Abstract
Activation of gene transcription in eukaryotes requires the cooperative assembly of an initiation complex containing many protein subunits. The necessity that these components contact each other and the promoter/enhancer in defined ways suggests that their spatial arrangement might influence the activation response. Indeed, growing evidence indicates that DNA architecture can profoundly affect transcriptional potency. Much less is known about the influence of protein architecture on transcriptional activation. Here, we examine the architectural dependence of activator function through the analysis of matched pairs of AP-1•DNA complexes differing only in their orientation. Mutation of a critical Arg residue in the basic-leucine zipper domain of either Fos or Jun yielded single point-mutant heterodimers that bind DNA in a single defined orientation, as determined directly by native chemical ligation/affinity cleavage; by contrast, the corresponding wild-type protein binds DNA as a roughly equal mixture of two isomeric orientations, which are related by subunit interchange. The stereochemistry of the point-mutant heterodimers could be switched by inversion of a C•G base pair in the center of the AP-1 site, thus providing access to both fixed orientational isomers. Yeast reporter gene assays consistently revealed that one orientational isomer activates transcription at least 10-fold more strongly than the other. These results suggest that protein architecture, especially the spatial relationship of the activation domain to the promoter, can exert a powerful influence on activator potency.
Keywords: transcriptional activation/orientational isomer/NFAT/bZip
Transcriptional initiation in eukaryotes requires the assembly of a multiprotein complex comprising RNA polymerase and its associated subunits, together with a variety of sequence-specific DNA binding and adaptor proteins (1). The latter appear to function by binding to specific sequences in the promoter/enhancer and establishing direct protein–protein contacts with components of the transcriptional machinery (1, 2), including chromatin-remodeling proteins (3). The DNA binding and activation functions of activator protein typically reside on distinct domains; a wealth of structural information is available regarding sequence-specific recognition by DNA-binding domains (4), but relatively little detailed structural information has been obtained on activation domain and their targets (5). One reason for this paucity of information stems from the fact that individual activation domains typically interact weakly with their targets, and activation usually requires a number of such interactions to be established cooperatively (1, 2). Because these cooperative ensembles involve multiple activation domains contacting multiple targets, it stands to reason that the entire system should be influenced strongly by geometrical constraints. Indeed, recent results on the cooperative assembly of enhanceosomes highlight the importance of DNA architecture in producing activation responses (6). The role of protein architecture in determining the strength of transcriptional responses, on the other hand, has received less attention (7).
Given the high degree of geometrical specificity expected to be inherent in transcriptional activation (6), it came as a surprise to discover that one of the most well-studied and ubiquitous activator proteins, AP-1, binds DNA as a roughly equal mixture of two distinct orientational isomers (8), which are related by interchange of the AP-1 subunits, Fos and Jun. Mixtures of orientational isomers also are observed in the solid state (9). Fos and Jun are homologous proteins having basic-leucine zipper (bZip) domains that pair to bind DNA as a Y-shaped heterodimer (Fig. 1A) (9). Just as intriguing was the discovery that the orientational degeneracy in AP-1•DNA recognition could be overcome through cooperative protein–protein interactions on DNA; namely, in the ternary DNA complex formed with AP-1 and the nuclear factor of activated T cells (NF-AT), AP-1 adopts a single fixed orientation (8). These findings thus raised the question as to whether the two orientational isomers of AP-1 might differ in their ability to activate transcription. To address this issue, we have devised a strategy to fix the orientation of AP-1 on DNA, thus allowing us to test the activation potential of the two orientational isomers directly. Here, we report that in the present experimental systems the two individual orientational isomers of the AP-1•DNA complex differ by an order of magnitude in the transcriptional response they produce. We propose a “stereochemical” model to explain the present observations: the distinctive protein architectures of the two oriented AP-1•DNA complexes give rise to spatial differences in the way the chemical functionality on the activation domain is presented to the transcriptional machinery, which in turn affects the strength or rate of interaction between the two. With natural promoters, wherein multiple activators simultaneously contact multiple targets, stereochemical issues seem likely to exert a profound influence on the generation and evolution of activation responses.
Figure 1.
