction (PCR), introduced in the PCR8/GW/TOPO vector, and then transferred to the pDEST12.2 vector. The latter plasmid was used to recombine the gfp:c-jun/c-fos cassette into the adenoviral vector pCAGAdDu-GFP (provided by Dr Vigo Heissmeyer).
Stable transfection and establishment of cell lines
Five micrograms of p4271 DNA was transfected into 5 × 106 Raji cells by electroporation in 250 µl medium at 230 V and 975 µF using a Biorad electroporation apparatus in 4-mm cuvettes. Immediately after electroporation, 400 µl of FCS was added to the cells, which were transferred to a flask containing 5 ml of cell culture medium. Cells were incubated at 37°C and 5% CO2 for one day. The cells were plated into 96-well plates in 200 µl of cell culture medium/well, supplemented with puromycin (1.2 µg/ml) until resistant cells grew out, which were further cultivated under selective pressure.
Native chromatin immunoprecipitations
A total of 5 × 107 Raji cells expressing the GFP:c-Jun/c-Fos protein were used for chromatin immunoprecipitation (ChIP) experiments, which were performed as described previously (5).
Electromobility shift assays
Electromobility shift assays (EMSAs) were performed as described previously [Bergbauer et al. (5)].
Enzyme-linked immunosorbent assay
Enzyme-linked immunosorbent assays (ELISAs) were performed with purified protein from HEK293 cells transiently transfected with Strep/FLAG:c-Jun/c-Fos (p4548). Two days post-transfection, cells from three 130-mm dishes were pooled and lysed in 2 ml of RIPA-buffer (50 mM Tris, 150 mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, pH 8.0). Cell lysates were sonicated and Strep/FLAG:c-Jun/c-Fos was affinity purified with Strep-Tactin sepharose (iba-biotagnology). Strep/FLAG:c-Jun/c-Fos was eluted in 500 µl of Strep elution buffer. Purified Strep/FLAG:c-Jun/c-Fos protein was coated on a 96-well ELISA plate at 100 nM in phosphate-buffered saline at 4°C overnight. Cy5-labeled oligonucleotides were provided by Metabion. After washing and blocking with 5% bovine serum albumin, 100 μl of serial dilutions of annealed double-stranded Cy5-labeled oligos (in 20 mM Hepes, 75 mM NaCl, 1 mM DTT, 1% glycerin, 2 mM MgCl2, 0.01 mg/ml bovine serum albumin and a 20× molar excess of polydIdC) were then added and incubated for 2 h at room temperature. Unbound oligo was removed with repeated washes in phosphate-buffered saline. Finally, the Cy5 levels/well were measured in the phosphoimager (FLA 5100, Fuji). The obtained data were fit to the Hill equation with single site-specific binding using the Prism 5 software to calculate the dissociation constant.
Luciferase reporter assays
Forty-eight hours post-transfection, the HEK293 cells were analyzed with the Dual-Luciferase Reporter Assay System (Promega). Luciferase activity was measured in a 96-well microplate luminometer (Victor2, Wallac).
Bisulfite sequencing
Bisulfite modification was done using the EpiTect Bisulfite Kit (Qiagen), and the regions of interest were amplified by suitably designed PCR primer pairs. Primer sequences are available on request.
Library construction
Sequence libraries were constructed with paired-end DNA sample preparation kits (Illumina) according to the manufacturer’s recommendations with minor modifications and as described previously (5). In vitro immunoprecipitations with non–CpG-methylated and fully CpG-methylated EBV DNAs were converted into the libraries ‘4271.unmethylated’ and ‘4271.methylated’, respectively. DNA obtained with ChIP of Raji cell chromatin was converted into the library ‘4427.Raji’. Wi-38 DNA used for ChIP and ChIP DNA were converted into the libraries ‘4673.Wi38.Input’ and ‘4673.Wi38.ChIP’, respectively.
Read mapping
Sequencing reactions were performed on an Illumina Genome Analyzer IIx or Illumina HiSeq 2000 machines. We generated 32 million and 31 million 36-bp single-end reads for libraries ‘4271.unmethylated’ and ‘4271.methylated’, respectively, 50 million 36-bp single-end reads for ‘4427.Raji’ and 50 and 47 million 100-bp pair-end reads for the ‘4673.Wi38.Input’ and ‘4673.Wi38.ChIP’ samples, respectively. Read mapping to the reference sequence and subsequent assembly was performed using the resequencing software Burrows-Wheeler Alignment (BWA) (v0.7.1; 21). We used a combination of the recombinant EBV strain 2089 (16) and the bovine genome (build bosTau4) as reference sequences for the two libraries ‘4271.unmethylated’ and ‘4271. methylated’. Calf thymus DNA was used as an unspecific competitor in the in vitro immunoprecipitation assays with GFP:c-Jun/c-Fos. The reference sequence for the in vivo samples ‘4427.Raji’, ‘4673.Wi38.Input’ and ‘4673. Wi38.ChIP’ was the human reference sequence hg19.
Transcription factor binding site detection
Genomic regions with a read depth above background level are considered as regions containing transcription factor binding sites. We used two different programs, Site Identification from Short Sequence Reads (SISSRs) (22) and QuEST (23), to identify these regions. Transcription factor binding sites are typically shorter than the sequenced DNA fragments. Binding sites for the sample libraries ‘4271.unmethylated’ and ‘4271.methylated’ were determined using SISSRs standard parameters, except for E (min number of directional reads), which was set to 3200 for the library ‘4271.unmethylated’ and to 1600 for ‘4271.methylated’, and F (average length of DNA fragments), which was set to 200 in both cases. Binding sites for the sample ‘4427.Raji’ were inferred using the default QuEST parameters, except for kernel density estimator and read count threshold (th), which were set to 30 and 160, respectively. In the case of the sample ‘4673.Wi38.ChIP’, it was analyzed by SISSRs with the results from‘4673. Wi38.Input’ as background file and setting th at 220. To identify consensus motifs within the discovered binding sites, the sequences containing possible binding sites were extracted and used as input for the motif finding algorithm Multiple EM for Motif Elicitation (MEME) (24).
ChIPs with an α-c-Jun antibody
ChIP experiments were conducted following standard protocols. Chromatin was cross-linked for 10 min at room temperature and incubated with the antibody α-c-Jun (sc-44, Santa Cruz Biotechnology).
Quantitative real time PCR
Quantitative real time PCR (qRT-PCR) analysis was performed with a LightCycler 480 II instrument (Roche), with software version 1.5.0 SP4, according to the manufacturer's instructions. Amplifications were monitored with the LightCycler FastStart reaction mix (SYBR green I; Roche). The amplification of PCR products was terminated after 45 cycles. The following touchdown PCR protocol was used: (i) initial denaturation at 95° for 10 min and (ii) 45 cycles of 95° (2 s), 65°–62° (0.6° decrement in annealing temperature/cycle; 10 s), 72° (10 s). For all primer pairs, a standard curve from serial dilutions of the PCR product with known concentration or the input DNA was assessed. Primer sequences are available on request.
