INTRODUCTION
The production of human monoclonal mAbs for research and clinical use is closely related to the development of phage display technology, initially described by Smith in 1985 and further developed by other groups (e.g., Winter, McCafferty, Lerner, Barbas). Antibody phage display (APD) is based on genetic engineering of bacteriophages (viruses that infect bacteria) and repeated rounds of antigen-guided selection and phage propagation (Barbas, 2001). This technique allows in vitro selection of mAbs of virtually any specificity, greatly facilitating recombinant production of reagents for use in research and clinical diagnostics, as well as for pharmaceuticals for therapeutic use in humans (e.g., adalimumab, the first fully human APD-derived mAb) (Lee et al., 2007).
Because of a physical connection established between the antibody fragment on the outside and the genetic information encoding the displayed protein within the phage, APD also allows comprehensive studies of genetics and function of antigen-specific mAbs. These characteristics make APD a powerful tool to better understand immunological processes and human diseases that involve formation of (auto-)antibodies against defined (self-)antigens.
ANTIBODY-LIBRARY PREPARATION
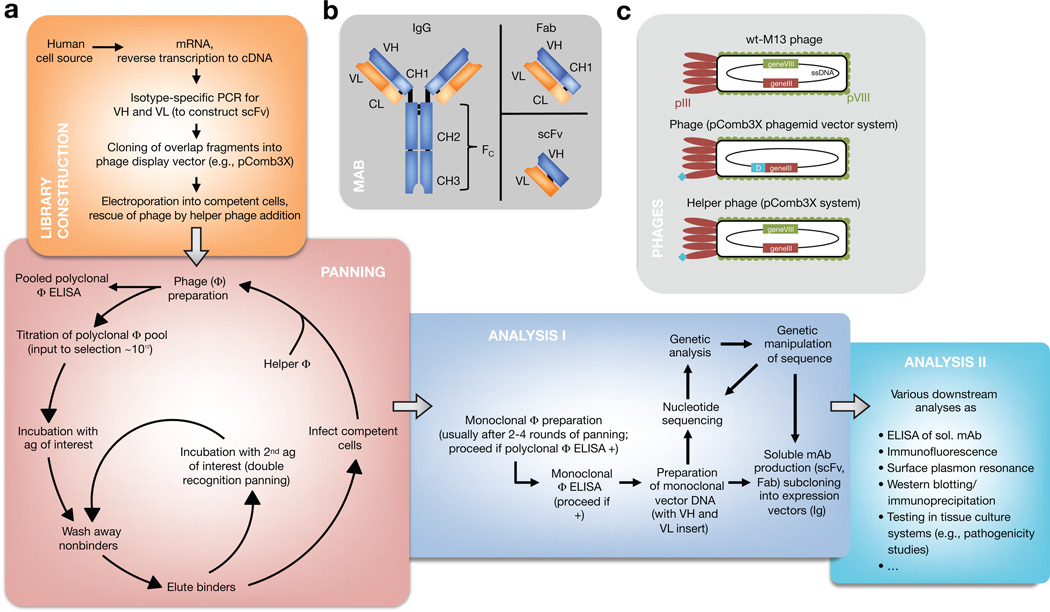
The APD process begins with antibody-library preparation, followed by ligation of the variable heavy (VH) and variable light (VL) PCR products into a phage display vector, culminating in analysis of clones of mAbs. A large antibody library and efficient selection are needed to isolate specific mAbs from a cloned immunoglobulin repertoire. The key to success is preparation of quality RNA from the cell source chosen (e.g., peripheral blood mononuclear cells). This RNA is reverse-transcribed into cDNA, which is used for PCR of the VH and VL chains of the encoded antibodies (Figure 1a, b). Defined sets of primers specific for the different VH and VL chain–region gene families then allow amplification of all transcribed rearranged variable regions within a given immunoglobulin repertoire for library construction, thus reflecting all antibody specificities in a particular individual. This immortalizes recombinant cDNA clones for expressed Igs. Variations on phage display libraries include (i) libraries constructed for Ig isotypes (e.g., IgG, IgA, and IgE) and (ii) libraries of mAbs expressed as Fab fragments or as single-chain variable fragments (scFv), the latter of which consist of the VH and VL joined by a linker (Figure 1b).