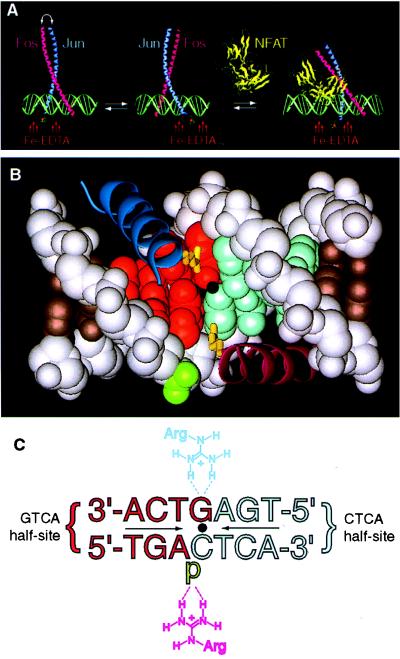
The stereochemistry of DNA-bound Fos and Jun alone and in the presence of NFAT. (A) AP-1 alone binds DNA as a mixture of two stereoisomers, which are related by subunit interchange (left and center). In the cooperative complex formed by AP-1 and NFAT on DNA (right), Fos•Jun is locked into a single stereochemical orientation. The stereochemistry of the protein•DNA complex is reported by a covalently attached Fe-EDTA moiety, which effects proximity-directed hydroxyl radical cleavage of DNA. The model is derived from the x-ray structure of AP-1•DNA (9) and NFATp•AP-1•DNA (34). (B) Contacts between the basic regions of Fos•Jun central base pair of a consensus AP-1 site (9). In the orientation shown, Jun (dark blue α-helix) is bound over the GTCA half-site (red space-filling model), and its residue Arg-279 (yellow tube) makes a base-specific bidentate hydrogen bond to the central guanine; Fos (purple α-helix) is bound over the CTCA half-site (light blue space-filling model), and its residue Arg-155 makes a nonspecific contact to the phosphate flanking the central cytosine (green). In the other orientational isomer (not shown), the locations of Fos and Jun are swapped, and the roles of Arg-155 and Arg-279 are interchanged. Dot denotes the center of pseudodyad symmetry. (C) Schematic representation of the contacts shown in Fig. 1B (p, phosphate). Coloring and orientation matched those of Fig. 1B. Note: the strand sense in Fig. 1C is written opposite to the usual convention (i.e., the sequence of the top strand is written in the 3′–5′ orientation going from left to right) to facilitate comparison of Figs. 1B and C; all other figures present the sequence in the conventional sense.
MATERIALS AND METHODS
In Vitro Experiments.
The bZip domains of Fos (residues 134–200, preceded by a N-terminal Cys) and Jun (residues 258–324, preceded by a N-terminal Cys) and the corresponding Ala mutant Fos (R155A) and Jun (R279A) proteins (9) were overexpressed, purified, and conjugated to EDTA by using native chemical ligation as described previously (10); electrophoretic mobility shift assays and affinity cleavage experiments were performed as described previously (10). All constructs contained an additional mutation, Fos (C154S) and Jun (C278S), which confers redox insensitivity but has no effect on dimerization or DNA binding (11).
Transcriptional Assays and Yeast Constructs.
The transcriptional assays, yeast strains, and reporter constructs are described in ref. 12. The B42 activation domain (tandem AP-1 site experiments) or stronger VP16 activation domain (single AP-1 site experiments) were fused to the N terminus (basic region) of the cFos bZip domain (residues 118–203) by standard constructions in the vector pJG4–5 (13). cJun (residues 242–327) was expressed under the control of a GAL1 promoter and ADH1 transcriptional terminator from the HIS3-selectable plasmid pAM423 (A. Mendelsohn and R. Brent, personal communication). The centers of dyad symmetry of the tandem consensus AP-1 sites studied in vivo were separated by an 18-bp spacer and were located at −250 (single and tandem AP-1 site constructs) and −269 bp (tandem AP-1 construct only) upstream of the transcription startpoint.
RESULTS
Strategy.
The consensus AP-1 site is an imperfect palindrome, having identical residues at all symmetry-related positions, except the central base pair (Fig. 1B, C). Because the site is not perfectly symmetric, Fos and Jun could in principle recognize the two half-sites (5′-GTCA and 5′-CTCA, Fig. 1C) differently by making nonidentical contacts to the central base pair. Indeed, the x-ray crystal structure of the AP-1•DNA complex (9) reveals that the AP-1 heterodimer does contact the central base pair asymmetrically, with one subunit making an Arg–G contact (GTCA half-site) and the other subunit making an Arg–phosphate contact (CTCA half-site, Fig. 1B, C). However, because these Arg residues (Arg-279 of Jun and Arg-155 of Fos) occupy equivalent positions in Fos and Jun, their roles can be freely interchanged. Furthermore, all other residues that contact DNA also are conserved in Fos and Jun. For these reasons, the Fos and Jun subunits are incapable of distinguishing between the two AP-1 half-sites, and consequently the Fos•Jun heterodimer binds the AP-1 site in two stereochemically isomeric orientations, which are related by subunit interchange (Fig. 1A). If and only if the Arg–G and Arg–phosphate contacts to the central base pair are energetically inequivalent, then removing one of these contacts should break the degeneracy of the half-site contacts, thereby yielding a single, stereochemically oriented heterodimer. The GCN4 homodimer, a yeast AP-1 homolog, has been shown to bind the AP-1 site in an energetically asymmetric manner, with the GTCA half-site being bound more strongly than the CTCA half-site (14–16).
