SUMMARY
Transitions between pluripotent and differentiated states are marked by dramatic epigenetic changes. Cellular differentiation is tightly linked to X-chromosome inactivation (XCI), whereas reprogramming to induced pluripotent stem cells (iPSCs) is associated with X-chromosome reactivation (XCR). XCR reverses the silent state of the inactive X, occurring in vivo in mouse blastocysts and the germline. In spite of its importance, little is known about underlying mechanisms. Here, we examine the role of the long noncoding Tsix RNA and the germline factor, PRDM14. In blastocysts, XCR is perturbed by mutation of either Tsix or Prdm14. In iPSCs, XCR is disrupted only by PRDM14-deficiency, which also affects iPSC derivation and maintenance. We show that Tsix and PRDM14 directly link XCR to pluripotency: First, PRDM14 represses Rnf12 by recruiting Polycomb repressive complex 2. Second, Tsix is required for PRDM14 to bind Xist. Thus, our study provides functional and mechanistic links between cellular and X-chromosomal reprogramming.
INTRODUCTION
With revolutionary technologies such as somatic cell nuclear transfer and induced pluripotency, it is now widely appreciated that the differentiated cell state is reversible through epigenomic reprogramming [reviewed in (Gurdon and Melton, 2008; Stadtfeld and Hochedlinger, 2010)]. In vivo, resetting of epigenetic memory occurs most prominently in the germline, where the production of gametes is preceded by the erasure of old and reestablishment of new parental marks to achieve totipotency in the zygote (Hackett et al., 2012; Saitou et al., 2012). The mammalian X-chromosome undergoes some of the most dramatic epigenetic changes during transitions between pluripotent and differentiated cell states, both in embryonic development as well as in cell culture systems (Barakat and Gribnau, 2012; Payer and Lee, 2008; Payer et al., 2011; Wutz, 2011). During differentiation of the pluripotent mouse epiblast in vivo and embryonic stem cells (ESCs) in vitro, one of the two X-chromosomes is randomly chosen for XCI in females, by which X-linked gene dosage parity is achieved with males. XCI is controlled by the long-noncoding RNA Xist, which coats the inactive X-chromosome (Xi) and recruits repressive complexes to initiate silencing (Brown et al., 1992; Zhao et al., 2008).
While XCI is a characteristic property of the differentiated cell state in females, de-differentiation towards pluripotency is associated with XCR. XCR is first observed during reversal of imprinted XCI in preimplantation embryos in the epiblast lineage of the inner cell mass (ICM) of mouse blastocysts (Mak et al., 2004; Okamoto et al., 2004). This reactivated state is also reflected in mouse ESCs, which are derived from the ICM. Epiblast stem cells (EpiSCs) on the other hand, are derived from a later postimplantation epiblast stage and represent “primed” pluripotency, rather than the “naïve” or “ground state” pluripotency of ESCs (Nichols and Smith, 2009). EpiSCs can differentiate into all three germ layers in teratoma assays and in vitro, but, in contrast to ESCs, cannot efficiently form germline chimeras when injected into blastocysts (Gillich and Hayashi, 2011). Interestingly, these relative degrees of pluripotency can be distinguished by X-chromosome states, with XCR characterizing the naive pluripotency of female ESCs, and XCI the primed pluripotency of EpiSCs, as well as cells further down the differentiation pathway. In vivo, a second XCR event occurs in female primordial germ cells (PGCs) (Chuva de Sousa Lopes et al., 2008; Sugimoto and Abe, 2007). PGCs are per se not pluripotent, but express pluripotency factors. Upon explantation, PGCs become pluripotent embryonic germ cells (EGCs), which continue to display two active X-chromosomes (Xa). Finally, in vitro, the reprogramming of somatic cells by fusion with pluripotent stem cells or by expression of defined transcription factors into iPSCs is accompanied by XCR (Maherali et al., 2007; Tada et al., 2001).
For regenerative medicine, the tight linkage between pluripotency and X-chromosomal states may be used to evaluate the epigenetic status of in vitro-derived human stem cells. Human ESCs have been grouped into three classes based on whether they retain two Xa (Class I), have undergone XCI (Class II), or lost the ability to maintain XCI (Class III) (Silva et al., 2008). This indicates that some hESC lines resemble epigenetically mouse EpiSCs and may not have full differentiation potential (Nichols and Smith, 2009). Likewise, human iPSCs rarely achieve XCR and may better resemble EpiSCs with only primed pluripotency (Anguera et al., 2012; Tchieu et al., 2010; Tomoda et al., 2012). Thus, XCR might have an important place in the qualitative assessment of stem cells.
However, remarkably little is known about how XCR is achieved. In general, XCR is accompanied by Xist-downregulation and removal of chromosome-wide repressive marks such as H3K27me3 and promoter-associated CpG methylation. The XCR pathway could involve steps opposite to those during XCI. Thus, candidate factors to repress XCI might be the antisense Tsix RNA (Lee and Lu, 1999) and various pluripotency factors (OCT4, SOX2, REX1, and NANOG) that bind Xist intron 1 (Navarro et al., 2008) and the Tsix locus (Donohoe et al., 2009; Gontan et al., 2012; Navarro et al., 2010). Control of XCR could also involve repression of Xist activators such as the noncoding Jpx RNA (Sun et al., 2013; Tian et al., 2010) and the E3 ubiquitin ligase RNF12/RLIM (Barakat et al., 2011; Jonkers et al., 2009; Shin et al., 2010). The relationships appear complex. For instance, deletion of Xist intron1, a binding site for many pluripotency factors, does not cause Xist de-repression during embryogenesis or iPSC reprogramming (Minkovsky et al., 2013). Also in undifferentiated ESCs, deletion of Xist intron 1 (Barakat et al., 2011), Tsix (Lee and Lu, 1999), or both (Minkovsky et al., 2013) is not sufficient to fully de-repress Xist.
Therefore, how XCR is controlled remains largely mysterious. Here we test two candidate genes, Tsix and Prdm14. While the noncoding RNA gene Tsix is a well-established Xist repressor for XCI, its role in XCR has been unclear. One study (Ohhata et al., 2011) demonstrated that induced expression of Tsix was sufficient to downregulate imprinted Xist expression in mice, but whether Tsix is necessary for XCR in the physiological context is unknown. PRDM14 caught our attention because it is a germline factor with spatiotemporal correlation with XCR in vivo and has been implicated in epigenetic reprogramming events in PGCs (Yamaji et al., 2008; Yamaji et al., 2013). In addition, PRDM14 has a potential function in repressing Xist in ESCs (Ma et al., 2010) and its overexpression accelerates XCR during conversion of EpiSCs to ESCs (Gillich et al., 2012). We demonstrate that both Tsix and PRDM14 play crucial roles during XCR in mice and perform in vitro analyses in iPSC and ESC models to study mechanism and relationship to pluripotency.