B cell isolation
Human primary B cells from adenoids were prepared and naïve B cells were sorted as described previously (14,25). Anonymized adenoid samples from routine adenoidectomies were provided by the Department of Otorhinolaryngology, Klinikum Grosshadern, Ludwig-Maximilians-University of Munich. The institutional review board, Ethikkommission of the Klinikum Grosshadern, approved the study and did not require prior informed patient consent.
Bioinformatic identification of meAP-1 sites in primary human B cells
In the reference human genome assembly GRCh37/hg19, we detected AP-1 and meAP-1 binding sites using the search algorithm Find Individual Motif Occurrences (FIMO) (26) based on our MEME output motifs. Called sites were mapped to promoter sequences of RefSeq genes contained in the UCSC Genome Browser −5 kb to +1 kb of their transcription start sites (TSS) (27). Predicted meAP-1 binding site within promoter sequences were further evaluated for CpG methylation in naïve B cells (28) (C04M7ACXX_XF, D088DACXX_XF, D08E5ACXX_XF). For further analysis, we selected only meAP-1 binding sites that show at least 80% methylation in two of three replicates. To identify genes upregulated in activated B cells, we used expression data obtained from unstimulated (GSM907730, GSM907738, GSM907746) and stimulated (GSM907731, GSM907739, GSM907747) B cell data sets available on Gene Expression Omnibus (GSE36975) (29). We selected transcripts that were significantly expressed in 5 of 6 samples. Transcripts showing significant differential expression between unstimulated and stimulated B cells were identified performing a one way ANOVA test using MATLAB.
RESULTS
c-Jun/c-Fos binding motifs in genomic EBV DNA
We started our search with a synthetic model, which builds on the entire DNA genome of EBV cloned in E. coli as a single plasmid (16). This DNA is large (∼180 kb), GC-rich (60%), free of methylated cytosine nucleotides (which can be easily methylated in vitro), contains ∼85 genes and diverse promoter elements and has been used previously for the analysis of Zta’s DNA binding characteristics (5). We predicted that the DNA’s complexity and size is suitable to identify putative DNA motifs, which are targeted by AP-1 if methylated. The second component in our model is a single-chain c-Jun/c-Fos protein designed to ensure the formation of a single, defined AP-1 heterodimer unaffected by other proteins that complex with either c-Jun or c-Fos. Bakiri et al. (17) showed that similar constructs with the murine sequences of c-Jun and c-Fos specifically bind and transactivate cellular promoters that contain the consensus AP-1 binding site.
We used in vitro pull-down experiments as described (5) with randomly sheared non–CpG-methylated or fully CpG-methylated EBV DNA and a GFP:c-Jun/c-Fos fusion protein purified from HEK293 cells. Immunoprecipitation of the fusion protein followed by deep sequencing of the enriched DNA fragments provided an extensive coverage of DNA sequences selectively bound by the AP-1 heterodimer (Supplementary Figure S1). A comparison of fully CpG-methylated versus non–methylated-DNA identified several regions preferentially bound by c-Jun/c-Fos if CpG-methylated (Supplementary Figure S1, bottom), three of which were located at EBV promoters: BFLF2, BBLF4 and BALF1 (Supplementary Figure S2A). Algorithms suitable to identify sequences of potential binding sites readily found the known palindromic AP-1 consensus binding site 5′-TGACTCA-3′ (Figure 1) in samples of unmethylated as well as methylated DNA, confirming the validity of our approach. Putative AP-1 binding motifs identified in CpG-methylated DNA were grouped according to whether they encompassed CpG pairs or not. The reanalysis revealed a second, novel motif, 5′-TGACG/TCG-3′, which is closely related to the consensus AP-1 but may contain up to two CpG dinucleotides. We termed this motif ‘meAP-1’ in analogy to meZRE motifs bound by Zta (5). Lists of the identified AP-1 binding sites are provided in Supplementary Tables S1–S3.
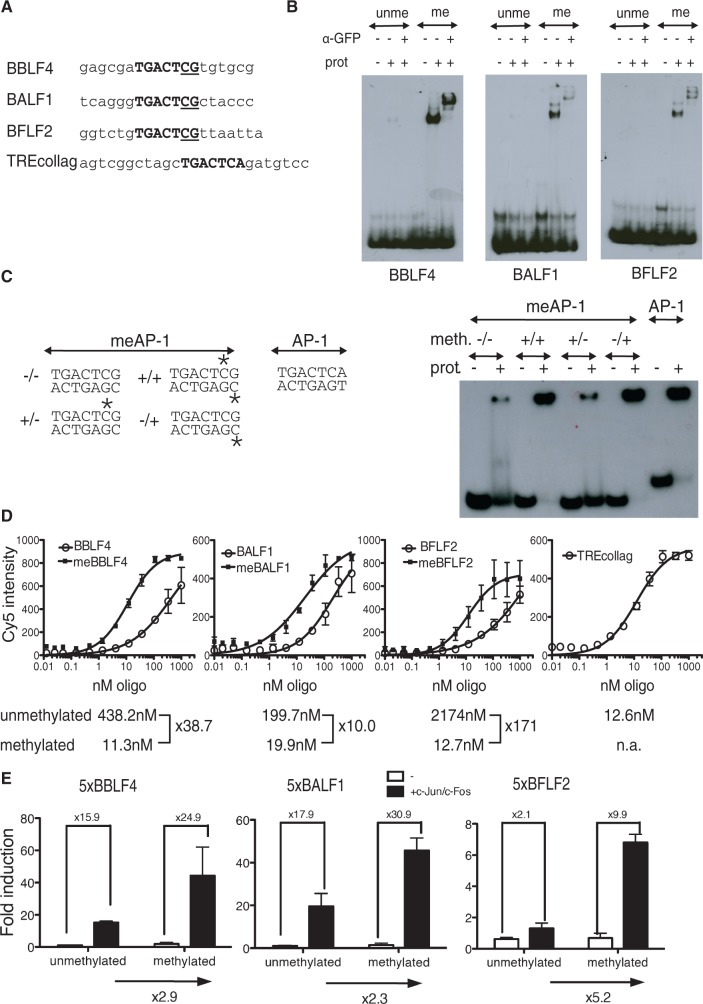
Figure 1.