Native Chemical Ligation/Affinity Cleavage Experiments.
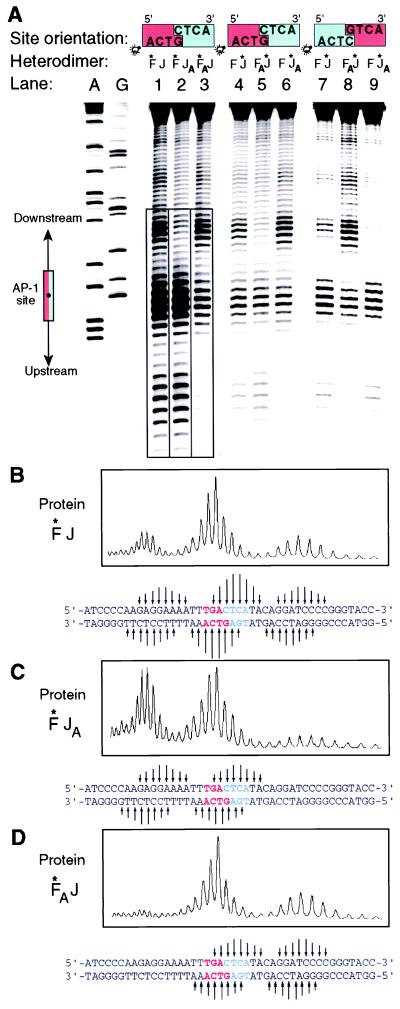
To test the effect of amino acid changes on AP-1 orientation, we mutated Arg-279 of Jun and Arg-155 of Fos to Ala, thus generating the single point-mutant forms JunAla and FosAla, respectively. These single point-mutant subunits were then heterodimerized with the corresponding wild-type Fos or Jun subunit. To determine the orientation of the Fos•JunAla and FosAla•Jun heterodimers on DNA, we attached the iron-chelating moiety EDTA onto one of the two subunits via the native chemical ligation procedure developed for use with recombinant proteins (10). This procedure permits the selective ligation of nonnative functionality—in this case, EDTA—onto the N terminus of a recombinant protein through amide bond formation (see also ref. 17). The EDTA moiety in our conjugated proteins effects proximity-directed hydroxyl radical cleavage of the DNA, thereby reporting the location of the tagged basic region with respect to the AP-1 site. As demonstrated previously (Fig. 1A) (8, 18), this procedure yields distinct DNA cleavage patterns for oriented heterodimers versus unoriented heterodimers and is thus a test for the orienting effects of the mutations. Consistent with earlier reports (8, 10), the heterodimer formed between wild-type EDTA-conjugated Fos and wild-type Jun (Fos*•Jun) yields a tripartite cleavage envelope (Fig. 2A, lane 1 and Fig. 2B), which is characteristic for a mixture of two nonoriented bZip heterodimers (8, 10, 18). As expected, the corresponding heterodimer having EDTA on Jun (Fos•Jun*) yields virtually identical tripartite cleavage envelope (Fig. 2A, lane 4). Inversion of the central C•G base pair has no effect on the affinity cleavage pattern produced by Fos•Jun* (Fig. 2A, lane 7) or Fos*•Jun (data not shown). Strikingly, Fos* heterodimerized with the point-mutant JunAla (Fos*•JunAla) produces a bipartite cleavage pattern (Fig. 2A, lane 2 and Fig. 2C), thus indicating that the removal of a single Arg side chain is sufficient to orient the AP-1 heterodimer on DNA. The observed orientation places the wild-type subunit over the GTCA half-site, thus allowing Arg-155 of Fos to make a bidentate hydrogen bond to the central base pair (refer to Fig. 1B, upper protein subunit and Fig. 1C, upper Arg). Switching the mutation from Jun to Fos results in inversion of the bipartite cleavage pattern (compare Fos*•JunAla vs. Fos*Ala•Jun; Fig. 2A, lanes 2 vs. 3 and Fig. 2C vs. Fig. 2D, respectively), again indicating conservation of the Arg–G contact. Inversion of the cleavage pattern also resulted from moving the Fe-EDTA label from one subunit to the other (Fig. 2A, lane 2 vs. 6 and 3 vs. 5). If the key Arg contact to the central G residue drives the preference for a particular oriented AP-1 stereoisomer, then flipping the central C•G pair in the AP-1 site should invert the affinity cleavage pattern. Such behavior is indeed observed (Fig. 2A, lanes 5 vs. 8 and 6 vs. 9).