Figure 1. Antibody phage display (APD).
(a) Diagram showing how APD is performed. (b) Interrelatedness of Ig (here, IgG), single-chain variable fragment (scFv), and Fab. Importantly, both scFv and Fab reflect the original specificity of the Ig they are derived from because variable heavy (VH) and variable light (VL) chains form the interface with the antigen. (c) Phage display systems are derived from wild-type bacteriophage of Escherichia coli. Monovalency of the displayed protein (turquoise) ensures selection for high-affinity mAbs in phagemid vector-based systems (e.g., pComb3X). Helper phages display a fusion protein as well after propagation from phage-infected cells and are also selected during panning (but they do not possess the sequence of the displayed protein within). A defective helper phage origin of replication ensures preferential production and packaging of the desired phage particles, requiring addition of new helper phage in every panning round.
PHAGE AND PHAGEMID BIOLOGY IN APD
The VH and VL PCR products, representing the antibody repertoire, are ligated into a phage display vector (e.g., the phagemid pComb3X) that is engineered to express the VH and VL as an scFv fused to the pIII minor capsid protein of a filamentous bacteriophage of Escherichia coli that was originally derived from the M13 bacteriophage. However, the phage display vector pComb3X does not have all the other genes necessary to encode a full bacteriophage in E. coli. For those genes, a helper phage is added to the E. coli that are transformed with the phage display vector library. The result is a library of phages, each expressing on its surface a mAb and harboring the vector with the respective nucleotide sequence within (Figure 1c).
In addition to the ability to produce phage displaying the mAb, the phage display vector can be used to produce the mAb itself (not attached to phage capsid proteins) in certain strains of E. coli. Additional cDNA is engineered, in the phage display vector, after the VL and VH sequences to allow characterization and purification of the mAb produced. Specifically, the recombinant antibody may have a hemag-glutinin (HA) epitope tag and a polyhistidine to allow easy purification from solution (Barbas, 2001).
FINDING THE NEEDLE(S) IN THE HAYSTAC K: SELECTION BY PANNING
Diverse APD libraries are produced from ~108 independent E. coli transformants infected with helper phage. A library is screened for phage binding to an antigen through its expressed surface mAb by a technique called (bio-)panning. Cyclic panning allows for pulling out potentially very rare antigen-binding clones and consists of multiple rounds of phage binding to antigen (immobilized on ELISA plates or in solution on cell surfaces), washing, elution, and reamplification of the phage binders in E. coli (Figure 1a). During each round, specific binders are selected out from the pool by washing away non-binders and selectively eluting binding phage clones. After three or four rounds, highly specific binding of phage clones through their surface mAb is characteristic for directed selection on immobilized antigen. For panning on eukaryotic cell surfaces, more rounds of panning are usually needed, and more sophisticated protocols involving cell-sorting techniques have been published (Barbas, 2001). Of note, it is also possible to perform double recognition panning to select for bispecific mAbs (i.e., mAbs that recognize two antigens), as demonstrated in a patient with active mucocutaneous pemphigus vulgaris (PV) and serum antibody reactivity against desmoglein (Dsg) 3 and Dsg1, yielding scFv specific for both Dsg3 and Dsg1 (Payne et al., 2005).