RESULTS AND DISCUSSION
XCR is perturbed in Tsix- and Prdm14-mutant blastocysts
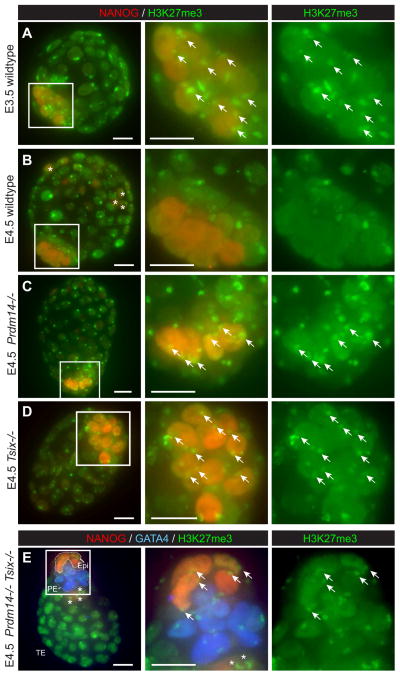
In female mouse blastocysts, reactivation of the imprinted paternal X (Xp) occurs specifically in the NANOG-positive epiblast cells of the ICM, while Xp silencing is maintained in extraembryonic lineages. A characteristic of XCR is erasure of the H3K27me3 mark, which occurs between embryonic days 3.5 and 4.5 (E3.5–4.5) of the blastocyst stage (Mak et al., 2004)(Figure 1A,B). To examine whether XCR was affected by a deficiency of either Prdm14 or Tsix, we took advantage of existing Prdm14 (Yamaji et al., 2008; Yamaji et al., 2013) and Tsix (Lee and Lu, 1999) knockout strains and investigated the effects on H3K27me3 erasure in mutant embryos. Intriguingly, while wildtype blastocysts had lost the H3K27me3 mark in the epiblast by E4.5 (Figure 1A,B; loss of green spots in NANOG+ [red] cells), Prdm14−/− and Tsix−/− mutants still retained H3K27me3 on XP in E4.5 epiblast cells (Figure 1C,D), indicating that a loss of function of either gene affected XCR. Prdm14−/−/Tsix−/− double mutants were similarly compromised (Figure 1E).
Figure 1. XCR assayed by H3K27me3 staining in blastocysts.
Z-series projections of E3.5 and E4.5 female blastocysts immunostained for H3K27me3 (green) and the lineage markers NANOG (red) and GATA4 (blue). (A,B) Wildtype, (C) Prdm14−/−, (D) Tsix−/−, (E) double mutant embryos. Cell lineage territories are outlined in (E) [Epiblast (Epi) = NANOG+ within ICM, primitive endoderm (PE) = GATA4+, trophectoderm (TE) = rest]. Middle and right panels show close-ups of the ICM (white boxes). Arrows, Xi in NANOG+ cells. NANOG+ cells outside the ICM never undergo XCR (stars). Scale bars = 20 μm. See also Figure S1.
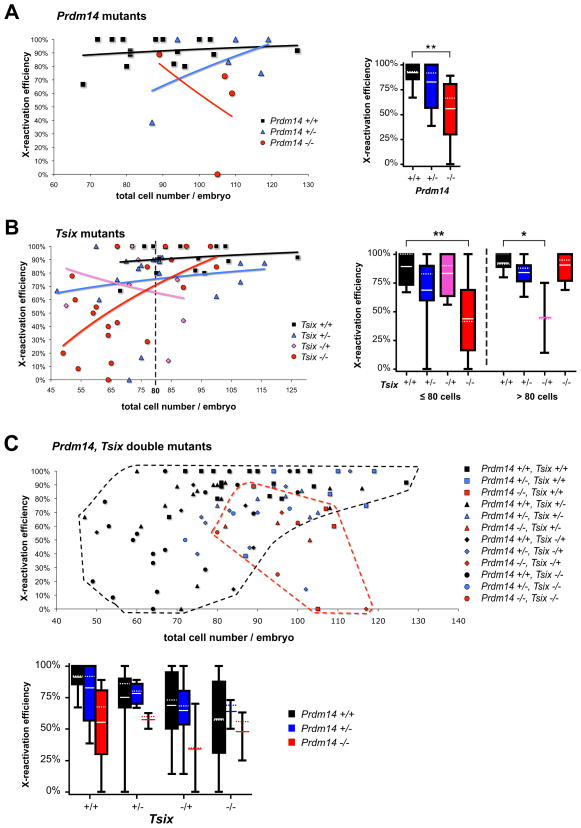
Further analysis of single-gene mutations revealed a reduction rather than absolute loss of XCR (Figure 2A,B). In late wildtype blastocysts, most epiblast cells (>90% mean average) lost H3K27me3 from the Xi. By contrast, significantly fewer cells (55%) erased the mark in Prdm14−/− blastocysts (Figure 2A), consistent with an XCR defect in Prdm14−/− embryos. Although the effects were only significant in homozygous mutants, Prdm14+/− mutants also showed a slight decrease on average, suggesting that XCR depends on the dose of PRDM14. Loss of XCR was not absolute in either case, as a fraction of epiblast cells in both Prdm14−/− and Prdm14+/− embryos still erased H3K27me3. Examination of more advanced embryos in the late blastocyst stage indicated that XCR seemed not merely delayed, as even advanced blastocysts (>80 cells) displayed a reduced XCR frequency (Figure 2A). We cannot rule out possible maturation delays in the Prdm14−/− epiblast, which would in turn compromise XCR. However, strong indicators of normal developmental progression (e.g., normal total cell numbers, normally sized NANOG+ epiblasts) argue against that possibility.
Figure 2. Quantification of XCR in Prdm14- and Tsix-mutant blastocysts.
XCR in Prdm14−/− (A), Tsix−/− (B) and double mutant (C) E4.5 blastocysts was scored as %NANOG+ epiblast cells without H3K27me3 foci per embryo. Every data point on the scatter plots represents reactivation efficiency plotted against total number of cells per embryo indicating developmental progression. Trend lines (A,B) follow logarithmic regression. The box plots summarize the data, with boxes demarcating the 25th–75th percentile (only shown if n>3), the median indicated by dotted and the mean by solid lines. Whiskers extend to minimum and maximum values. Statistical significances have been calculated using 1-way ANOVA (** P<0.01). Note: For genotypes, maternal allele is represented first by convention. In (B), black dashed lines separate delayed embryos with low cell number (≤80 cells) from advanced embryos (>80 cells). (C) All embryos including Prdm14/Tsix double-mutants. The highlighted areas encompass Prdm14+/+ (black) or Prdm14−/− (red) embryos of all Tsix genotypes.
In Tsix−/− embryos, we observed a similarly significant reduction in XCR (Figure 2B). We noticed that many Tsix−/− embryos, for unknown reasons, had fewer than 80 cells, suggesting a developmental delay when compared to wildtype. To adjust for differences in developmental progression, we compared XCR efficiencies of less developed (≤80 cells) and more advanced (>80 cells) E4.5 blastocysts. While wildtype embryos with low cell number (≤80) lost H3K27me3 from the Xi in almost all epiblast cells (mean average 89%), significantly fewer (44% of epiblast cells) did so in Tsix−/− embryos. However, Tsix−/− embryos with over 80 cells showed XCR rates nearly equivalent to wildtype. The ability to recover XCR in older Tsix-mutant embryos contrasted with the inability to do so in Prdm14-mutants. Thus, Tsix−/− embryos showed slower kinetics of both XCR and developmental progression, consistent with a role of Tsix in XCR timing in the physiological state in mice. Furthermore we noticed that the previously described defects in imprinted XCI in Tsix-null male and female embryos (Lee, 2000, 2002; Sado et al., 2001) could be observed as early as E5.5 (Figure S1). In extraembryonic cells near the epiblast we saw one H3K27me3-spot per nucleus in male Tsix−/Y and two spots in female Tsix−/− embryos. This suggests that cell lineage (PE and/or TE) or the vicinity to the epiblast might play a role in the timing when the XCI-defect in Tsix-mutant embryos begins to emerge.