Figure 2.
Affinity cleavage of DNA by Fe(III)-EDTA-modified peptides. (A) Autoradiogram of a DNA sequencing gel analyzing the products of affinity cleavage. The orientation of the site is depicted at the top, with the GTCA half-site in red and the CTCA half-site in blue. The end bearing the 33P label is indicated by a light bulb. The diagram on the far left depicts the location of consensus AP-1 site. A and G refer to the Maxam-Gilbert A- and G-specific reactions, respectively, used as sequence markers. Boxed lanes on the autoradiogram indicate those that were phosphorimaged to produce B-D. (B-D) Phosphorimage analysis of affinity cleavage reactions. Arrows under the panels denote the locations of cleaved bases, with the length being roughly proportional to the relative intensity of the cleavage.
Impact of Arg Mutations on the Affinity of AP-1.
To assess the thermodynamic consequences of mutating a DNA contact residue in AP-1, we determined the equilibrium dissociation constant (Kd) of AP-1 single and double point-mutant proteins for a consensus AP-1 site in DNA. The results of this analysis are presented in Table 1. The Fos•JunAla heterodimer bound DNA with equivalent or perhaps slightly greater affinity than that of Fos•Jun. On the other hand, transposition of the Ala mutation from Jun to Fos (FosAla•Jun) resulted in a 2-fold decrease in affinity relative to Fos•Jun and Fos•JunAla. Mutation of the key Arg residue in both Fos and Jun yielded a double point-mutant (FosAla•JunAla) that is virtually devoid of sequence-specific DNA-binding activity. Combining these thermodynamic data with the orientational results described above, we conclude that the Arg–phosphate contact made by either Arg-279 of Jun or Arg-155 of Fos contributes little if any binding energy to formation of the protein–DNA complex; on the other hand, the Arg–G bidentate hydrogen bond contributes significantly to the overall energetics of complexation.
Table 1.
Binding affinities in nM
| Fos•Jun | 10.8 ± 0.7 |
| FosAla•Jun | 22.6 ± 0.9 |
| Fos•JunAla | 7.1 ± 0.3 |
| FosAla•JunAla | >500 |
Stereochemical Effects in Transcriptional Activation.
The observation that AP-1 itself binds DNA as a mixture of orientational isomers (8, 9), but binds together with NFAT in a single orientation, first raised the question of whether heterodimer orientation might influence the potency of transcriptional activation. Addressing this issue requires a comparison of the transcriptional activation response generated by the two individual orientational isomers. Even though NFAT could in principle be used to generate complexes having AP-1 in two different orientations, the interpretation of these experiments would be rendered ambiguous by the presence of NFAT. Instead, we deemed it more attractive to engineer orientationally isomeric complexes containing only AP-1 and differing only in the orientation of the heterodimer on DNA. Having demonstrated the feasibility of producing defined, orientationally locked AP-1•DNA complexes, we then assayed the transcriptional potency of the two individual stereoisomers. Matched pairs of AP-1•DNA stereoisomers can be produced in either of two ways: (i) by using two different heterodimers—Fos•JunAla vs. FosAla•Jun—on the same DNA site; or (ii) by using a single-point mutant heterodimer on two DNA sites that differ only in the orientation of the central (C•G or G•C) base pair.