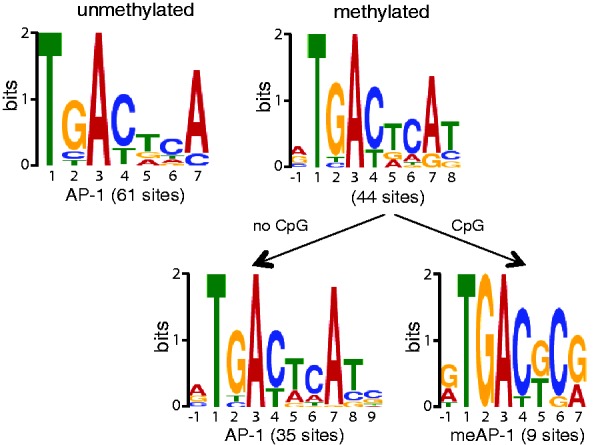
Bioinformatic identification of c-Jun/c-Fos binding motifs in the EBV genome. Deep sequencing data were analyzed with the SISSRs algorithms for putative stretches of DNA, which c-Jun/c-Fos binds. The output files of SISSRs were used as training sets for MEME, which identifies gapless, local and multiple sequence motifs. A total of 61 motifs were identified in the DNA free of CpG methylation and 44 motifs were identified in the fully CpG-methylated EBV DNA. The identified motifs in methylated DNA were selected at the level of the SISSRs training set data and grouped into motifs with and without CpG dinucleotides followed by MEME analysis.
To determine whether the biochemically identified meAP-1 sites in EBV DNA are also bound by c-Jun/c-Fos in cells in vivo, we engineered a derivative of the latently EBV-infected cell line Raji with a constitutively expressed GFP:c-Jun/c-Fos chimera. ChIP followed by deep sequencing analysis (ChIP-seq) of DNA fragments specifically bound to GFP:c-Jun/c-Fos protein was performed, mapped to the EBV genome and analyzed bioinformatically in conjunction with methylation data (7). Confirming our results in vitro, the analyzed promoters in Raji cells were only enriched if the meAP-1 sites were highly CpG-methylated (Supplementary Figure S2).
c-Jun/c-Fos binds CpG-methylated meAP-1 sites with high affinity mediating transcriptional activation
The different meAP-1 motifs identified in the EBV genome may contain up to two CpG pairs (Figure 1), but the three more prominent meAP-1 sites (Supplementary Figure S1), located in three EBV promoters, encompass the sequence 5′-TGACTCG-3′ with one CpG pair, only (Figure 2A). We addressed this uncertainty together with the question of whether 5′-methylcytosine residues present in the top, bottom or both strands of the meAP-1 DNA sequence affect the affinity of c-Jun/c-Fos binding to DNA.
Figure 2.
The c-Jun/c-Fos binds preferentially to and activates gene transcription through CpG-methylated meAP-1 motifs. (A) Oligonucleotides containing meAP-1 binding sites identified in the viral BBLF4, BALF1 and BFLF2 promoters. The TREcollag oligo containing a consensus AP-1 binding site was used as a control for the KD measurements. The meAP-1 sites and AP-1 site are indicated in bold capital letters, and the CpG pairs within the meAP-1 sites are underlined. (B) Preferential binding of c-Jun/c-Fos to methylated meAP-1 sites from the EBV genome. EMSAs were performed with affinity-purified GFP-tagged c-Jun/c-Fos fusion protein transiently expressed in HEK293 cells. Supershifts with a GFP antibody confirmed the identity of the protein–DNA complexes. EMSAs of typical experiments are shown as examples. (C) Only one methyl group is responsible for the binding of c-Jun/c-Fos to methylated DNA. The meAP-1 and AP-1 binding sites contained in the oligonucleotide used in EMSAs are shown; the asterisks represent a 5′-methylcytosine residue. EMSAs using BBLF4 and TREcollag oligonucleotides with the sites depicted in the left panel and GFP:c-Jun/c-Fos-purified protein are shown. One representative experiment out of three is provided. (D) Determination of the apparent KD of c-Jun/c-Fos bound to meAP-1. ELISAs were performed with affinity-purified Strep/FLAG-tagged c-Jun/c-Fos fusion protein transiently expressed in HEK293 cells. Serial dilutions of Cy5 oligonucleotides were used to determine the KD values. The obtained data were fitted to the Hill equation with one site-specific binding to determine the dissociation constants, which are indicated. Each data point indicates the mean and standard deviation of three independent experiments. (E) Functional identification of single meAP-1 binding sites. Pentamers of single meAP-1 sites present in the promoters of BBLF4, BALF1 and BFLF2 EBV genes as in (A) were introduced into a basic luciferase reporter plasmid with a minimal EF1α promoter and free of CpG dinucleotides. Unmethylated and fully CpG-methylated reporter constructs were analyzed in the presence or absence of a cotransfected VP16:c-Jun/c-Fos expression plasmid. Each experiment was performed three times and the means and standard deviations are depicted.
Unmethylated or CpG-methylated synthetic oligonucleotides containing the meAP-1 binding sites in the BBLF4, BFLF2 and BALF1 promoters (Figure 2A) and three promoters identified later in ChIP-seq experiments with Raji cell chromatin (Supplementary Figure S3) were analyzed in EMSAs. As shown in Figure 2B, c-Jun/c-Fos bound almost exclusively to the CpG-methylated meAP-1 motif 5′-TGACTCG-3′ that contains a single CpG pair, but methylation of a CpG pair in a more central position of the motif decreased the binding of c-Jun/c-Fos (Supplementary Figure S3B). Therefore, we concentrated our analysis on meAP-1 binding sites with a CpG pair at the distal position. The consensus meAP-1 site consists of the sequence motif 5′-TGACTCG-3′, and only the methyl group in 5′ position of the cytosine in the bottom strand is necessary for the methylation-specific binding of c-Jun/c-Fos to DNA (Figure 2C).
To quantify DNA binding, the apparent dissociation constants (KD) of c-Jun/c-Fos were measured with unmethylated or methylated meAP-1 binding sites. The KD values of BBLF4, BALF1 and BFLF2 meAP-1 sites were lower by a factor of 38.7, 10.0 and 171, respectively, if methylated, indicating that the presence of 5′-methylcytosine increases the binding affinity of c-Jun/c-Fos to DNA (Figure 2D). The consensus AP-1 binding site from the human ‘collagenase’ promoter (TREcollag) with a published KD for c-Jun/c-Fos of 10.8 nM (30) was used as a positive control.
The transactivation potential of the c-Jun/c-Fos heterodimer was investigated in luciferase reporter assays. Reporter plasmids containing pentamers of the three selected meAP-1 sites were transiently transfected into HEK293 cells together with an expression plasmid encoding VP16:c-Jun/c-Fos, in which the VP16 transactivator domain was fused to the single-chain AP-1 heterodimer. VP16:c-Jun/c-Fos induced the CpG-methylated reporter plasmids by a factor of 2–5 as compared with their non-methylated controls (Figure 2E).