FUNCTIONAL AND GENETIC ANALYSIS OF SELECTED MABs
After cyclic panning, the resultant (polyclonal) phage pools (i.e., a mixture of all the phages that bind to the antigen chosen) are tested by phage ELISA, in which the phage pool is incubated on an antigen-coated plate and then, after being washed, developed with an enzyme-conjugated anti-phage antibody directed against the major capsid protein and a chromogenic substrate specific for the enzyme chosen. If these pools bind the desired antigen (compared with control nonpanned phage libraries), the enzyme renders a change in color, indicating enrichment of phage binding of the specific antigen. In that case, E. coli infected with polyclonal phage is plated out and individual colonies are picked and expanded for monoclonal phage production. These are each tested again by phage ELISA to confirm antigen binding. The phage display vector, isolated from each clone, is then subjected to sequencing to determine the nucleotide sequence of VL and VH encoding the mAb that bound to the antigen. Furthermore, soluble scFv (or Fab) from clones of interest can easily be produced in bacteria that have been transformed with the phage display vector of interest. These mAb are then purified by metal chelation (e.g., through polyhistidine) or affinity purification (e.g., through a HA tag). To further analyze these soluble mAbs, a vast array of methods exists (Figure 1a). Obtained nucleotide sequences can be analyzed and grouped (e.g., by heavy- or light-chain gene usage and shared “finger-prints,” known as complementarity-determining region 3, indicating common B-cell clonal origin) with tools available online (e.g., VBASE2 and IMGT/V-QUEST).
APPLICATIONS OF APD IN INVESTIGATIVE DERMATOLOGY
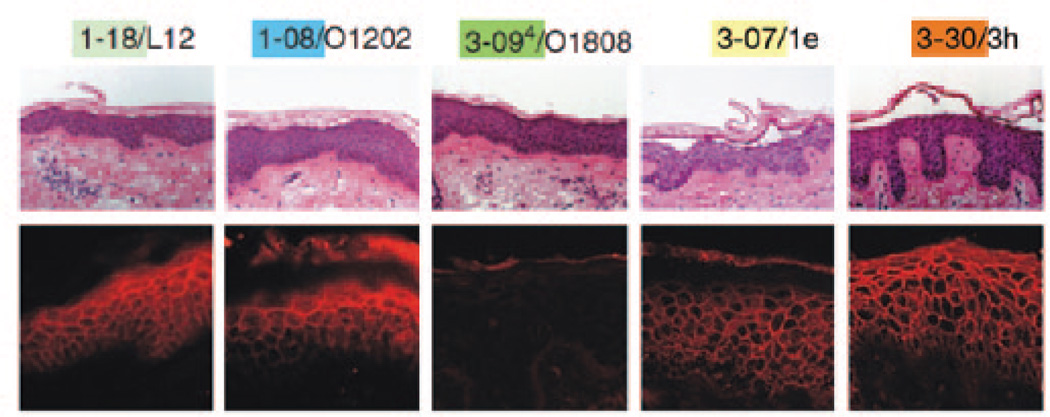
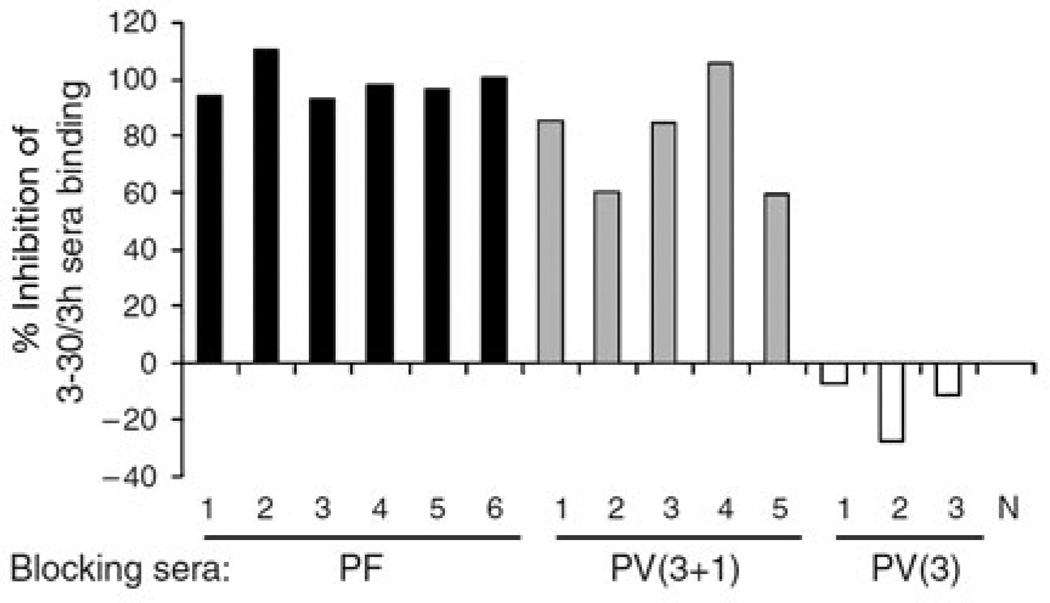
Despite the power to genetically and functionally characterize antibody responses, APD has been used in only a few studies to mechanistically dissect human skin diseases, perhaps because it is a demanding technology. Ishii et al. (2008) applied APD to characterize the IgG coding sequences from a pemphigus foliaceus (PF) patient