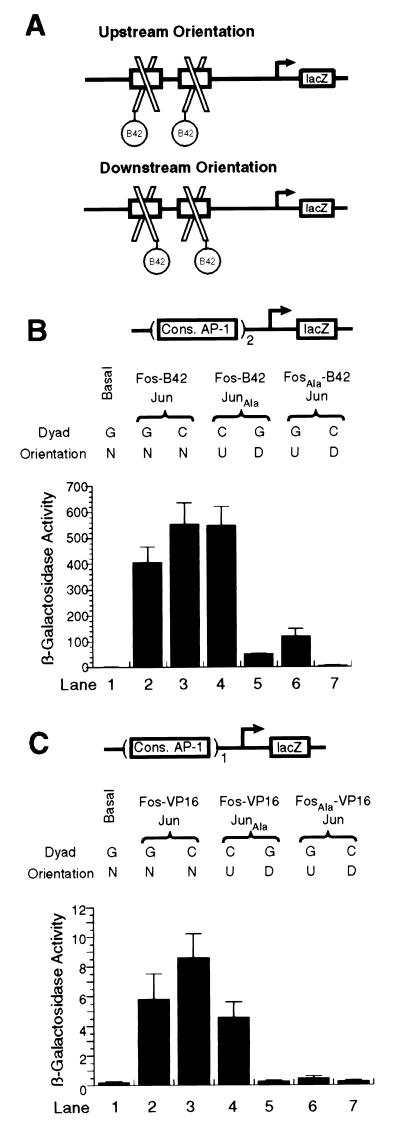
To compare the activation response generated by those matched pairs of orientational isomers, we expressed them in yeast and compared their ability to drive expression from reporter plasmids in which either a single consensus AP-1 site or two tandemly arrayed sites control the activation of the lacZ gene. For each stereochemically defined heterodimer, matched pairs of reporter plasmids, differing only in the orientation of the central C•G pair in the AP-1 site, were analyzed (the orientation is the same in both copies of the tandemly arrayed sites). To ensure that these assays were specifically responsive to the Fos•Jun heterodimer, we fused an activation domain only onto Fos, which is incapable of homodimerizing (19). To drive expression of the lacZ reporter from tandem AP-1 consensus sites, we used the B42 activation domain (wild-type Fos: Fos-B42; R155A point mutant Fos: FosAla-B42), whereas the stronger VP16 activation domain was used with single sites (below).
With the tandem AP-1 reporter, wild-type AP-1 protein (Fos-B42•Jun) furnished high levels of lacZ expression (Fig. 3B, lanes 2 and 3). Inversion of the central C•G base pair in the AP-1 site had a modest effect, bordering on statistical significance (compare lanes 2 and 3). In this case, C•G inversion was expected to have little or no effect on activation because wild-type AP-1 protein binds either site as a mixture of stereoisomers (8). By contrast, C•G inversion had a marked effect on the transcriptional activity of the oriented heterodimers Fos-B42•JunAla (compare lanes 4 and 5) and FosAla-B42•Jun (compare lanes 6 and 7). In each case, the defined stereoisomer that positions the activation domain over the two upstream-facing AP-1 half sites activates transcription much more strongly than the corresponding downstream facing isomer (lane 4 vs. 5, 10-fold difference; lane 6 vs. 7, 18-fold difference; also refer to Fig. 3A). Even though the relative difference in transcriptional potency between the two orientational isomers was found to be similar, the absolute magnitude of activation for a given isomer was dependent on which subunit bore the point mutation. Thus, for example, Fos-B42•JunAla on the C•G site activated reporter gene expression ≈4-fold more strongly than FosAla-B42•Jun, despite the fact that both complexes possess the same stereochemistry (Fig. 3B, compare lanes 4 and 6). Indeed, in these and other experiments (see below), we noted a consistent difference in transcriptional activity of AP-1 heterodimers having the same stereochemistry but differing in whether the Ala point mutation resides in the Jun or Fos subunit, with the latter invariably being less potent. A likely source of this isoform-dependent but orientationally independent effect lies in the fact that FosAla•Jun binds DNA ≈3-fold more weakly than Fos•JunAla (see Table 1).
Figure 3.
(A) Schematic illustration of the two DNA-bound stereoisomers of AP-1 bZip heterodimers bearing the B42 activation domain linked to Fos. (B and C) β-galactosidase gene reporter assays using constructs having either two or one consensus AP-1 sites, respectively. Dyad refers to the oreintation of the central G•C base pair, which is the same for both sites in Fig. 3B. The identity of the base on the coding strand is denoted. Orientation is as given in Fig. 3A: n, mixture of both orientations; U, upstream; D, downstream.