Effects on XCR were also observable in heterozygotes, as might be expected because of imprinted Tsix silencing on XP [normally enables the paternal Xist allele to be expressed (Lee, 2000; Lee and Lu, 1999; Sado et al., 2001)]. Indeed, embryos with paternal Tsix mutation (Tsix+/−) appeared to show a delay in XCR with only 69% of epiblast cells in less developed E4.5 embryos (≤80 total cell number) having reactivated the Xp (Figure 2B). However, embryos with a mutation on the maternal allele (Tsix−/+) showed no reactivation defect. The effect seen later (>80 cells) may be due to the low sample size for this particular genotype (n=3, note large variance). In neither heterozygous case was the defect as pronounced as in early Tsix−/− homozygous mutants. These epigenetic and genetic differences may be explained in several ways. First, there may be dose-dependency on Tsix. Alternatively, Tsix’s function might not be restricted to action exclusively in cis. This would be consistent with the observation that deleting both Tsix alleles showed a lower XCR efficiency than deleting either allele. Indeed, trans effects at Tsix have been reported (Lee, 2002), which might differ in action from the cis effect on XCR.
We then investigated whether combining the Prdm14 and Tsix mutations had additive effects on XCR efficiency. To our surprise, Prdm14−/−, Tsix−/− double-mutant blastocysts showed only a mildly, but not significantly stronger phenotype (48% mean H3K27m3 erasure in epiblast) than Prdm14−/− (55%) or Tsix−/− (58%) single-mutant embryos (Figure 2C) suggesting that Prdm14 and Tsix might act through a common genetic pathway during XCR. We conclude that both Tsix and Prdm14 are negative regulators of Xist and thereby positive regulators of XCR. Because some Tsix-mutant embryos could catch up and eventually progress through XCR, Tsix may not be an absolute requirement. However, we note that the TsixΔCpG allele (Lee and Lu, 1999) used in our study may be a hypomorph rather than a complete null, as 5–29% of wildtype Tsix RNA levels remain (Shibata and Lee, 2004; Sun et al., 2006), and may therefore contribute to the incomplete XCR phenotype. The remaining low-level Tsix transcription might allow TsixΔCpG-mutant embryos to undergo XCR partially. A similar effect has been observed for the role of Tsix during imprinted XCI, where a maternally inherited TsixΔCpG mutation resulted in incomplete lethality (Lee, 2000) compared to the almost absolute lethality of more severe Tsix null animals (Sado et al., 2001). Regardless, Tsix clearly regulates the efficiency and timing of XCR in vivo.
Prdm14 plays a role for survival of postimplantation embryos independent of its function during XCR
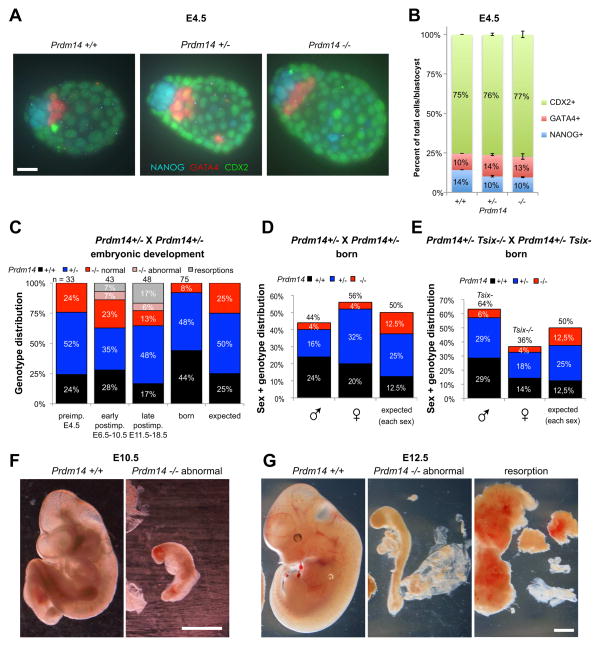
Given the defects in XCR in Prdm14-null female embryos, we asked if this had an impact on their survival in a sex-specific manner. Furthermore, given that knocking down Prdm14 in mouse ESCs has been reported to lead to a cell fate conversion into extraembryonic endoderm lineage (Ma et al., 2010), we also wished to determine, if mutation of Prdm14 showed similar effects in vivo. To address these questions, we intercrossed Prdm14+/− mice and analyzed the resulting embryos and newborn litters (Figure 3). E4.5 Prdm14−/− blastocysts showed no abnormalities in cell type distribution (Figure 3A,B), with normal numbers of epiblast (NANOG+), primitive endoderm (GATA4+), and trophectoderm (CDX2+) cells. In particular, we did not see a significant reduction in epiblast cell number in favor of primitive endoderm, as might have been predicted from the Prdm14 knockdown in ESCs (Ma et al., 2010). This suggests that Prdm14 is not essential in vivo to protect the epiblast from inappropriate differentiation into primitive endoderm. This was the case for both male and female Prdm14−/− mutants.
Figure 3. Prdm14−/− embryos show normal cell fates and Mendelian ratios during preimplantation stages, but abnormal postimplantation development.
(A, B) Analysis of cell lineage distribution in E4.5 blastocysts from Prdm14+/− intercrosses (2 litters, 11 embryos) by immunostaining (A) for the cell lineage markers NANOG (cyan, Epi), GATA4 (red, PE) and CDX2 (green, TE). Scale bar = 20 μm. Counts in (B) are given in % of cells of each lineage per total blastocyst cell number (error bars = SEM).
(C) Genotype distribution of embryos and pups from Prdm14+/− heterozygous intercrosses. The right column represents the expected Mendelian ratio. Resorptions were not genotyped due to lack of embryonic material. n = number of embryos.
(D, E) Sex- and genotype distribution from Prdm14+/− het-crosses (D; 16 litters, 75 pups) and Prdm14+/− Tsix−/− female with Prdm14+/− Tsix− male intercrosses (E; 14 litters, 49 pups).
(F, G) Examples for abnormal Prdm14−/− E10.5 (F) and E12.5 (G) embryos compared to wildtype littermates. The resorption in (G) consists of decidual tissue with no apparent embryonic material. Scale bars = 1 mm.
