Abstract
Regenerative medicine for heart failure seeks to replace lost cardiomyocytes. Chemical approaches for producing ample supplies of cells, such as pluripotent stem cells and cardiomyocytes, hold promise as practical means to achieve safe, facile cell-based therapy for cardiac repair and regenerative medicine. In this review, we describe recent advances in the application of small molecules to improve the generation and maintenance of pluripotent stem cells. We also describe new directions in heart repair and regeneration in which chemical approaches may find their application.
PLURIPOTENT STEM CELLS FOR CARDIAC REGENERATIVE MEDICINE
Heart failure is usually accompanied with severe loss of cardiomyocytes, the beating cells of heart tissue.1 Cell transplantation might be a way to rebuild damaged heart tissue but it requires ample sources of cells.2 Pluripotent stem cells (PSCs) differentiate into any cell type, including cardiomyocytes, and thus hold tremendous promise for regenerative medicine and heart repair.3 The therapeutic potential of pluripotent, human embryonic stem cells (ESCs) has long been recognized.4 Their derivation, however, inevitably involves manipulation of human embryos and thus is controversial.
Takahashi and Yamanaka began a new era of stem cell biology with their revolutionary reprogramming technology. They demonstrated that murine somatic cells can be “reprogrammed” into induced pluripotent stem cells (iPSCs) with a specific set of transcription factors (TFs), namely Oct4, Sox2, Klf4 and c-Myc (OSKM).5 The same strategy was soon proven applicable to reprogram human somatic cells and the human iPSCs thus generated can differentiate into cells in the three germ layers.6, 7 The emergence of iPSC technology circumvented the ethical and political controversies associated with human ESCs and provides an exciting potential autologous cell source for cell-based regenerative therapy.8 Notably, human iPSCs have started to take root in disease modeling and drug development. 9, 10
Despite its groundbreaking success, the TF-based method to generate iPSCs has significant drawbacks that limit its application in therapies. The involvement of oncogenic TFs and genetic modifications imposes clinically unacceptable risks such as carcinogenicity and genomic instability of iPSCs.11 In addition, the efficiency and speed of cell reprogramming must be significantly improved to render the process more useful in practice.
Small molecules are appealing substitutes for genetic materials. The former can exert their cellular effects in a transient and dose-dependent manner, and allow the timing and the magnitude to be precisely controlled and fine-tuned. The essentially unlimited possibilities for structural variations in small molecules allow for ample opportunities to improve their potencies, selectivities, and pharmacological properties. Bioactive small molecules with high specifities can potentially serve as valuable chemical probes to investigate biological processes.12 In addition, those advantages also renders small molecules particularly suitable for translational development of drugs.
The search of small molecules to improve and/or enable cell reprogramming towards pluripotency has been most fruitful. Progress in this approach has been comprehensively reviewed elsewhere.13, 14, 15 In this review, we want to focus on the efforts to replace TFs with small molecules to generate iPSCs from somatic cells. We will highlight the insights drawn from the most recent, significant advances in murine and human cell reprogramming. Special attention will be paid to the connections between the molecular functions of small molecules and their roles in establishing pluripotency, as such knowledge will eventually lead to the realization of chemically induced, therapeutically useful human PSCs (hPSCs). The development of chemically defined conditions to maintain hPSCs will also be summarized. Another focus of the review is the applications of small molecules in cardiac regenerative therapy. Chemical approaches to boost the generation and transplantation of cardiac cells derived from PSCs will be highlighted. Potential opportunities for small molecule-based strategies in in situ heart repair will also be discussed.
Inducing PSCs with Small Molecules
Although they share essentially identical genomes, PSCs differ from somatic cells most distinctively in gene expression. The identities of the PSCs and all cells are largely established by their gene expression and epigenetic signatures.16, 17 During reprogramming, somatic cells must undergo significant epigenetic changes (i.e., histone modifications and DNA methylation) to adopt the ESC-like patterns.18, 19 On the other hand, epigenetic modifications allow for proper changes of the chromatin structure and thus influence the expression of genes crucial for cell reprogramming.20 Small molecules modulating activities of enzymes involved in epigenetic modifications can, therefore, exert profound effects on cell reprogramming.