Because reporter constructs having lacZ driven by a single AP-1 site did not furnish measurable levels of β-galactosidase expression (data not shown), we replaced the B42 activation domain with the stronger VP16 activation domain for this series of experiments. Even under these conditions, the activation signal of isoforms having the Ala mutation in Fos was too low to permit accurate comparison. Nevertheless, the results obtained with the constructs that could be measured were consistent with the observations made by using tandem sites: (i) nonoriented wild-type complexes activated transcription to a similar extent (Fig. 3C, compare lanes 2 and 3); (ii) the isomer that orients the VP16 activation domain upstream stimulates transcription much more strongly than the downstream-facing orientation (16-fold difference; Fig. 3C, compare lanes 4 and 5).
In control experiments, it was found that removal of either the activation domain from Fos, or the AP-1 sites from the reporters, resulted in complete loss of the activation response (ref. 20 and data not shown).
DISCUSSION
The AP-1 heterodimer has been shown to bind DNA in solution (8) as a roughly equal mixture of two stereoisomers, which are related by interchange of the Fos and Jun subunits (refer to Fig. 1A). X-ray crystallographic studies on AP-1 have illuminated the molecular basis for the stereochemical degeneracy of AP-1; Fos and Jun possess identical residues at all positions that make sequence-specific DNA contacts (9). Thus, the DNA contact interface of AP-1 has the symmetry of a homodimer, even though the protein overall has the asymmetry of a heterodimer. In the absence of any external influence, AP-1 is therefore incapable of binding DNA in a single orientation. The speculation has been raised that AP-1 might form oriented complexes through differential contacts to bases outside the canonical recognition site (21). This speculation is presently unsupported by any direct experimental data and furthermore is difficult to reconcile with the x-ray structure of the AP-1 DNA-binding domain bound to a specific recognition site (9). Namely, the α-helical structure of the bZip domain largely restricts the reach of the residues to the 7-bp consensus site. The cluster of basic residues at the extreme N-terminal end of the bZip domains are potentially near enough to the base pairs −1 and 8 to make base-specific contacts, but these residues instead contact the DNA backbone nonspecifically, and differ only in one conservative replacement (Arg-143 in Fos vs. Lys-267 in Jun). Thus, although the possibility that wild-type AP-1 can bind some sequences in a stereochemically oriented manner cannot be rigorously excluded, the data presently available suggest that AP-1 alone usually binds DNA as a mixture of stereoisomers.
Using a native chemical ligation/affinity cleavage procedure to detect directly the orientation of the AP-1 bZip on DNA, here we have shown that a single amino acid change in one subunit of AP-1 is sufficient to orient the heterodimer on DNA. This residue occupies an equivalent position in both Fos (Arg-155) and Jun (Arg-279), and ordinarily the equivalent Arg side chains in the heterodimer make interchangeable contacts to the base pair that straddles the dyad axis; one makes a bidentate Arg–G hydrogen-bonding contact, whereas the other makes an electrostatic Arg–phosphate contact (refer to Fig. 1B). The finding that the AP-1•DNA complex becomes oriented upon removal of one Arg side chain, so as to preserve the Arg–G contact, clearly establishes that this interaction is energetically dominant over the Arg–phosphate contact. The relative insignificance of the Arg–phosphate contact is borne out by the fact that its loss has no detectable effect on the affinity of Fos•JunAla for DNA and has only a 2-fold effect on the affinity of FosAla•Jun. The essential nature of the Arg–G contact, on the other hand, is evidenced by the fact that its loss abolishes sequence-specific DNA binding (refer to Table 1). Given these results, it is noteworthy that bidentate Arg-G hydrogen-bonding contacts are the single most commonly observed sequence-specific interaction observed in protein•DNA complexes (4).
Leonard et al. (21) have reported that Arg to Ile mutations at positions 155 or 279 of Fos and Jun, respectively, generate point-mutant AP-1 heterodimers that exhibit oppositely phased mobility changes in electrophoresis-based phasing assays. These data alone were taken as evidence that the Ile mutation orients AP-1. This conclusion and any that follow from it should be viewed with caution, however, because (i) no experiments that directly analyze orientation were carried out on the Ile mutant AP-1 heterodimer; and (ii) the physical basis for electrophoretic mobility shifts is a subject of ongoing controversy (21–28). The present experiments neither confirm nor conflict with those of Leonard et al. (21) because the two studies involve different mutations (Ala vs. Ile). Our system does, however, provide a direct and straightforward means of testing whether the Ile mutant AP-1 heterodimers form oriented complexes on DNA.