The human genome contains meAP-1 binding sites
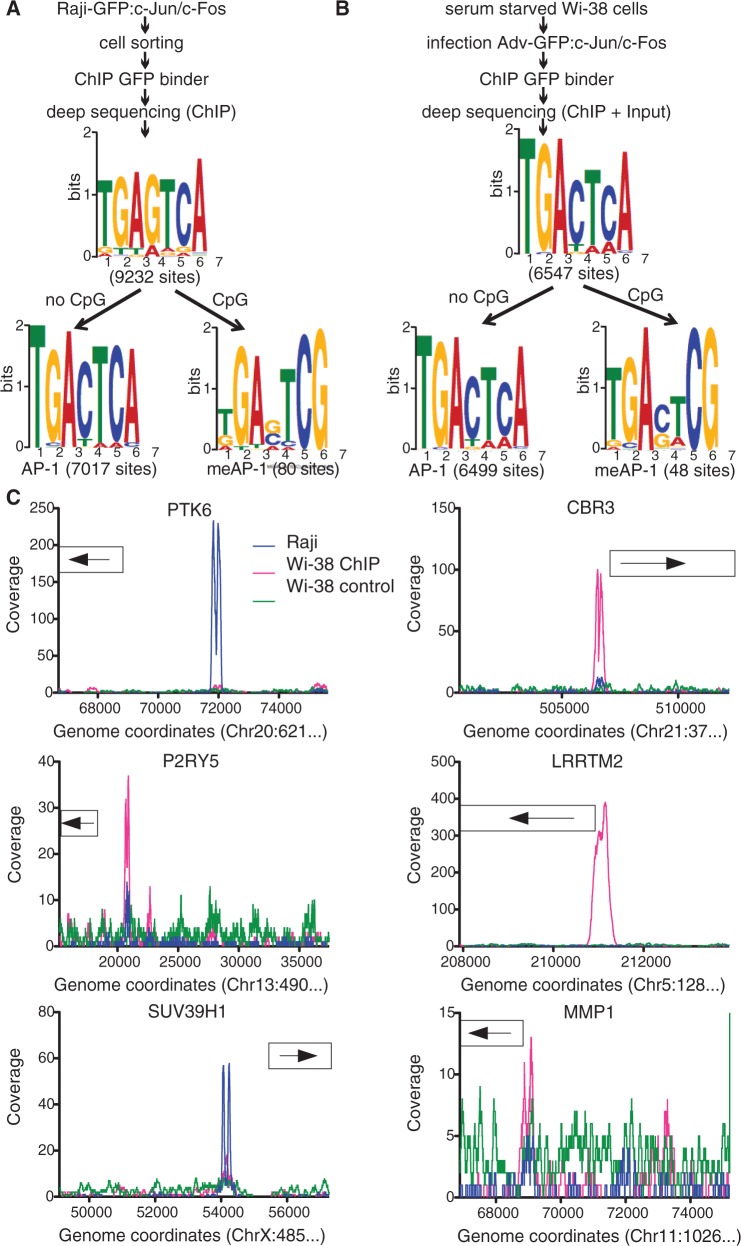
Two different cell lines were used to identify AP-1 and meAP-1 binding sites in vivo, the Burkitt lymphoma B-cell line Raji and the human fibroblast cell line Wi-38. As explained earlier in the text, a Raji cell line expressing the GFP:c-Jun/c-Fos chimera was used in ChIP-seq experiments. In a second experimental setting, serum-starved Wi-38 cells were infected with a GFP:c-Jun/c-Fos-expressing adenovirus. Chromatin from Wi-38 (two days post infection) and Raji cells was prepared, immunoprecipitated with an α-GFP antibody to enrich DNA fragments specifically bound by GFP:c-Jun/c-Fos protein and analyzed with next-generation sequencing (Figure 3).
Figure 3.
Identification of AP-1 binding motifs in the human genome. Identification of c-Jun/c-Fos binding motifs in Raji (A) and Wi-38 (B) cells. The samples used for deep sequencing were prepared as indicated. Data obtained from Raji and Wi-38 cell ChIPs were analyzed with the algorithms QuEST and SISSRs, respectively. MEME analysis was conducted as described in detail in the legend of Figure 1. In chromatin of Raji and Wi-38 cells, 9232 and 6547 motifs, respectively, were discovered, which were subsequently split into two groups. One group contains all motifs identified with the aid of the MEME algorithm harboring a CpG pair in positions 6 and 7 of the consensus motif. In all, 80 and 48 motifs were identified in Raji and Wi-38 cells, respectively, termed meAP-1. The remaining motifs were reanalyzed by MEME, which identified 7017 and 6499 sites that correspond to the known consensus AP-1 site lacking a CpG dinucleotide. (C) Visualization of selected human promoters with meAP-1 sites that bind c-Jun/c-Fos in vivo. The ‘collagenase’ (MMP1) promoter served as a positive control. Depicted is the read depth (coverage) at single base pair resolution (y-axis). The boxes indicate the position of the transcripts. Genomic coordinates correspond to the human genome assembly GRCh37/hg19.
In accordance with our first functional analysis shown in Supplementary Figure S3B and discussed earlier in the text, we adjusted the bioinformatic parameters and refined the characteristics of the two AP-1 motifs found previously: consensus AP-1 motifs lacking a CpG dinucleotide and putative meAP-1 motifs with a single distal CpG pair in positions six and seven (Figure 3A and B). The two predominant motifs revealed in both cellular models confirm the known AP-1 consensus (5′-TGACTCA-3′) and identified an infrequent second meAP-1 motif (5′-TGAC/GTCG-3′) (Figure 3A and B). Supplementary Tables S4 through S7 provide the location of the AP-1 and meAP-1 sites in both cell types.
The majority of all consensus AP-1 sites are located in intergenic regions (31,32) with no assigned functions. To concentrate our further analysis on meAP-1 sites in proximity of promoter elements, meAP-1 sites located within 5 kb upstream of annotated TSS were considered only if no other conventional AP-1 was located within a window of ±1 kb. Only a small number of cellular genes fulfilled our criteria. From ChIP-seq experiments of Raji cells, we included the promoters of three genes in our study, PTK6, P2RY5 and SUV39H1. In Wi-38 cells, we concentrated on two genes, CBR3 and LRRTM2. A positive control, the collagenase promoter MMP1 containing two AP-1 binding sites (33) was also included.
Visualization of the coverage of these promoters after ChIP-seq analysis indicated that they are differentially bound by c-Jun/c-Fos in the two cell types (Figure 3C), suggesting that the extent of cytosine methylation might also differ. Bisulfite sequencing of the five promoters revealed only one promoter element, P2RY5, that was fully CpG-methylated in both cell lines, but CpG pairs in other promoters with meAP-1 sites differed in a cell type-specific manner (Supplementary Figure S4).