and obtained, after cyclic panning against Dsg1, five Dsg1-specific IgG heavy-chain clones with restricted VH gene usage. Two of these five anti-Dsg1 clones proved pathogenic, meaning that the antibodies recombinantly produced from their nucleotide sequences caused typical PF blister formation in human skin (Figure 2). Inhibition ELISA studies using a pathogenic scFv derived from these clones and multiple PF sera suggested that the pathogenic antibody response in other PF patients is directed at similar or identical Dsg1 epitopes as defined by the clones’ scFv from this patient (Figure 3), also illustrating the biological validity of studying human disease with monoclonal scFv. Yamagami et al. (2009) reported similar findings with another PF patient, and comparable results have also been obtained by APD of PV patients’ peripheral blood mononuclear cells (Payne et al., 2005).
Figure 2. Monovalent mAbs (in the form of scFv) cause blister formation typical of pemphigus foliaceus in human skin.
After injection of purified antidesmoglein 1 scFv, human skin specimens were cultured for 24 hours. Histology and direct immunofluorescence (IF) are shown. 3-07/1e and 3-30/3h caused superficial epidermal blisters. All scFv, except 3-094/O1808, bound to the cell surface of epidermal keratinocytes. Immunoprecipitation and indirect IF experiments suggested that 3-094/O1808 binds to the proprotein but not to the mature protein (data not shown). mAb, monoclonal antibody; scFv, single-chain variable fragment. Reprinted with permission from Ishii et al. (2008).
Figure 3. Multiple PF and PV sera compete for the same or nearby epitopes targeted by an anti-Dsg-1 monoclonal scFv (3-30/3h) derived from a PF patient.
6 PF sera, 5 mucocutaneous PV sera, and 3 mucosal PV sera (and a normal control, N) were used to block binding of the 3-30/3h to Dsg1. All PF and mucocutaneous PV sera inhibited binding of 3-30/3h, but mucosal PV patients’ sera (featuring anti-Dsg3 antibodies only) did not. Dsg, desmoglein; PF, pemphigus foliaceus; PV, pemphigus vulgaris; scFv, single-chain variable fragment. Reprinted with permission from Ishii et al. (2008).