We next addressed whether Prdm14 had any other role in survival and development of early mouse embryos. At the blastocyst stage (E4.5), Prdm14-mutants were present at expected Mendelian ratios (Figure 3C) without any significant sex ratio distortion with 55% male and 45% female embryos (total E4.5 embryos n=33). However, at birth (Figure 3C,D), the distribution of genotypes deviated very significantly from the expected (P=0.0002, two-tailed χ2-test), with Prdm14−/− embryos being strongly underrepresented (4% instead of expected 12,5%) both in males and females. On the other hand, Prdm14+/+ mice were overrepresented at birth. This indicates that Prdm14 plays a role for survival of embryos during postimplantation development, though in previous work embryonic lethality has not been observed (Grabole et al., 2013; Yamaji et al., 2008). Therefore it is likely that strain background affects this phenotype. Regardless, in the context of our C57/BL6 genetic background, no sex ratio distortion was observed. These data imply that the developmental phenotype is independent of Prdm14’s role during XCR. It is possible that the reduced XCR efficiency observed in Prdm14−/− embryos impacted survival of some female embryos; however, the equivalent birthrates of Prdm14−/− male and female pups suggest larger, sex-independent defects. When we intercrossed Prdm14+/− mice, which were at the same time Tsix-mutants (Figure 3E), we also observed a very significant deviation from the expected Mendelian ratios (P=0.0076, two-tailed χ2-test), with very few Prdm14−/− Tsix−/− double-mutant mice being born (6% and 4% for males and females, respectively). In this case however we also observed a sex ratio distortion with fewer females (36%) born than males (64%), which is similar to Tsix-single mutants and can be explained by chaotic choice during XCI in Tsix−/− females (Lee, 2002). In summary, Prdm14−/− Tsix−/− double-mutants are born as rarely as Prdm14−/− single mutants and display a similar sex-ratio-distortion as Tsix−/− single-mutants. This shows that the Prdm14 and Tsix mutations despite having similar defects in XCR in blastocysts have distinctive effects on viability during post-implantation development.
We then investigated the developmental stage at which the Prdm14 mutation caused lethality in our strain background. We dissected litters throughout postimplantation development and grouped them into early (E6.5–E10.5) and late stage (E11.5–E18.5) postimplantation embryos (Figure 3C). Lethality did not occur at one particular stage. Rather, between E6.5–E18.5, there was a gradual increase in abnormal and developmentally retarded Prdm14−/− embryos and of empty deciduae (resorbed embryos) (Figure 3C,F,G). The observed Prdm14 postimplantation phenotype was surprising and intriguing, as Prdm14 expression is thought to be restricted to the ICM of the blastocyst and to PGCs (Grabole et al., 2013; Yamaji et al., 2008; Yamaji et al., 2013). As Prdm14 seems not to be expressed and in later embryos, the phenotype in Prdm14−/− embryos must be a knock-on effect of PRDM14 deficiency in the ICM and suggests that this early expression primes the epiblast epigenetically for later postimplantation development. Further investigations will be needed to clarify, if for example other epigenetic (re)programming steps in the ICM besides XCR are facilitated by PRDM14, which might have an influence on viability later on.
Prdm14, but not Tsix, is required for self-renewal of iPSCs
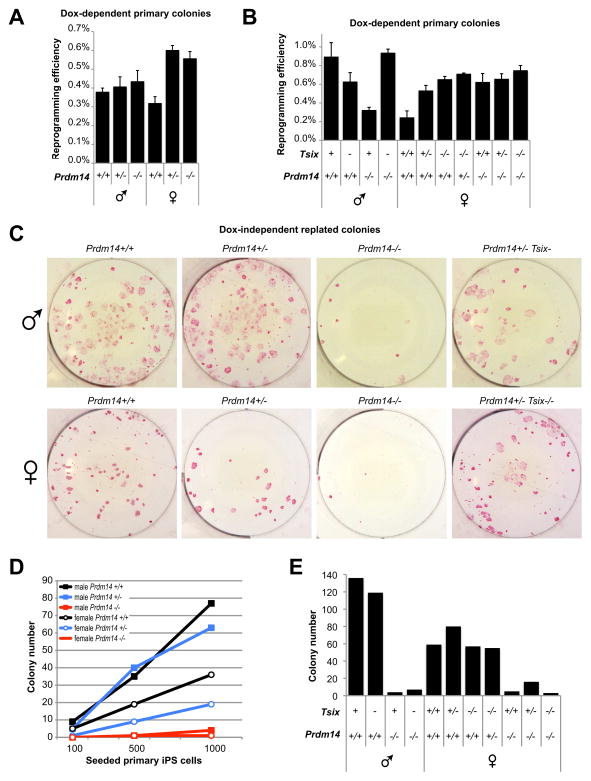
The occurrence of XCR during reprogramming of somatic cells (Maherali et al., 2007) makes female iPSCs an excellent ex vivo model for mechanistic studies of Prdm14 and Tsix. We reprogrammed tail-tip fibroblasts (TTFs) from newborn mice of different Prdm14 and Tsix genotypes into iPSCs using a lentiviral doxycycline(Dox)-inducible Oct4, Klf4, Sox2, and c-Myc (OKSM) delivery system (Sommer et al., 2009). Formation of primary iPSC colonies (% iPSC colonies formed per input TTFs) did not differ between different Prdm14, Tsix, or double-mutant genotypes (Figure 4A,B), suggesting that neither Prdm14 nor Tsix were absolutely required for formation of primary colonies.
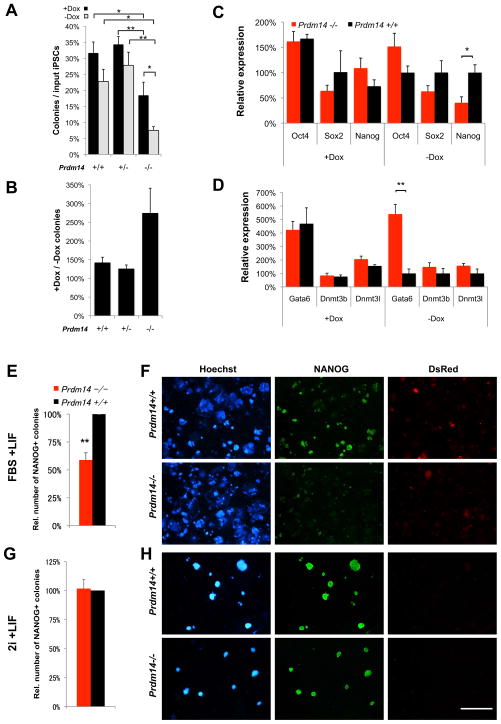
Figure 4. Prdm14 is important for self-renewal of iPSCs.
(A, B) Primary iPSC colonies of different Prdm14 and Tsix genotypes. Reprogramming efficiency was scored as % primary colonies formed per input TTFs after 10 days of Dox-induction. Each graph depicts data from one experiment performed in triplicate (error bars = SEM).
(C) AP-stainings of replated iPSC colonies. 1000 primary iPSCs for each genotype have been reseeded and grown for 14 days without Dox (independent of viral 4-factor expression).
(D) Quantification of AP-positive replated colonies of different Prdm14 genotypes. 100, 500 or 1000 primary iPSCs were seeded and grown without Dox for 7 days.
(E) Quantification of AP-positive replated colonies of different Prdm14 and Tsix genotypes grown from 1000 primary iPSCs without Dox for 11 days.
However, important characteristics of true iPSCs are reactivation of endogenous pluripotency genes and independent self-renewal without continued exogenous OKSM expression (Stadtfeld et al., 2008). To test whether the primary colonies had become OKSM-independent, we replated primary iPSCs (100, 500 or 1000 cells per 6-well) without further induction of OKSM. Under these conditions, alkaline phosphatase(AP)-positive replated colonies rarely emerged from male or female Prdm14−/− cells (Figure 4C,D). By contrast, colonies from Tsix−/− and Prdm14+/− cells appeared at similar frequencies as wildtype controls (Figure 4D,E).