5′-methylcytosine-dependent binding of c-Jun/c-Fos to meAP-1 sites in the human genome induces transcriptional activation
Only 5% of meAP-1 sites identified in both cell types are located in putative promoter regions, and therefore presumably involved in directly regulating gene expression. Three candidate meAP-1 motifs with no AP-1 consensus binding sites in their close proximity were analyzed further: the meAP-1 sites in the PTK6, P2RY5 and SUV39H1 promoters (Figure 4). As shown in Figure 4B, c-Jun/c-Fos bound almost exclusively to the CpG-methylated meAP-1 motif. In ELISAs, the apparent dissociation constants (KD) of PTK6 and P2RY5 meAP-1 sites could only be determined with methylated oligonucleotides because KD values of the unmethylated probes were too high to be estimated (Figure 4C). The KD for the SUV39H1 meAP-1 site could not be calculated, but the results confirm that the binding affinity is higher when the meAP-1 site is methylated (Figure 4B and C). These results indicate that the presence of 5′-methylcytosine favors the binding of c-Jun/c-Fos to the meAP-1 sequences identified in the human genome.
Figure 4.
Functional analysis of human meAP-1 sites. (A) Oligonucleotides containing meAP-1 binding sites of the PTK6, P2RY5 and SUV39H1 promoters. The meAP-1 sites are indicated in bold capital letters and the CpG pairs within the meAP-1 sites are underlined. (B) EMSAs showing the preferential binding of c-Jun/c-Fos to the CpG-methylated meAP-1 sites of the PTK6, P2RY5 and SUV39H1 promoters. EMSAs were performed with affinity-purified GFP-tagged c-Jun/c-Fos fusion protein transiently expressed in HEK293 cells. EMSAs of typical experiments are shown. (C) Determination of the apparent KD of c-Jun/c-Fos bound to meAP-1. ELISAs were performed with affinity-purified Strep/FLAG-tagged c-Jun/c-Fos fusion protein transiently expressed in HEK293 cells. Serial dilutions of Cy5 oligonucleotides were used to determine the KD values. The data were fitted to the Hill equation with one site-specific binding to determine the dissociation constants, which are indicated. Each data point indicates the mean and standard deviation of three independent experiments. (D) The c-Jun/c-Fos activation of promoters with meAP-1 binding sites is increased on CpG methylation. Pentamers of short oligonucleotides shown in (A), which contain the meAP-1 sites present in the promoters of the human PTK6, P2RY5 and SUV39H1 genes were introduced into a basic luciferase reporter plasmid as described in Figure 2E. Unmethylated and fully CpG-methylated reporter constructs were analyzed in the presence or absence of a cotransfected VP16:c-Jun/c-Fos expression plasmid. Each experiment was performed three times and means and standard deviations are depicted.
The ability of c-Jun/c-Fos to transactivate gene transcription through the binding to the methylated meAP-1 sites from the human genome was tested in reporter assays. Reporter plasmids containing pentamers of selected meAP-1 sites in the human promoters were cotransfected with an expression plasmid encoding the transactivator VP16:c-Jun/c-Fos or a negative control DNA. Luciferase activities with CpG-methylated reporter plasmids were significantly higher as compared with unmethylated reporter plasmid DNAs (2.0–2.7-fold, Figure 4D).
Promoters with methylated meAP-1 sites are bound by c-Jun/c-Fos in vivo
So far our studies were based on the ectopic expression of artificial c-Jun/c-Fos dimers to identify potential meAP-1 sites in cellular DNA. In contrast, Raha et al. (31) mapped the location of endogenous c-Jun and c-Fos protein in two sets of ChIP-seq experiments and identified >40 000 binding sites in the established human myeloblastic cell line K562. To correlate their findings and ours, we performed ChIP assays in this cell line with a c-Jun specific antibody and investigated by quantitative PCR analysis the enrichment of the five previously identified promoter elements PTK6, P2RY5, SUV39H1, CBR3 and LRRTM2 along with negative and positive control regions (34). As shown in Supplementary Figure S5, the previously identified five promoter elements, which contain only meAP-1 but no consensus AP-1 sites, were enriched at levels similar to or exceeding those of the positive controls in K562 chromatin indicating that endogenous AP-1 can target meAP-1 sites in the context of cellular chromatin.
Next, we analyzed the state of CpG methylation in K562 cell DNA by bisulfite sequencing. The repetitive meAP-1 site within the P2RY5 promoter was fully methylated (Supplementary Figure S6A) similar to the situation in Raji and Wi-38 cells (Supplementary Figure S4A and B). In contrast, two other promoters were only moderately methylated (Supplementary Figure S6A). If c-Jun/c-Fos preferentially bound to methylated meAP-1 sites, ChIP experiments with an α-c-Jun antibody should enrich methylated DNA. A clear enrichment of methylated CpGs could be detected in the two promoters of the PTK6 and LRRTM2 genes sequenced after bisulfite treatment (Supplementary Figure S6B). The single functionally identified meAP-1 binding site in the PTK6 promoter in Raji cell chromatin (Figure 3C) was enriched 3-fold after ChIP in K562 cells, whereas the remaining three potential meAP-1 sites in this promoter, which were only predicted with the aid of the search algorithm FIMO (26), could not be confirmed in this functional analysis (Supplementary Figure S6B), supporting the validity of our approach.
In vivo, c-Jun binds meAP-1 motifs in promoters of genes upregulated in activated primary human B cells
Our detailed characterization of the newly discovered meAP-1 motif provided evidence that c-Jun/c-Fos binds these sequences in vivo. To substantiate our findings, we wanted to assess if meAP-1 sites are also capable of conferring transcriptional regulation in the context of cellular chromatin in a relevant model. AP-1 signaling cascades play an important role in physiological B cell activation and differentiation. Comprehensive data sets are publicly available that address key aspects of the biology of primary lymphocytes comparing genes expressed in resting and activated naïve human B cells (29). In addition, the methylome of naïve human B cells has been determined recently (28).
Individual AP-1 and meAP1 sites recruit the heterodimer c-Jun/c-Fos in a highly cell specific manner as exemplified in Figure 3C comparing Raji B cells and Wi-38 fibroblasts. Therefore, we turned to the search algorithm FIMO (26) to detect potential AP-1 and meAP-1 binding sites in the reference human genome assembly GRCh37/hg19 (35) (Supplementary Tables S8 and S9, respectively). Next, we identified those genes in human DNA that (i) contain potential meAP-1 binding sites within their promoter regions, from −5 kb to +1 kb relative to the TSS, and (ii) were upregulated by a factor of two or more comparing activated with resting human naïve B cells 16 h post induction with CD40 ligand, CpG stimulation and B cell receptor cross-linking (29). Finally, we excluded from this list those meAP-1 sites in which the degree of cytosine methylation of the CpG pair was <80% in naïve B cells (28). Applying these bioinformatic selection criteria, we identified ∼280 genes (Supplementary Table S10), which were upregulated in activated B cells (29) and contain methylated meAP-1 sites in their promoters. To focus our attention on the most promising candidates, we picked 10 genes with presumed functional meAP-1 sites located within the first 2 kb upstream of the TSS that are at least 1 kb apart from a consensus AP-1 site, if any.