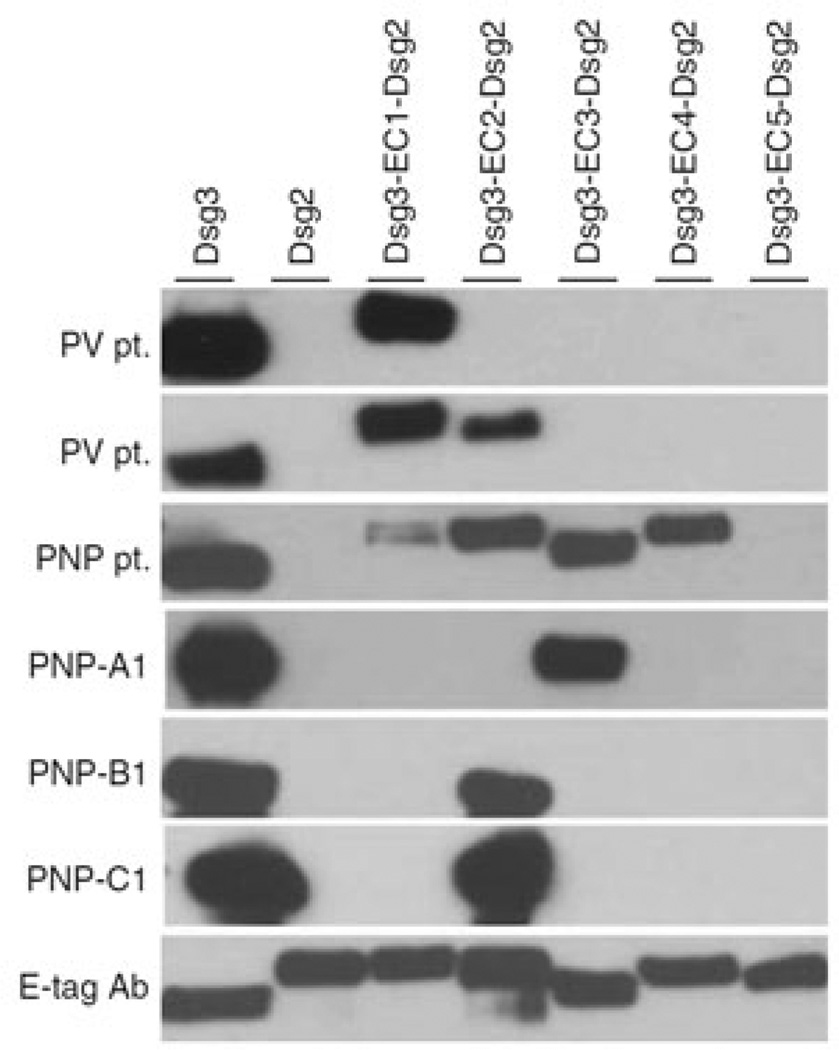
Similarly, Saleh et al. (2012) cloned pathogenic anti-Dsg3 mAbs from a patient with paraneoplastic pemphigus and found four Dsg3-specific clones (profoundly restricted to the VH1 family), of which three were pathogenic. Further characterization of those scFv (and the patients’ serum antibodies) using a series of Dsg3/Dsg2 domain-swapped molecules and immunoprecipitation demonstrated that the epitopes covered extracellular domains (EC) 1–4, in contrast to classical PV in which pathogenic abs mostly bind to EC1–2 (Figure 4).
Figure 4. Three anti-Dsg3 scFvs (PNP-A1/B1/C1) isolated from a PNP patient bind to the EC2 and EC3 domains of Dsg3 in immunoprecipitation/immunoblot analysis of the patient’s and a PV patient’s sera.
The PNP patient’s sera recognized Dsg3 epitopes EC1–4, but the mAbs derived by phage display recognized only EC2 and EC3. In classic PV patients, the main epitopes are EC1 and EC2. These findings also demonstrate that antibody phage display may miss some pathologic clones (here, such mAbs that target EC1 and EC4) for various reasons, as discussed in the last paragraph of this article. Dsg, desmoglein; PNP, paraneoplastic pemphigus; PV, pemphigus vulgaris; scFv, single-chain variable fragment. Reprinted with permission from Saleh et al. (2012).