To test if prolonged exogenous OKSM expression could mitigate this effect, we compared replating efficiencies of Prdm14+/+, +/− and −/− iPSCs with or without further Dox treatment. Although continued OKSM induction increased colony number to some degree even for Prdm14+/+ and +/− cells, the most significant increase was observed for Prdm14−/− cells (Figure 5A,B). Nevertheless, even continuous Dox administration could not fully rescue the self-renewal defect of Prdm14−/− iPSCs, which still formed significantly fewer colonies after replating. To ask how endogenous pluripotency gene expression was affected, we tested Prdm14−/− iPSCs with and without prolonged Dox treatment (Figure 5C) and observed that both endogenous Oct4 and Sox2 were reactivated and expressed at wildtype levels in the absence of PRDM14 regardless of exogenous OKSM expression. Nanog, however, was significantly reduced in Prdm14−/− iPSCs without Dox-treatment. As PRDM14 has been previously described as a repressor of the extraembryonic endoderm marker, Gata6 (Ma et al., 2010), and DNA methyltransferases (Grabole et al., 2013; Yamaji et al., 2013), we also tested their expression in our system (Figure 5D). Gata6 was significantly de-repressed and Dnmt3b and Dnmt3l were upregulated after Dox withdrawal in Prdm14−/− cells. Thus, continued OKSM expression can partially mask the defects in Prdm14−/− iPSCs.
Figure 5. The self-renewal defect of Prdm14−/− iPSCs is partially rescued by maintained exogenous pluripotency factor expression or by derivation and culture in 2i+LIF.
(A, B) Replating efficiency of iPSCs depending on exogenous pluripotency factor expression. Primary iPSCs were derived in 11 days with Dox before reseeding and culture for 14 days with (+Dox) or without (−Dox) induction of the lentiviral reprogramming cassette. Replating efficiency was measured as AP-positive colonies per seeded input iPSCs (A). * P<0.05, ** P<0.01 2-way ANOVA with Bonferroni post-tests to compare individual means. In (B), the relative colony number increase of continuous Dox-treatment over Dox-withdrawal is shown. Error bars = SEM.
(C, D) Quanititative (q)PCR measurements of gene expression (C) in Prdm14+/+ (black) and Prdm14−/− (red) iPSCs cultured with (+Dox) or without (−Dox) exogenous OKSM factors. (C) Endogenous pluripotency gene expression. * P=0.034 two-tailed Student’s t-test. (D) Gata6 (endodermal differentiation marker) and de novo DNA-methyltransferase expression. ** P=0.00097. Error bars = SEM.
(E–H) Comparison of derivation efficiency of Prdm14+/+ vs. Prdm14−/− iPSCs under FBS+LIF (E, F) or 2i+LIF conditions (G, H) using retroviral vectors. (E, G) Relative number of NANOG+ colonies from reprogrammed Prdm14−/− cells (red) compared to Prdm14+/+ (black, set to 100%). ** P=0.008 two-tailed Student’s t-test. Error bars = SEM. (F, H) NANOG-activation and silencing of retroviral vectors (DsRed-negative) in iPSC-colonies as indicators of successful reprogramming (Hoechst = DNA). Scale bar = 4 mm.
As Prdm14−/− ESCs could only be derived under dual inhibition of MAPK/ERK and GSK3 pathways in 2i medium (Grabole et al., 2013; Yamaji et al., 2013; Ying et al., 2008), we asked if 2i medium would be beneficial during generation and maintenance of Prdm14−/− iPSCs. For this assay, we chose a retroviral reprogramming approach, in which the exogenous pluripotency genes in the viral vectors would be silenced after successful reprogramming (Maherali et al., 2007). Indeed, in FBS+LIF conditions, we observed significantly fewer and smaller NANOG-positive iPSC colonies when reprogramming Prdm14−/− fibroblasts than in wildtype (Figure 5E,F) and we could not maintain them as undifferentiated iPSCs during prolonged culture. On the other hand, 2i+LIF conditions gave rise to nearly equal numbers of NANOG+ iPSC colonies from Prdm14−/− and Prdm14+/+ cells (Figure 5G,H) and these colonies were efficiently maintained thereafter.
Taken together, these findings indicate that Prdm14−/− iPSCs are compromised during vector-independent self-renewal. They are iPSC-like in character, but cannot be called bona fide iPSCs. Continued expression of exogenous pluripotency factors and 2i-conditions can partially rescue this defect. Thus, while Tsix appears to be dispensable for iPSC reprogramming, PRDM14 is a critical factor for the generation of fully reprogrammed self-renewing iPSCs.
Prdm14 regulates XCR during iPSC reprogramming
Given that Tsix and Prdm14 mutations affect XCR in vivo (Figure 1,2) and that Prdm14 is necessary for proper iPSC formation (Figure 4,5), we asked whether the ability to undergo XCR during iPSC reprogramming was dependent on Prdm14 and Tsix. To analyze Tsix’s effects, we made use of an X-linked GFP marker gene (XGFP) (Hadjantonakis et al., 2001) (Figure 6A,B) and FACS-sorted TTFs that bear the XGFP transgene on the Xi (GFP-negative cells). We reprogrammed this population and determined XCR efficiency by scoring for XGFP reactivation. Regardless of Tsix genotype (+/+, +/−, or −/−), reactivation of XGFP occurred with similar kinetics and efficiency (Figure 6A) and was complete when primary colonies were replated during the OKSM-independent phase (Figure 6B). RNA immunoFISH confirmed these results, as the Xist RNA cloud and H3K27me3 foci (not shown) characteristic of the Xi were both lost in iPSCs regardless of Tsix genotype (Figure 6C,D). Thus, whereas Tsix mutation caused delayed XCR in vivo, it did not have a measurable XCR phenotype when assayed in the iPSC system. We cannot exclude the possibility that effects were masked as the TsixΔCpG allele is a hypomorph (Shibata and Lee, 2004; Sun et al., 2006) or that we missed a phenotype due to the asynchronous nature of XCR in the iPSC system relative to embryos. It is also plausible that XCR in vivo within the context of the blastocyst is mechanistically different from the forced XCR that occurs in the iPSC system. In blastocysts, imprinted XCI is reversed by XCR; by contrast, iPSC reprogramming involves reversal of random XCI. Therefore, it is possible that Tsix is more crucial to oppose imprinted Xist expression. Indeed, one study suggests that Tsix is not expressed during XCR in PGCs (Sugimoto and Abe, 2007), which similar to iPSCs need to overcome random XCI.