Table 1 lists the selected genes that fulfill all these stringent criteria and contain the consensus meAP-1 motif 5′-TGACTCG-3′. Next, we isolated naïve primary human B cells, which were activated with CD40-ligand and IL-4 for 5 days to induce their proliferation and activate cellular AP-1 pathways. ChIP assays with a c-Jun-specific antibody were performed and the enrichment of the in silico selected promoter elements was assessed by quantitative PCR analysis. As shown in Figure 5, the 10 promoter elements were enriched at levels similar to those of the positive controls in activated B cells. The selected meAP-1 sites were confirmed to be methylated by analyzing the state of CpG methylation in genomic DNA of non-activated B cells by bisulfite sequencing (Table 1 and Supplementary Figure S8). Supporting our finding in primary B cells, the 10 promoter regions were also bound by c-Jun in the K562 cell line (Supplementary Figure S7).
Table 1.
Selection criteria and experimental results of promoters of meAP-1-regulated genes in naïve human B cells and the cell line K562
| Gene name | Fold upregulationa | CpG methylationb (%) | Distance meAP-1 to TSS (nt) | Distance AP-1 to TSS (nt) | ChIP in primary B cellsc | BS-seqd (%) | ChIP in K562 cellse |
|---|---|---|---|---|---|---|---|
| DCTPP1 | 14.03 | >98 | −678 | n.a. | 2.5 | >98 | 3.0 |
| SLAMF1 | 6.13 | 100 | −1698 | n.a. | 2.6 | >90 | 4.8 |
| UQCRB | 3.89 | >90 | −1244 | n.a. | 3.0 | >98 | 4.0 |
| SCRIB | 3.04 | >80 | −1625 | n.a. | 3.3 | >90 | 3.6 |
| VAT1 | 2.01 | >90 | −1735 | n.a. | 2.7 | >98 | 4.2 |
| SAE1 | 5.79 | >90 | −718 | −2831 | 2.3 | >98 | 5.3 |
| PSMC1f | 4.69 | >80 | −1418 | −2532 | 3.0 | 100 | 6.0 |
| >90 | −929 | 2.6 | n.d. | 4.1 | |||
| ITFG2 | 3.09 | >90 | −1007 | −4336 | 2.7 | >90 | 4.4 |
| LARP4B | 2.85 | >80 | −1272 | −4707 | 2.6 | >98% | 3.6 |
| FGR | 2.06 | >90 | −744 | −2004 | 2.5 | >90% | 7.0 |
n.a., not applicable; n.d., not done.
aTranscriptional activation in naïve B cells 16 h post stimulation (29).
bmethylome data from resting naïve B cells (28).
cdata derived from Figure 5, fold enrichment compared with the mean of negative controls.
dBS-seq; bisulfite sequencing; data derived from Supplementary Figure S8.
eData derived from Supplementary Figure S7, fold enrichment compared with the mean of negative controls.
fPSMC1 promoter encompasses two meAP1 and one AP-1 site.
Figure 5.
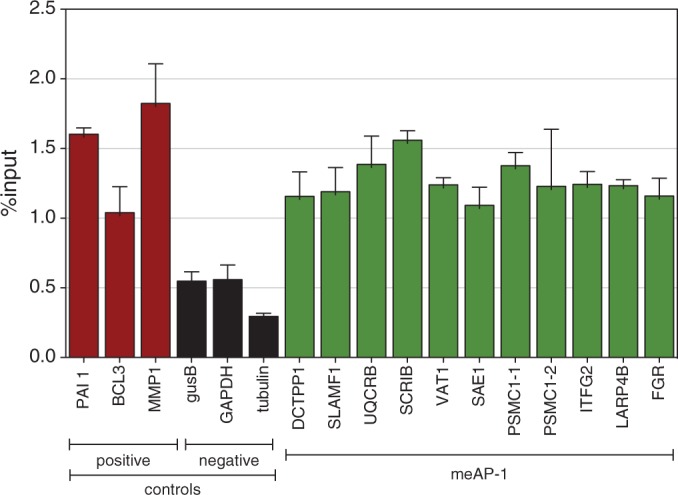
In vivo binding of c-Jun/c-Fos to cellular promoters with meAP-1 sites in activated primary human B cells. After immunoprecipitation (ChIP) of chromatin from activated naïve B cells with an α-c-Jun antibody, the enrichment of the indicated promoter regions was assayed by quantitative PCR. Shown are the results obtained from three promoter regions indicated in red that contain conventional previously identified AP-1 sites (34) and three promoter regions indicated in black, which were identified in this study and are devoid of conventional AP-1 or meAP-1 sites. Both sets of promoters served as positive and negative controls, respectively. Ten promoters that were found to contain meAP-1 sites (and lack conventional AP-1 sites in proximity) were analyzed by ChIP and the results are shown in green. The PSMC1 promoter contains two separate regions with one meAP-1 site each depicted as PSMC1-1 and -2, which were analyzed separately. Details of all promoters investigated here are listed in Table 1. Means and standard deviations of three independent experiments are shown.
Table 1 summarizes the original data that led to the in silico identification of the 10 genes and the experimental results obtained in this work. Our data indicate that the predicted meAP-1 binding sites are CpG-methylated and targeted by c-Jun in activated B cells in vivo, suggesting that individual meAP-1 sites contribute to transcriptional activation of highly CpG-methylated, hence epigenetically repressed genes in naïve B lymphocytes on their activation. One caveat remains that cannot be addressed experimentally: mitogenic physiological signals in B cell proliferation and development-like B cell receptor cross-linking, CD40 engagement and interleukin receptor signals induce the MAPK cascades leading to AP-1 activation but also engage additional signaling cascades that might also act on meAP-1-regulated genes assisting their transcriptional reactivation (36).
DISCUSSION
The aforementioned results confirm our starting hypothesis and indicate that AP-1 has the capacity to bind a class of previously unknown discrete DNA sequence motifs with methylated cytosine residues in vivo. It thus appears that Zta is not only a closely related member of the large AP-1 family because it shares a highly conserved DNA binding domain with c-Fos (6), but also may have evolved from a predecessor of the AP-1 protein family, which can specifically make contacts with the 5′-methyl group of cytosine nucleotides. We propose that additional members of the vast AP-1 family can also bind to methylated DNA in a sequence-specific manner.