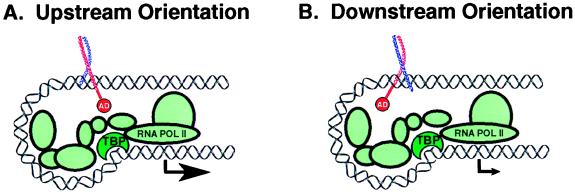
Reporter gene assays reported herein reveal not only that orientation affects transcriptional potency, but that the magnitude of this effect is striking. Regardless of reporter construct (tandem vs. single site), heterodimeric form (Fos•JunAla vs. FosAla•Jun) or configuration of the AP-1 site central base pair (C•G vs. G•C), the orientational isomer that positions the Fos-AD subunit over the upstream-facing half-site(s) was found to activate transcription more strongly (10- to 18-fold) than the corresponding opposite orientation. One model that could in principle explain these findings is based on the proposal that the two subunits of AP-1 induce differential DNA bending, thus giving rise to a bend-specific transcriptional response (22). However, this model has been called seriously into question by independent results in two different assay systems, which indicate that neither Jun•Jun nor Fos•Jun induce significant levels of DNA bending. The latter results are consistent with the x-ray structure of the AP-1 bZIP•DNA complex in which the DNA is unbent (9). Furthermore, gel electrophoresis-based-phasing assays have been shown to be susceptible to hydrodynamic anomalies that can be misinterpreted as having resulted from DNA bending (26–28). Regardless of whether Jun and Fos alter the conformation of DNA, we believe an alternative model can explain the orientation dependence of transcriptional activity by AP-1 and perhaps other proteins as well. This model centers on the proposal that transcription activation is strongly dependent on the stereochemical relationship between activation domains and the transcriptional apparatus, both of which are anchored to the promoter through protein–DNA interactions (Fig. 4). Because activation domains appear to function by establishing direct protein–protein contacts with proteins that associate with either RNA polymerase II (1, 2) or chromatin remodeling factors (3), it stands to reason that the strength of these interactions depends on the spatial and chemical complementarity between the interacting partners as they reach one another on the DNA scaffold. The two orientational isomers of AP-1 present the activation domain in distinctly different ways on DNA, and hence produce different transcriptional responses.
Figure 4.
Cartoon depicting a stereochemical model to explain the orientation-dependent activation of transcription by AP-1 bZip fusion proteins containing a single activation domain. The level of transcription activation is dependent on the strength or speed of interaction between the activation domain and the transcriptional apparatus, which is influenced by the stereochemistry of the protein–DNA complex.
It is interesting to note that the levels of activation produced by the Fos-B42•JunAla heterodimer in its most favored orientation are nearly the same as those afforded by the unoriented Fos-B42•Jun heterodimer (Fig. 3B, compare lanes 3 and 4). These data may suggest that the transcriptional machinery is itself capable of orienting AP-1 on the promoter, thereby selecting through equilibration for the orientation having the strongest protein–protein contacts; this very mechanism is used by NFAT to orient AP-1 (8).
The transcriptional stereochemistry model proposed here is consonant with recent studies of enhancesome assembly on the intact interferon-β and T cell antigen receptor-α enhancers, the results of which indicate that even relatively subtle effects on DNA architecture can exert a powerful influence on transcriptional synergy between multiple transcription factors (6). That protein architecture plays an important role in enhanceosome assembly has been clearly suggested by Kim et al. (7) who showed that inversion of the Jun/ATF2 or NF-κB sites in the interferon-β enhancer dramatically decreased synergistic activation in vitro. These studies leave open the question of whether site inversion alters direct cooperative contacts between proteins bound on the enhancer or affects synergy by interfering with cooperative contacts between activation domains and components of the transcriptional apparatus (29). The simplicity of the model transcriptional system analyzed here has the virtue of minimizing the influence of cooperativity between the activator proteins themselves on DNA, thus increasing the confidence that the orientation-dependent effects can be ascribed to targeting of the transcriptional apparatus; however, this simplification precludes any quantitative extrapolation to the behavior of a native promoter. Nonetheless, our results clearly suggest that the stereochemistry of activation domain presentation is likely to be a key determinant of transcriptional responses for all inducible promoters. Thus, whereas it has been widely recognized that DNA architecture is a critical determinant of enhanceosome assembly, these and related results (7) point to the equally important role of protein architecture. Orientational degeneracy could serve a useful biologic purpose in the case of AP-1, allowing this single protein to present two markedly different interaction surfaces to the transcriptional machinery. Access to such geometric diversity may explain why AP-1 is able to regulate such a large number of target genes from widely varying positions within promoter/enhancers. Perhaps the stereoisomer most responsible for activation differs depending on the location of the site and its relationship to other trans-acting proteins that synergize with AP-1 to recruit the transcriptional apparatus. These considerations raise the interesting possibility that the transcriptional apparatus can select one of the two AP-1 stereoisomers during preinitiation complex formation, as has been suggested for TATA box-binding protein (30). Indeed, stereochemical locking of AP-1 through cooperative protein–protein contacts with the transcriptional apparatus is fundamentally related to the locking observed through contacts with NFAT (8). The potential advantages of stereochemical diversity in enhanceosome assembly may explain why so many heterodimeric transcriptional activator proteins, including NF-κB (31), Myc-Max (32) and certain nuclear hormone receptors (33), can bind at least some sites in an orientationally degenerate manner.