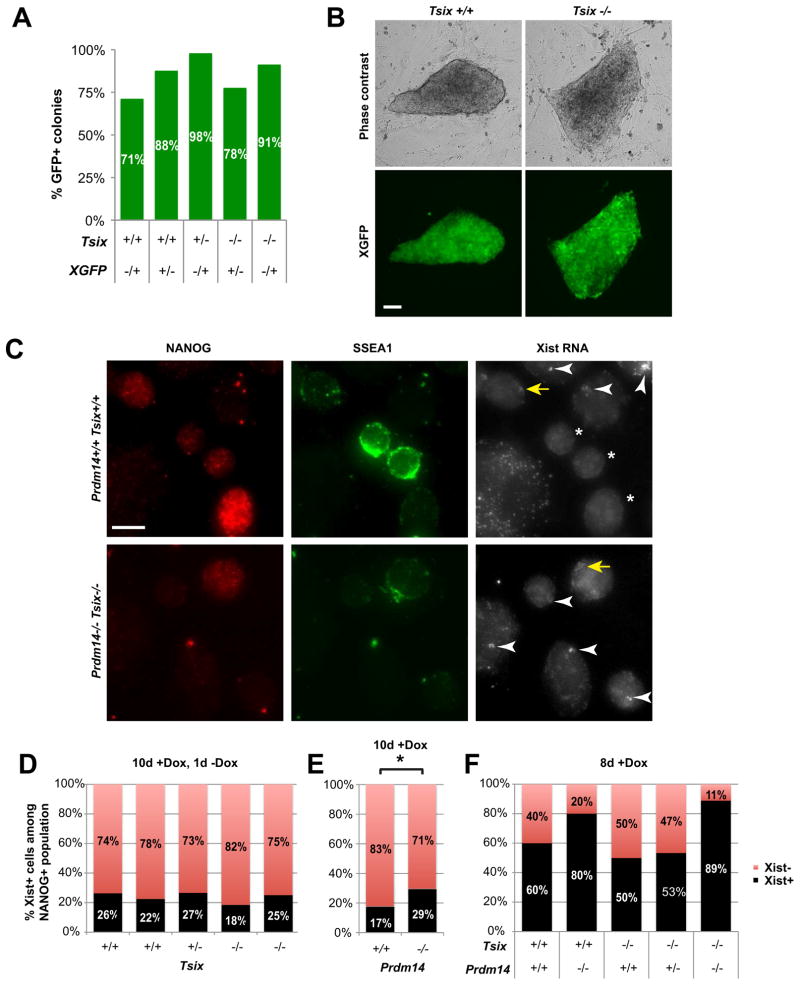
Figure 6. XCR during iPSC reprogramming is perturbed in Prdm14−/− but not in Tsix−/− cells.
(A) % iPSC colonies of different Tsix genotypes with XGFP-reactivation during reprogramming (10 days +Dox).
(B) Tsix+/+ and Tsix−/− iPSC colonies showing complete XGFP-reactivation during Dox-independent culture after replating (7 days −Dox). Scale bar = 100 μm.
(C) RNA immunoFISH of fibroblasts/iPSC during reprogramming (8 days +Dox). Some NANOG/SSEA1 double-positive cells have downregulated Xist (stars), while others (yellow arrows) still show Xist expression similar to NANOG-negative cells (white arrowheads), which were consistently Xist-positive. Scale bar = 10 μm.
(D–F) Xist RNA-FISH of NANOG-positive iPSCs of different Tsix- (D, 10d +Dox, 1d −Dox), Prdm14- (E, 10d +Dox), or Prdm14/Tsix- (F, 8d +Dox) genotypes. * P=0.0463 two-tailed Fisher’s exact test.
In contrast to Tsix, mutating Prdm14 resulted in perturbed XCR. When comparing emerging Prdm14+/+ with Prdm14−/− iPSCs, we observed a significantly lower efficiency but not absolute abolition of Xist downregulation in Prdm14−/− cells (Figure 6C,E). This reduced XCR efficiency appears to decrease slightly further by deleting Tsix (Figure 6F), suggesting a minor additive effect of deleting both genes. Combined, these data indicate that Prdm14, but not Tsix, affects XCR in the ex vivo iPSC model. Thus, there seem to be context-dependent differences - in vivo versus ex vivo - in the requirement for Prdm14 and Tsix, with the in vivo model being more sensitive.
Molecular convergence of Tsix and PRDM14 at the X-inactivation center
To define a molecular basis of the effects on XCR and cellular reprogramming, we then examined expression of genes at the X-inactivation center (Xic) in female Prdm14−/− ESCs (Yamaji et al., 2013). Expression of neither Xist nor Tsix was significantly affected (Figure 7A), which was surprising, as increased Xist levels were previously reported in Prdm14-knockdown ESCs (Ma et al., 2010). Further, with PRDM14 acting as a positive regulator of XCR (Figure 1,2,6), we might have expected its deficiency to affect Xist. However, it is known that Xist expression not only depends on downregulation of repressors like Tsix or pluripotency factors, but also on upregulation of the activators Rnf12/Rlim and Jpx (Jonkers et al., 2009; Tian et al., 2010). In Prdm14−/− cells, we did consistently observe a significant 4-fold increase in Rnf12/Rlim RNA (Figure 7A), which is upregulated during XCI and encodes an ubiquitin ligase (Barakat et al., 2011; Jonkers et al., 2009; Shin et al., 2010). On the other hand, we did not see a statistically significant expression increase of Jpx RNA, which normally increases >10-fold as a prerequisite for XCI (Sun et al., 2013; Tian et al., 2010). Thus, Xist upregulation may not occur in Prdm14−/− cells because persistently low levels of the Jpx activator and high levels of the Tsix repressor remain.
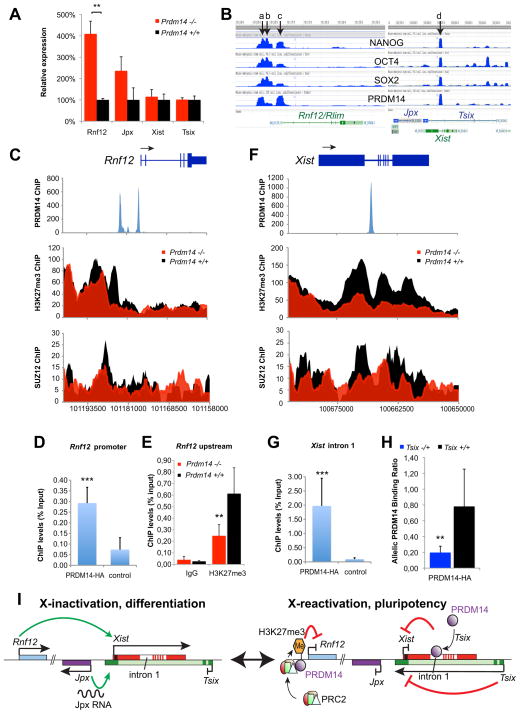
Figure 7. PRDM14 promotes XCR through repressing the Xist-activator Rnf12 and through Tsix-dependent binding to Xist intron 1.
(A) Expression analysis of X-inactivation regulator genes at the Xic by qPCR in undifferentiated Prdm14+/+ and Prdm14−/− ESCs. Expression levels are normalized by Gapdh relative to Prdm14+/+ cells (=100%) ** P=0.0026 two-tailed Student’s t-test. Error bars = SEM.
(B) ChIP-Seq data for binding of NANOG, OCT4, SOX2 (Marson et al., 2008) and PRDM14 (Ma et al., 2010) along the Xic, retrieved via the NCBI epigenomics database (http://www.ncbi.nlm.nih.gov/epigenomics). Arrows indicate co-bound pluripotency factor binding sites 5kb (a), 4kb (b) or immediately (c) upstream of Rnf12 and in Xist intron 1 (d). See also Figure S2.
(C) ChIP-Seq analysis of the Rnf12 locus. PRDM14 binding (blue) and comparison of SUZ12 and H3K27me3 occupancy between Prdm14+/+ (black) and Prdm14−/− (red) ESCs (Yamaji et al., 2013).
(D) ChIP-qPCR with HA-tagged PRDM14 expressed in undifferentiated ESCs at the Rnf12 promoter (*** P=0.0008). Control = non-transfected ESCs. Error bars (D, E, G, H) = SD.
(E) ChIP-qPCR for H3K27me3 upstream of Rnf12 in Prdm14+/+ and −/− ESCs. (** P=0.002).