A close inspection of ZRE/meZRE and AP-1/meAP-1 pairs (Figure 6) suggests that they are related but have evolved differently. (i) In general, nucleotide transitions (but not transversions) cause the differences within the ZRE/meZRE and AP-1/meAP-1 pairs (Figure 6A) reflecting only minor differences in the molecular structures of thymine and 5′-methylcytosine (Figure 6B). (ii) The position of the methylated cytosine in the consensus sequences of meZRE and meAP-1 is offset by one nucleotide (Figure 6A). (iii) The consensus AP-1 motif is almost palindromic but the meAP-1 motif is not, suggesting that either c-Jun or c-Fos preferentially make contact with the 5′-methylcytosine group. This view is supported by two detailed reports indicating that c-Fos and c-Jun are unequal siblings (37,38). Interestingly, Risse et al. (38) delineated the importance of individual thymine residues in the AP-1 consensus sequence and found that all thymines including the two distal positions crucially contribute to DNA–protein interactions.
Figure 6.
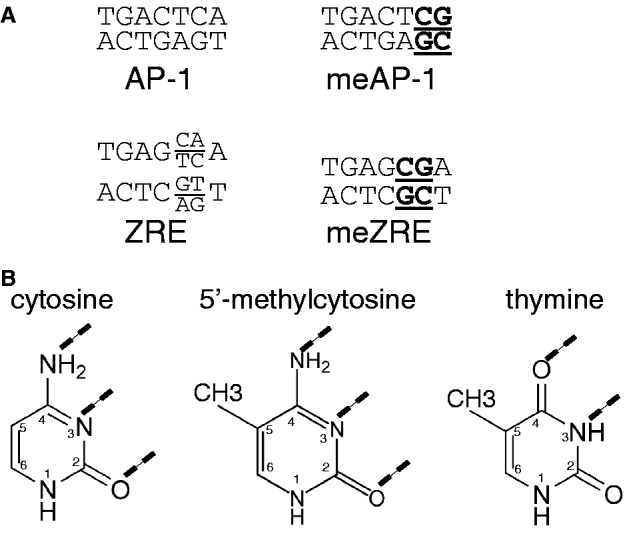
Sequence motifs of AP-1 and ZRE binding sites and their molecular features. (A) Comparison of AP-1, ZRE, meAP-1 and meZRE binding sites. The CpG pairs in the meAP-1 and meZRE motifs are underlined. (B) Structure of the DNA bases cytosine, 5′-methylcytosine and thymine. The dashed lines indicate the hydrogen bonds with the bases of the complementary DNA strand.
Zta is the first example of a sequence-specific transcription factor, which preferentially recognizes and selectively binds DNA sequence motifs with methylated CpG residues and activates gene transcription through these distinct motifs (4,5). Previously, other mammalian factors (RXF1, RBP-J, ZBTB4) have been shown to bind DNA sequences encompassing 5′-methylcytosine nucleotides in vitro, only, and no evidence for methylation-dependent binding of these factors exists in cells in vivo (39–42). Retrospectively, this failure is not surprising because consensus meAP-1 motifs are rarely targeted by c-Jun/c-Fos in cellular chromatin. Accessible and therefore functional meAP-1 sites account for <2% of the 45 000 c-Jun/c-Fos binding sites that are predicted in human genomic DNA (Figure 3A and B; Supplementary Table S9). This is in contrast to the high number of consensus AP-1 sites predicted in silico (32), which are readily bound by factors of the AP-1 family in human chromatin (31,34).
Interestingly, a recent systematic in vitro analysis of a large panel of transcription factors identified their frequent binding to sequence motifs with methylated CpG dinucleotides (43). The search did not identify the non-canonical meAP-1 motif reported here. Rather, it revealed distinct and therefore unrelated motifs either free of CpG dinucleotides or with methylated CpGs targeted by the same mammalian transcription factor. This observation is surprising and clearly in contrast to our findings, which led to the discovery of the meAP-1 motif, a close relative of the canonical AP-1 site (Figure 6).
Our results indicate that the 5′-methylcytosine residue can functionally substitute for the crucial distal thymine in one strand of the AP-1 consensus motif providing similar molecular structures for AP-1 binding (Figures 2C and 6). The structural resemblance of thymine to 5′-methylcytosine suggests that the c-Jun/c-Fos heterodimer binds similarly to AP-1 and meAP-1 sites (39,44). As c-Jun/c-Fos heterodimers target meAP-1 sites in vivo (Figures 3 and 5, Supplementary Figures S5 and S7), the status of DNA methylation determines AP-1 binding and consequently responsiveness to AP-1-mediated transcriptional activation. In this context, DNA methylation adds a new layer to gene regulation and presumably also cell type- and tissue-specific functionality of signaling pathways that activate AP-1 family members.
The majority of consensus AP-1 sites that were identified in cellular chromatin are located in intergenic regions or are distant from transcriptional start sites and have unknown functions (31,34). We see a comparable distribution of meAP-1 sites. Of the 45 000 potential meAP-1 sites (Supplementary Table S9), only a small fraction resides within 5 kb upstream of transcriptional start sites suggesting that these meAP-1 sites might serve an important and direct role in regulating genes that are governed by epigenetic inhibition including DNA methylation. The functions of the many consensus AP-1 and non-canonical meAP-1 site localized in intergenic regions remain an enigma that needs to be addressed.
Owing to the nature and frequency of potentially functional meAP-1 binding sites in the cell lines and conditions used in this study, we had to restrict our search in established cell lines to promoters that are bound by c-Jun/c-Fos heterodimers and contain meAP-1 sites but lack consensus AP-1 sites in proximity for obvious technical reasons. The strict filtering criteria resulted in a small set of promoter candidates that likely do not reflect profound or even decisive functions of the identified genes in human cells. The situation is different in primary naïve B cells of human origin, in which we concentrated on meAP-1 in the promoters of genes upregulated after induced B cell activation. The fact that AP-1 is binding these meAP-1 binding sites suggests their having functional roles in metazoan gene regulation in certain cell types and certain situations, e.g. during the transition from quiescent, resting B lymphocytes to proliferating B blasts. In this context, c-Jun/c-Fos differs from Zta, which frequently binds to many viral meZRE-containing promoters and efficiently reverses their epigenetic repression (5,7,14,45). These differences could be explained by the fact that the key functions of Zta in the EBV model are understood, whereas the c-Jun/c-Fos transcription factor might have yet to be uncovered functional roles in different settings. Although the functionality of the meAP-1 binding sites cannot be fully uncovered with the experiments presented here, the experiments document the in vivo existence of these elements, which are bound preferentially if their DNA motif carries a 5′-methylcytosine. It is likely that these elements play a critical role in the transcriptional activation of their target genes. These findings now allow the investigation of other experimental settings such as different cell lines or primary cell types as well as distinct stimuli, stress signals or developmental cues that activate cells, induce their immediate differentiation or trigger cell fate decisions. The goal is now to discover more examples of meAP-1 sites that regulate the expression of critical genes to unveil the full range of meAP-1 elements and their biological functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Alexander von Humboldt Foundation [Humboldt-Forschungsstipendium für Postdoktoranden to M.G.]; intramural grants, the Deutsche Forschungsgemeinschaft [SPP1230, SFB 1054, SFB 1064, SFB-TR36]; National Institutes of Health [CA70723]. Funding for open access charge: Helmholtz Zentrum München.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Christine Göbel for technical advice with the luciferase assays, Dagmar Pich for her support with cell sorting and Vigo Heissmeyer for his adenoviral vector constructs, advice and helpful discussions. They also thank Reiner Siebert (Institut für Humangenetik, Universitätsklinikum Schleswig-Holstein, Christian-Albrechts-Universität zu Kiel) and Steve Hoffmann (Transcriptome Bioinformatics, LIFE Research Complex, University Leipzig) for the valuable access to the methylome data of primary naïve B lymphocytes of three human donors and helpful suggestions and discussions.