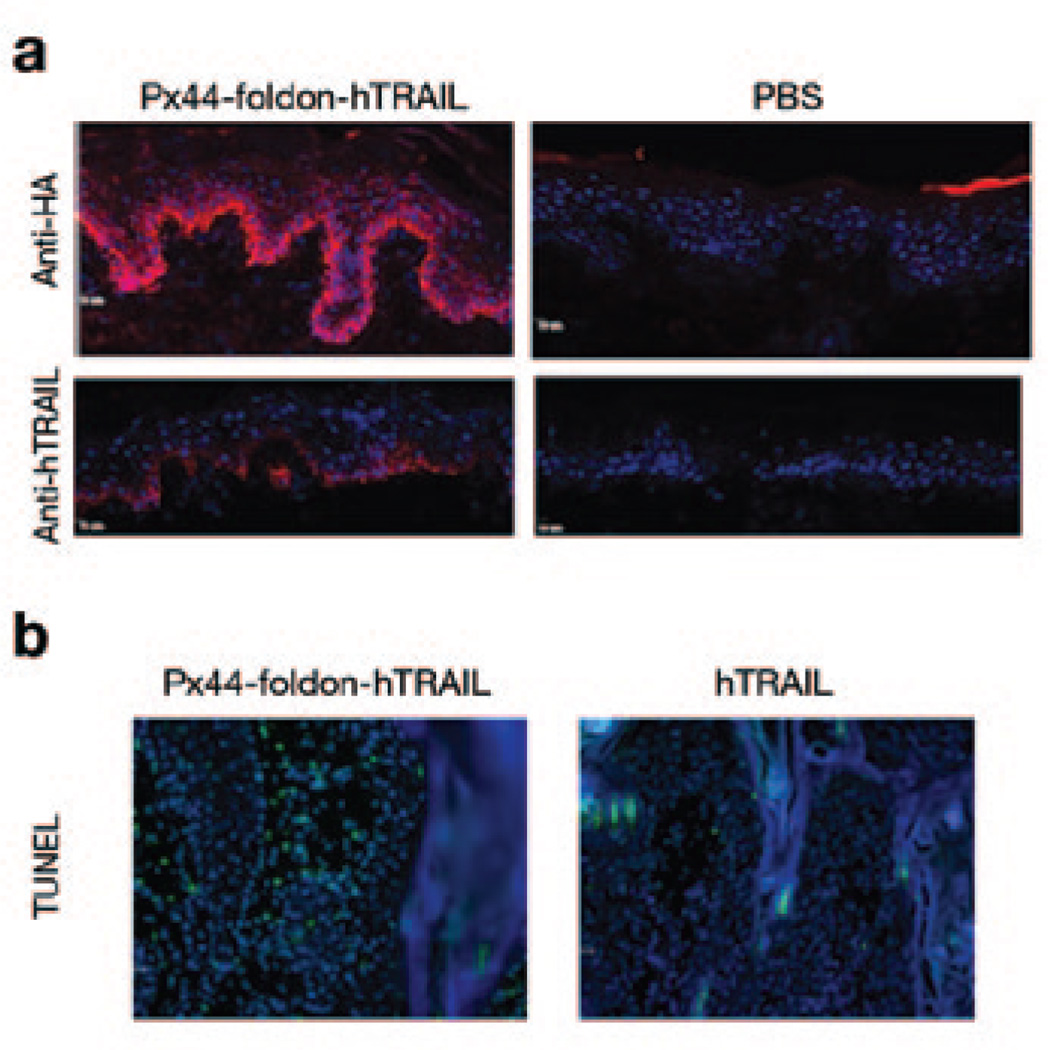
In a translational research approach designed to use a nonpathogenic cloned scFv from a pemphigus patient to deliver a biologically active agent to the epidermis, Kouno et al. (2013) created a fusion protein containing Px44, a nonpathogenic anti-Dsg-scFv domain (Payne et al., 2005), linked to an active domain of human tumor necrosis factor–related apoptosis-inducing ligand (hTRAIL). The recombinantly produced Px44-hTRAIL fusion protein bound and delivered TRAIL to the cell surface of human keratinocytes (Figure 5a). The TRAIL component maintained its known biological activity both to inhibit secretion of γ-interferon by activated CD4+ T cells and to cause apoptosis of rapidly proliferating lymphocytes and keratinocytes. In a preliminary study Px44–hTRAIL caused apoptosis in a mouse model of actinic keratosis and early squamous-cell carcinoma (Figure 5b). These studies suggest that mAbs cloned from pemphigus patients may be useful as agents to deliver biologically active proteins to the epidermis for therapy of various immunological and neoplastic diseases.
Figure 5. Px44-hTRAIL binds to human epidermal keratinocytes and causes apoptosis of tumor cells.