(F) ChIP-Seq analysis of the Xist locus (as in B).
(G) ChIP-qPCR with PRDM14-HA at Xist intron 1 (as in C). *** P=0.0009
(H) Mus/Cas allelic binding ratio of PRDM14-HA in undifferentiated hybrid Tsix−/+ (purple; Mus = Tsix-, Cas = Tsix+) and Tsix+/+ (black) ESCs (** P=0.0013).
(I) Model for the roles of PRDM14 and Tsix during XCR. In differentiated cells and before XCR (left), PRDM14 and Tsix are absent while RNF12 and Jpx RNA activate Xist leading to XCI. During XCR in pluripotent stem cells and during embryogenesis (right), PRDM14 is expressed and binds upstream of Rnf12. In turn PRC2 is recruited and methylates H3K27, which leads to Rnf12 repression. Furthermore, Tsix is expressed facilitating PRDM14-binding to Xist intron 1. The lack of Xist-activators (Rnf12 and Jpx) and repressive effects of Tsix and PRDM14 on Xist lead to Xist repression, an important step for XCR. A more comprehensive model (Figure S3) includes other key pluripotency factors binding at the Xic implicated in Xist-repression (e.g. NANOG, REX1, OCT4, SOX2).
To determine whether Xic regulators might be direct targets of PRDM14, we analyzed chromatin immunoprecipitation sequencing (ChIP-Seq) data for binding sites within the region (Ma et al., 2010) and detected binding of PRDM14 in undifferentiated ESCs, both to the upstream regulatory region of Rnf12 and to Xist intron 1 (Figure 7B). PRDM14 was shown previously to recruit Polycomb repressive complex 2 (PRC2) to target genes, resulting in gene silencing (Chan et al., 2012; Yamaji et al., 2013). As Rnf12 is upregulated in Prdm14−/− ESCs, we asked if Rnf12 could be such a target by examining PRC2 binding (via the SUZ12 subunit) and H3K27me3 occupancy in Prdm14+/+ versus Prdm14−/− ESCs (Figure 7C)(Yamaji et al., 2013). Indeed, when PRDM14 was absent, SUZ12 binding and H3K27me3 decreased in the Rnf12 promoter region near the PRDM14 binding site, suggesting that PRDM14 represses Rnf12 directly by recruitment of PRC2. ChIP-qPCR confirmed these findings (Figure 7D,E). At Xist intron 1, loss of PRDM14 resulted in decreased SUZ12 binding and H3K27-trimethylation around the PRDM14 binding site and much of the Xist gene body (Figure 7F,G). However, no gross H3K27me3 changes occurred at the Xist promoter, suggesting a PRC2-independent role of PRDM14 at Xist. In contrast, binding of OCT4, SOX2 or NANOG, did not change within Rnf12 or Xist in the absence of PRDM14 (Figure S2B). Taken together, these data argue that PRDM14 regulates the Xic by facilitating recruitment of PRC2 to Rnf12 and by binding to Xist intron 1.
We next asked how Tsix intersects this pathway. In ESCs, Tsix is not bound by PRDM14 (Figure 7B), consistent with absence of an effect on Tsix expression in Prdm14−/− ESCs (Figure 7A). We therefore asked whether Tsix might influence binding of PRDM14 to Xist. Intriguingly, in ESCs with a Tsix truncation (TsixTST)(Ogawa et al., 2008), PRDM14 was no longer efficiently recruited to Xist intron 1 in cis (Figure 7H), but normally recruited to Xist on the chromosome harboring the Tsix wildtype allele. This contrasted with patterns in wildtype cells, in which PRDM14 was bound equally to both Xist alleles. We therefore conclude that Tsix expression facilitates binding of PRDM14 to Xist intron 1. Interestingly, previous studies had shown that a deficiency of Tsix expression results in an increased recruitment of PRC2 and H3K27me3 to Xist (Navarro et al., 2009; Shibata et al., 2008; Sun et al., 2006). This suggests that Tsix-dependent PRDM14 binding to Xist intron 1 may not be the dominant recruiting mechanism of PRC2 to Xist in ESCs, which would be in agreement with unchanged H3K27me3 occupancy at the Xist promoter in Prdm14-mutant cells (Figure 7F). PRC2 is likely to be recruited by multiple mechanism, including (for example) RepA, a distinct RNA transcribed from exon 1 of Xist (Zhao et al., 2008).
Our data directly link PRDM14 and Tsix to the XCR pathway in pluripotent stem cells. PRDM14 thereby may have a dual role in repressing Xist during XCR (Figure 7I, Figure S3): First, an indirect role by silencing the Xist-activator, Rnf12, via recruitment of PRC2 to lay down the repressive H3K27me3 mark. In turn, the lack of RNF12 would prevent turnover of the REX1 transcription factor that normally binds to the Xist promoter (Gontan et al., 2012). REX1 stabilization might also dysregulate Tsix, where REX1 binds as well (Gontan et al., 2012; Navarro et al., 2010). Interestingly, Rex1 mutant mice have been reported to show postimplantation abnormalities due to epigenetic defects acquired in the blastocyst, where REX1 is normally expressed (Kim et al., 2011). This also might suggest a potential mechanism as to why our Prdm14−/− embryos showed defects during postimplantation development (Figure 3). A second role for PRDM14 during XCR might be direct repression of Xist by binding to Xist intron 1 (Ma et al., 2010), a region proposed to function as Xist repressor element (Navarro et al., 2008). We demonstrated that recruitment of PRDM14 to this region depends on Tsix expression. Thus, PRDM14 and Tsix RNA intersect the XCR pathway functionally and mechanistically within Xist. PRDM14 is, in addition, important for reprogramming to bona fide self-renewing iPSCs. Our work provides a framework in which the epigenetic stability and quality of iPSCs can be studied in the future.
EXPERIMENTAL PROCEDURES
Embryo isolation
Tsix- and Prdm14-mutant (RIKEN CDB Acc. No.: CDB0985K: http://www.cdb.riken.jp/arg/mutant%20mice%20list.html) mice were maintained in a predominant C57/BL6 strain background. Blastocysts were harvested at E3.5 and cultured in KSOM+AA medium (Millipore, MR-121-D) until E4.5, or flushed at E4.5 and E5.5 after diapause induction to delay implantation. Diapause was induced by intraperitoneal injection at E2.5 with Tamoxifen (Sigma, T5648; 10 μg/mouse) dissolved in Corn oil (Sigma, C8267) and subcutaneous injection of Medroxyprogesterone 17-Acetate (Sigma, M1629; 3 mg/mouse), dissolved in phosphate buffered saline (PBS). Mouse experiments were conducted under oversight of the Institutional Animal Care and Use Committee (IACUC) at Massachusetts General Hospital.
Derivation and culture of iPSCs and ESCs
MEFs from E13.5 embryos or TTFs from 2–4 day old mice were derived in DMEM with 15% FBS under physiological low oxygen (4%) conditions. For lentiviral reprogramming, fibroblasts co-infected with polycistronic Dox-inducibe 4-factor STEMCCA (Sommer et al., 2009) and rtTA lenitivirus (Maherali et al., 2008). Then cells were split onto 6-well plates with or without feeders and reprogrammed in ambient oxygen (21%) by adding 1 μg/ml doxycycline, 1000 U/ml LIF and 25 μg/ml L-ascorbic acid. Reprogramming (colonies/input fibroblasts) and XCR efficiency (% colonies containing X-GFP signal) was assessed after 10 days of Dox-induction. After 10–11 days, ascorbic acid was withdrawn and 1–2 days later iPSCs were trypsinized, panned (for removing feeders/non-reprogrammed fibroblasts) and replated in defined numbers (100, 500, 1000 cells) on 6-well plates with feeders. After 14 days of Dox-dependent or -independent growth, colonies were stained for alkaline phosphatase activity (Alkaline phosphatase substrate kit I, Vector laboratories) and counted.