REFERENCES
- 1.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 2.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 3.Prokhortchouk E, Defossez PA. The cell biology of DNA methylation in mammals. Biochim. Biophys. Acta. 2008;1783:2167–2173. doi: 10.1016/j.bbamcr.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 2004;36:1099–1104. doi: 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]
- 5.Bergbauer M, Kalla M, Schmeinck A, Gobel C, Rothbauer U, Eck S, Benet-Pages A, Strom TM, Hammerschmidt W. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathog. 2010;6:e1001114. doi: 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woellmer A, Arteaga-Salas JM, Hammerschmidt W. BZLF1 governs CpG-methylated chromatin of Epstein-Barr virus reversing epigenetic repression. PLoS Pathog. 2012;8:e1002902. doi: 10.1371/journal.ppat.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes. Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 9.Hantusch B, Kalt R, Krieger S, Puri C, Kerjaschki D. Sp1/Sp3 and DNA-methylation contribute to basal transcriptional activation of human podoplanin in MG63 versus Saos-2 osteoblastic cells. BMC Mol. Biol. 2007;8:20. doi: 10.1186/1471-2199-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl Acad. Sci. USA. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes. Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 12.Holzerlandt R, Orengo C, Kellam P, Alba MM. Identification of new herpesvirus gene homologs in the human genome. Genome Res. 2002;12:1739–1748. doi: 10.1101/gr.334302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinclair AJ. bZIP proteins of human gammaherpesviruses. J. Gen. Virol. 2003;84:1941–1949. doi: 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- 14.Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl Acad. Sci. USA. 2010;107:850–855. doi: 10.1073/pnas.0911948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiesner M, Zentz C, Mayr C, Wimmer R, Hammerschmidt W, Zeidler R, Moosmann A. Conditional immortalization of human B cells by CD40 ligation. PLoS One. 2008;3:e1464. doi: 10.1371/journal.pone.0001464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl Acad. Sci. USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 2002;22:4952–4964. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gloeckner CJ, Boldt K, Schumacher A, Roepman R, Ueffing M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics. 2007;7:4228–4234. doi: 10.1002/pmic.200700038. [DOI] [PubMed] [Google Scholar]
- 19.Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 20.Kalla M, Gobel C, Hammerschmidt W. The lytic phase of Epstein-Barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J. Virol. 2012;86:447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 29.Le Gallou S, Caron G, Delaloy C, Rossille D, Tarte K, Fest T. IL-2 requirement for human plasma cell generation: coupling differentiation and proliferation by enhancing MAPK-ERK signaling. J. Immunol. 2012;189:161–173. doi: 10.4049/jimmunol.1200301. [DOI] [PubMed] [Google Scholar]
- 30.Chytil M, Peterson BR, Erlanson DA, Verdine GL. The orientation of the AP-1 heterodimer on DNA strongly affects transcriptional potency. Proc. Natl Acad. Sci. USA. 1998;95:14076–14081. doi: 10.1073/pnas.95.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc. Natl Acad. Sci. USA. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H, Zarubin T, Ji Z, Min Z, Zhu W, Downey JS, Lin S, Han J. Frequency and distribution of AP-1 sites in the human genome. DNA Res. 2005;12:139–150. doi: 10.1093/dnares/12.2.139. [DOI] [PubMed] [Google Scholar]
- 33.Benbow U, Brinckerhoff CE. The AP-1 site and MMP gene regulation: what is all the fuss about? Matrix Biol. 1997;15:519–526. doi: 10.1016/s0945-053x(97)90026-3. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Ge Q, Wang W, Wang J, Lu Z. c-Jun binding site identification in K562 cells. J. Genet. Genomics. 2011;38:235–242. doi: 10.1016/j.jgg.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards S, Watanabe C, Santos L, Craxton A, Clark EA. Regulation of B-cell entry into the cell cycle. Immunol. Rev. 2008;224:183–200. doi: 10.1111/j.1600-065X.2008.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard DA, Kerppola TK. DNA bending determines Fos-Jun heterodimer orientation. Nat. Struct. Biol. 1998;5:877–881. doi: 10.1038/2316. [DOI] [PubMed] [Google Scholar]
- 38.Risse G, Jooss K, Neuberg M, Bruller HJ, Muller R. Asymmetrical recognition of the palindromic AP1 binding site (TRE) by Fos protein complexes. EMBO J. 1989;8:3825–3832. doi: 10.1002/j.1460-2075.1989.tb08560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartels SJ, Spruijt CG, Brinkman AB, Jansen PW, Vermeulen M, Stunnenberg HG. A SILAC-based screen for methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein. PLoS One. 2011;6:e25884. doi: 10.1371/journal.pone.0025884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niesen MI, Osborne AR, Yang H, Rastogi S, Chellappan S, Cheng JQ, Boss JM, Blanck G. Activation of a methylated promoter mediated by a sequence-specific DNA-binding protein, RFX. J. Biol. Chem. 2005;280:38914–38922. doi: 10.1074/jbc.M504633200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang XY, Jabrane-Ferrat N, Asiedu CK, Samac S, Peterlin BM, Ehrlich M. The major histocompatibility complex class II promoter-binding protein RFX (NF-X) is a methylated DNA-binding protein. Mol. Cell. Biol. 1993;13:6810–6818. doi: 10.1128/mcb.13.11.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, Shin J, Cox E, Rho HS, Woodard C, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 45.Karlsson QH, Schelcher C, Verrall E, Petosa C, Sinclair AJ. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog. 2008;4:e1000005. doi: 10.1371/journal.ppat.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.