Acknowledgments
M.C. was supported by the Alfred Bader Fellowship and Eli Lilly Fellowship. B.R.P. was supported by a postdoctoral fellowship from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRG-1337). This work was supported by a grant from the Hoffmann-La Roche Institute of Chemistry and Medicine.
ABBREVIATIONS
- AP-1
activator protein-1
- bZip
basic-leucine zipper
Note Added in Proof
Effects on transcriptional activation that may be due to the stereochemistry of activation domain presentation have been noted elsewhere (35).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 3.Kadonaga J T. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 4.Pabo C O, Sauer R T. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 5.Uesugi M, Nyanguile O, Lu H, Levine A J, Verdine G L. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 6.Carey M. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim T K, Mainiatis T. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Oakley M G, Glover J N, Jain J, Dervan P B, Hogan P G, Rao A, Verdine G L. Curr Biol. 1995;5:882–889. doi: 10.1016/s0960-9822(95)00178-3. [DOI] [PubMed] [Google Scholar]
- 9.Glover J N, Harrison S C. Nature (London) 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 10.Erlanson D A, Chytil M, Verdine G L. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 11.Abate C, Patel L, Rauscher F J d, Curran T. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 12.Sun L J, Peterson B R, Verdine G L. Proc Natl Acad Sci USA. 1997;94:4919–4924. doi: 10.1073/pnas.94.10.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 14.Stanojevic D, Verdine G L. Nat Struct Biol. 1995;2:450–457. doi: 10.1038/nsb0695-450. [DOI] [PubMed] [Google Scholar]
- 15.Oliphant A R, Brandl C J, Struhl K. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzamarias D, Pu W T, Struhl K. Proc Natl Acad Sci USA. 1992;89:2007–2011. doi: 10.1073/pnas.89.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muir T W, Sondhi D, Cole P A. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oakley M G, Dervan P B. Science. 1990;248:847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- 19.Halazonetis T D, Georgopoulos K, Greenberg M E, Leder P. Cell. 1988;55:917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- 20.Peterson B R, Sun L J, Verdine G L. Proc Natl Acad Sci USA. 1996;93:13671–13676. doi: 10.1073/pnas.93.24.13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leonard D A, Rajaram N, Kerppola T K. Proc Natl Acad Sci USA. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerppola T K, Curran T. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola T K. Proc Natl Acad Sci USA. 1996;93:10117–10122. doi: 10.1073/pnas.93.19.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerppola T K, Curran T. EMBO J. 1997;16:2907–2916. doi: 10.1093/emboj/16.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajaram N, Kerppola T K. EMBO J. 1997;16:2917–2925. doi: 10.1093/emboj/16.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitlani A, Crothers D M. Proc Natl Acad Sci USA. 1996;93:3248–3252. doi: 10.1073/pnas.93.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitlani A, Crothers D M. Proc Natl Acad Sci USA. 1998;95:1404–1409. doi: 10.1073/pnas.95.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick R J, Badalian T, Fisher D E. Proc Natl Acad Sci USA. 1996;93:14434–14439. doi: 10.1073/pnas.93.25.14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vashee S, Melcher K, Ding W V, Johnston S A, Kodadek T. Curr Biol. 1998;8:452–458. doi: 10.1016/s0960-9822(98)70179-4. [DOI] [PubMed] [Google Scholar]
- 30.Cox J M, Hayward M M, Sanchez J F, Gegnas L D, van der Zee S, Dennis J H, Sigler P B, Schepartz A. Proc Natl Acad Sci USA. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 32.Sommer A, Bousset K, Kremmer E, Austen M, Luscher B. J Biol Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- 33.Glass C K, Rose D W, Rosenfeld M G. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Glover J N, Hogan P G, Rao A, Harrison S C. Nature (London) 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 35.Merika M, Williams A J, Chen G, Collins T, Thanos D. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]