Retroviral generation of iPSCs was performed as described (Nakagawa et al., 2008). Briefly, MEFs were seeded in 6-well plates at 1×105 cells/well and infected the next day with retroviruses containing Oct4, Sox2, L-Myc, Klf4 and DsRed. Four days later, the cells were replated onto feeders with 10- or 200-fold dilution in ES medium. The transduced cells were grown in ES medium containing LIF or N2B27 medium containing LIF and 2i as described (Yamaji et al., 2013; Ying et al., 2008). 21 days after infection, the number of colonies was counted and expression of NANOG was examined by immunostaining.
Prdm14+/+ and Prdm14−/− ESCs are described elsewhere (Yamaji et al., 2013) and were cultured under 2i+LIF on feeders.
Immunostainings and RNA-FISH
Embryos were fixed in 4% paraformaldehyde for 15 min, washed 3x with PBS, and blocked/permeabilized in AB buffer (1% Triton X-100, 0.2% SDS, 10 mg/ml BSA in PBS), which was also used for antibody incubations and washes. They were then incubated with primary antibodies overnight at 4°C, washed 3x, and incubated with secondary antibodies for 1–2 hours at room temperature (antibodies listed in Supplemental Information). After 3 washes, embryos were stained for 10 min in PBS + Dapi (1 mg/ml) and mounted on slides in Vectashield + Dapi (Vector Labs).
Immunostaining of retroviral iPSCs (Yamaji et al., 2013) and combined immunostaining/RNA-FISH of iPSCs were performed as described (Namekawa and Lee, 2011). For Xist RNA-FISH, single stranded Cy5-labeled locked nucleic acid (LNA) or fluorescein-12-dUTP-labeled single stranded DNA probe cocktails were used. Images were collected using a Nikon 90i microscope and analyzed with Volocity software (Perkin Elmer). Statistical analyses were performed using Microsoft Excel and GraphPad Prism.
Supplementary Material
HIGHLIGHTS.
PRDM14 and Tsix regulate X-reactivation during mouse development
PRDM14 is required for iPSC self-renewal and maintenance
PRDM14 represses the X-inactivation regulator, Rnf12, by recruitment of PRC2
Tsix facilitates PRDM14 binding to Xist intron 1
Acknowledgments
We are grateful to all members of the Lee Lab for valuable discussions and suggestions. We would like to thank M. Stadtfeld, J. Polo, N. Maherali, E. Apostolou, M. Anguera, and K. Hochedlinger for critical advice on iPSC reprogramming; L. Prickett-Rice, S. Lahiri, M. Weglarz and K. Folz-Donahue for cell sorting; H. Sunwoo for Xist-LNA probe and Y. Ogawa for Xist-DNA probe. B.P. was funded by postdoctoral awards from the Human Frontier Science Program and the Charles A. King Trust. J.T.L. is supported by the NIH (R37-GM58839). J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Gribnau J. X chromosome inactivation in the cycle of life. Development. 2012;139:2085–2089. doi: 10.1242/dev.069328. [DOI] [PubMed] [Google Scholar]
- Barakat TS, Gunhanlar N, Pardo CG, Achame EM, Ghazvini M, Boers R, Kenter A, Rentmeester E, Grootegoed JA, Gribnau J. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 2011;7:e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Chan YS, Goke J, Lu X, Venkatesan N, Feng B, Su IH, Ng HH. A PRC2-dependent repressive role of PRDM14 in human embryonic stem cells and induced pluripotent stem cell reprogramming. Stem Cells. 2012;31:682–692. doi: 10.1002/stem.1307. [DOI] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Hayashi K, Shovlin TC, Mifsud W, Surani MA, McLaren A. X Chromosome Activity in Mouse XX Primordial Germ Cells. PLoS Genet. 2008;4:e30. doi: 10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillich A, Bao S, Grabole N, Hayashi K, Trotter MW, Pasque V, Magnusdottir E, Surani MA. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillich A, Hayashi K. Switching stem cell state through programmed germ cell reprogramming. Differentiation. 2011 doi: 10.1016/j.diff.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IW, Grootegoed JA, Gribnau J. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, Magnusdottir E, Surani MA. Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 2013;14:629–637. doi: 10.1038/embor.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811–1815. doi: 10.1126/science.1160810. [DOI] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28:164–174. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Cox LL, Tam PP, Nagy A. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. Genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Kim JD, Kim H, Ekram MB, Yu S, Faulk C, Kim J. Rex1/Zfp42 as an epigenetic regulator for genomic imprinting. Hum Mol Genet. 2011;20:1353–1362. doi: 10.1093/hmg/ddr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT. Homozygous Tsix mutant mice reveal a sex-ratio distortion and revert to random X-inactivation. Nat Genet. 2002;32:195–200. doi: 10.1038/ng939. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol. 2010;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkovsky A, Barakat TS, Sellami N, Chin MH, Gunhanlar N, Gribnau J, Plath K. The pluripotency factor-bound intron 1 of Xist is dispensable for X chromosome inactivation and reactivation in vitro and in vivo. Cell Rep. 2013;3:905–918. doi: 10.1016/j.celrep.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Lee JT. Detection of nascent RNA, single-copy DNA, and protein localization by immunoFISH in murine germ cells and pre-implantation embryos. Nat Protoc. 2011 doi: 10.1038/nprot.2010.195. in press. [DOI] [PMC free article] [PubMed]
- Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- Navarro P, Chantalat S, Foglio M, Chureau C, Vigneau S, Clerc P, Avner P, Rougeulle C. A role for non-coding Tsix transcription in partitioning chromatin domains within the mouse X-inactivation centre. Epigenetics Chromatin. 2009;2:8. doi: 10.1186/1756-8935-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P, Oldfield A, Legoupi J, Festuccia N, Dubois A, Attia M, Schoorlemmer J, Rougeulle C, Chambers I, Avner P. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468:457–460. doi: 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA Interference and X-Inactivation Pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhata T, Senner CE, Hemberger M, Wutz A. Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice. Genes Dev. 2011;25:1702–1715. doi: 10.1101/gad.16997911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT, Namekawa SH. X-inactivation and X-reactivation: epigenetic hallmarks of mammalian reproduction and pluripotent stem cells. Hum Genet. 2011;130:265–280. doi: 10.1007/s00439-011-1024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–1286. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Shibata S, Lee JT. Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr Biol. 2004;14:1747–1754. doi: 10.1016/j.cub.2004.09.053. [DOI] [PubMed] [Google Scholar]
- Shibata S, Yokota T, Wutz A. Synergy of Eed and Tsix in the repression of Xist gene and X-chromosome inactivation. Embo J. 2008 doi: 10.1038/emboj.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, et al. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA Activates Xist by Evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell. 2012;11:91–99. doi: 10.1016/j.stem.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.