(a) When injected intradermally into human skin, the fusion protein binds to the cell surface of epidermal keratinocytes as detected by immunofluorescence with anti-hemagglutinin (HA) and anti-hTRAIL antibodies. (b) TUNEL staining of actinic keratosis-like lesions in K14-activated Fyn transgenic mice after intratumoral injection of the fusion protein shows apoptosis of tumor cells (green fluorescence signal). hTRAIL, human tumor necrosis factor–related apoptosis-inducing ligand. Reprinted with permission from Kouno et al. (2013).
LIMITATIONS AND PECULIARITIES
Compared to other methods used to dissect human immune responses (Table 1), APD offers sufficient depth of coverage to find antigen-specific mAbs even if they are rare in the ab-coding repertoire of circulating cells. The relative ease of constructing and screening antibody libraries comes from many well-established published protocols, and there are various easily accessible systems that facilitate production of soluble mAbs. Still, there are limitations to APD. One example is that pairing of VH and VL chains during phage display library construction may not represent the in vivo antibody pairing in patients. However, scFvs derived from APD bind the same epitopes on Dsg as polyclonal patient IgGs do (Figure 3), and some data indicate that in both heterohybridoma and APD the same genes for VH and VL are detected (unpublished observation), suggesting that the mAbs derived from APD accurately represent the antibodies in patients. A second caveat is that APD libraries may not fully represent all antibodies, because not all phage clones of a given library will display a protein because of inherent toxicity or interference with phage assembly. A third issue is that clones of interest may be missed as a result of poor recovery of RNA from cells or from loss of DNA during library construction, which requires repetitive gel purifications. Finally, undersized sampling of monoclonals after panning may result in the loss of some relevant clones. However, even given these limitations, APD—by virtue of its ability to screen up to 1012 phages to select those with antigen binding—is thought to be much more likely to detect relevant antibodies than are cell-based methods, whose numbers are much more limited. Combination of APD with other established high-throughput methods such as next-generation sequencing and/or proteomics may offer the most powerful approach for future molecular deciphering of antigen-specific serum responses in disease and health (Cheung et al., 2012; DeKosky et al., 2013; Wine et al., 2013).
Table 1.
Other typical methods used for studies characterizing the genetics of human (auto-)antibody responses
| Method | Production of mAb | Sequencing | Selection bias1 | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Antibody phage display | scFv, Fab (Ig) | Directly from phage display vector | + | Relative ease of screening, sequencing, and production of soluble protein; depth of coverage; ease of adapting isotype-specific applications | Random pairing of VH and VL, tedious Ig production (subcloning required) |
| Heterohybridoma | Ig | RT-PCR | + | In vivo VH/VL pairing, production of full-length Ig | EBV/unselected fusions often yield IgM, instability phenomena and inefficient isolation, expensive culturing |
| Single B-cell PCR | (Ig) | RT-PCR | + | In vivo VH/VL pairing | Difficult mAb production (subcloning), difficult for rare clones |
| Deep sequencing | None | From any nucleotide source | – | No selection bias, calculation of frequencies | Necessary trade-off of sequencing length and read number; no pairing of VH and VL for analysis |
Advancements and combinations of the methods detailed above may pave the way for future studies of unparalleled depth and accuracy (see DeKosky et al., 2013, and others).
EBV, Epstein–Barr virus; scFv, single-chain variable fragment; VH, variable heavy; VL, variable light.
Selection bias may result from differences in phage growth rates, instability of cell fusions, or limited numbers of B cells analyzed by single B-cell PCR. These effects may lead to nonrepresentative (biased) distributions of the parameters analyzed.
Supplementary Material
WHAT ANTIBODY PHAGE DISPLAY DOES
Antibody phage display (APD) allows in vitro selection of human mAbs of virtually any specificity and affinity.
Owing to its design, this technique permits both genetic and functional analyses of the mAb selected, thus facilitating studies on mechanisms of the human immune system.
Translational research approaches embracing APD may converge on new future therapies.
LIMITATIONS
Phage display may not recover all antigen-specific mAbs present in a given antibody library.
The heavy- and light-chain pairing may not reflect that of the in vivo immunoglobulin.
APD is a complicated, demanding, and time-consuming technology.
QUESTIONS.
This article has been approved for 1 hour of Category 1 CME credit. To take the quiz, with or without CME credit, follow the link under the “CME ACCREDITATION” heading.
-
What of the following is required to obtain antigen-specific mAbs by antibody phage display?
A source containing antibody-expressing cells.
A phage display vector.
A defined antigen for selection.
Molecular tags for protein purification.
All of the above.
-
Which technique(s) mentioned below reflect(s) accurately the in vivo pairing of variable heavy and light chain?
Deep sequencing.
Heterohybridoma.
Antibody phage display
Single B-cell PCR.
-
Which of the following points is limiting for the use of antibody phage display technology for an experiment planned?
Using surface antigens on eukaryotic cells for panning.
Known toxicity of the displayed protein for E. coli.
Low frequency of cells producing the antibody of the desired specificity in the initial cell source.
Desired production of full-length immuno-globulins for further analyses.
ACKNOWLEDGMENTS
This work was supported by a research fellowship from the DFG (HA6736/1-1) to C.M.H., and by grants R01-AR052672 and P30-AR057217 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health to J.R.S. We thank Aimee S. Payne for providing us with an earlier version of Table 1.
Footnotes
CONFLICT OF INTEREST
J.R.S. holds a US patent (8,470,323) on parts of the technologies described herein.
CME ACCREDITATION
This CME activity has been planned and implemented in accordance with the Essential Areas and Policies of the Accreditation Council for Continuing Medical Education through the Joint Sponsorship of ScientiaCME and Educational Review Systems. ScientiaCME is accredited by the ACCME to provide continuing medical education for physicians. ScientiaCME designates this educational activity for a maximum of one (1) AMA PRA Category 1 Credit™. Physicians should claim only credit commensurate with the extent of their participation in the activity.
To take the online quiz, follow the link below: http://www.classmarker.com/online-test/start/?quiz=kja529668ecdcb7e
SUPPLEMENTARY MATERIAL
A PowerPoint slide presentation appropriate for journal club or other teaching exercises is available at http://dx.doi.org/10.1038/jid.2013.521.
REFERENCES
- Barbas CF. Phage display: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Cheung WC, Beausoleil SA, Zhang X, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol. 2012;30:447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- DeKosky BJ, Ippolito GC, Deschner RP, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Lin C, Siegel DL, et al. Isolation of pathogenic monoclonal antidesmoglein 1 human antibodies by phage display of pemphigus foliaceus autoantibodies. J Invest Dermatol. 2008;128:939–948. doi: 10.1038/sj.jid.5701132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouno M, Lin C, Schechter NM, et al. Targeted delivery of tumor necrosis factor–related apoptosis-inducing ligand to keratinocytes with a pemphigus mAb. J Invest Dermatol. 2013;133:2212–2220. doi: 10.1038/jid.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Iorno N, Sierro F, et al. Selection of human antibody fragments by phage display. Nat Protoc. 2007;2:3001–3008. doi: 10.1038/nprot.2007.448. [DOI] [PubMed] [Google Scholar]
- Payne AS, Ishii K, Kacir S, et al. Genetic and functional characterization of human pemphigus vulgaris monoclonal autoantibodies isolated by phage display. J Clin Invest. 2005;115:888–899. doi: 10.1172/JCI24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MA, Ishii K, Yamagami J, et al. Pathogenic anti-desmoglein 3 mAbs cloned from a paraneoplastic pemphigus patient by phage display. J Invest Dermatol. 2012;132:1141–1148. doi: 10.1038/jid.2011.449. [DOI] [PubMed] [Google Scholar]
- Wine Y, Boutz DR, Lavinder JJ, et al. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci USA. 2013;110:2993–2998. doi: 10.1073/pnas.1213737110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami J, Kacir S, Ishii K, et al. Antibodies to the desmoglein 1 precursor proprotein but not to the mature cell surface protein cloned from individuals without pemphigus. J Immunol. 2009;183:5615–5621. doi: 10.4049/jimmunol.0901691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.