Posttranslational modifications to histones are one of the most common epigenetic features. Acetylated histones have generally been associated with transcriptional activation.21 Histone deacetylase (HDAC) inhibitors presumably help to maintain a high level of acetylation of histones and thus facilitate the expression of pluripotency-related genes crucial for the reprogramming process.22 As an HDAC inhibitor, valproic acid (VPA) was demonstrated to enhance reprogramming of mouse embryonic fibroblasts (MEFs) 23 in the absence of exogenously expressed c-Myc, which has been known to recruit multiple histone acetylase complexes to the genome and thus presumably converts the chromatin structure of somatic cells to an opened, active state24 characteristic of PSCs.25 Although viable, reprogramming under c-Myc-free conditions was inefficient.26, 27 VPA significantly improved the efficiency of this sluggish process. VPA was also reported to promote the reprogramming of human fibroblast in the absence of Klf4 and c-Myc.28 Small molecules modulating histone and/or DNA methylations were also used to replace TFs in the reprogramming of somatic cells. BIX-01294, an inhibitor of the H3K9 histone methyltransferase G9a,29 when used in conjunction with either Bayk-8644 (an L-type calcium channel agonist) or RG108 (a DNA methyltransferase inhibitor), enabled Oct4/Klf4 (OK)-mediated reprogramming of MEFs.30 BIX-01294 could even compensate the absence of ectopic Oct4 in the Sox2/Klf4/Myc (SKM)-mediated conversion of neural progenitor cells to iPSCs.31
During TF-mediated reprogramming, the coordinating orchestra of signal transduction pathways is crucial to establish pluripotency.32 Small molecules modulating signaling pathways have been identified to enhance reprogramming efficiency or even functionally replace TFs in iPSC reprogramming. Activation of the transforming growth factor-β (TGF-β) signaling pathway inhibits mesenchymal-to-epithelial transition (MET), a cellular process indispensible for induction of pluripotency. 33 During the early stage of reprogramming, murine fibroblasts typically undergo MET and are characterized with the adoption of epithelial-like morphology, upregulation of epithelial genes, such as E-cadherin, and simultaneous downregulation of mesenchymal genes, such as Snail. E-cadherin is also expressed at high levels in human ESCs that resemble epithelial cells.2 Blocking TGFβ signaling with small molecules may facilitate reprogramming towards pluripotency.34 SB431542 (a TGFβ signaling inhibitor) and PD0325901 (PD) (an inhibitor of MEK) significantly accelerated the rate and enhance the efficiency of human iPSCs generation. E-616542, a small-molecule inhibitor of TGFβ signaling, can functionally replace Sox2 in MEF reprogramming (Table 1 and 2).35 TGFβ signaling inhibitors can functionally replaced c-Myc as well as Sox2.36 More recently, iPSCs were generated from MEFs transduced with Oct4 and treated with small molecules A83-01, a TGFβ receptor inhibitor, and AMI-5, a protein methyltransferase inhibitor (Table 1 and 2).37
Table 1.
Representative Small Molecules Used to Reprogram Cells and Maintain hPSCs
| Name | Structure | Known function(s) |
|---|---|---|
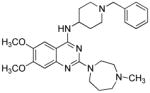
| A83-01 |
|
TGFβ receptor ALK4/5/7 inhibitor |
| AMI-5 |
|
Protein arginine N-methyltransferase inhibitor |
| (±)BayK 8644 (BayK) |
|
L-type calcium channel agonist |
| BIX-01294 (BIX) |
|
Histone methyltransferase G9a inhibitor |
| CHIR99021 (CHIR) |
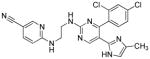
|
Glycogen synthase kinase 3β inhibitor |
| 3-Deazaneplanocin A (DZNep) |
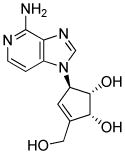
|
Histone methylation inhibitor |
| Forskolin |
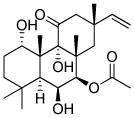
|
PKA activator |
| Kenpaullone |
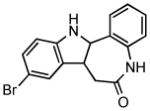
|
GSK3 and CDK1/cyclin B inhibitor |
| Parnate |
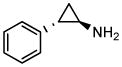
|
Lysine specific demethylase 1 inhibitor |
| PD0325901 (PD) |
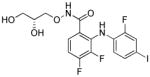
|
MEK inhibitor |
| PS48 |
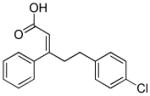
|
PDK1 activator |
| E-616452 |
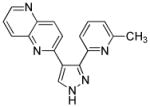
|
TGFβ Receptor I kinase inhibitor |
| RG108 |
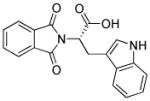
|
DNA methyltransferase inhibitor |
| SB431542 |
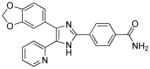
|
Activin receptor-like kinase 4/5/7 inhibitor |
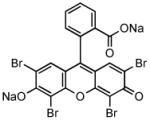
| Sodium butyrate (NaB) |
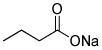
|
Histone deacetylase inhibitor |
| Thiazovivin (Tzv) |
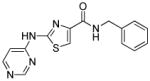
|
ROCK inhibitor |
| Pyrintegrin (Ptn) |
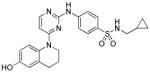
|
Unknown |
| Valproic acid (VPA) |
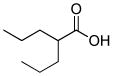
|
Histone deacetylase inhibitor |
Table 2.
Small Molecules Replacing TFs in the Reprogramming of Somatic Cells
| Small molecules combination | Starting cells | TFs required | Ref. |
|---|---|---|---|
| VPA | MEFs | OSK | Huangfu et al., 2008 |
| BIX, BayK or RG108 | MEFs | OK | Shi et al., 2008 |
| Kenpaullone | MEFs | OSM | Lyssiotis et al., 2009 |
| E-616542 | MEFs | OKM | Ichida et al., 2009 |
| A83-01, AMI-5 | MEFs | O | Yuan et al., 2011 |
| VPA, CHIR, E-616542, Parnate | MEFs | O | Li et al., 2011 |
| VPA, CHIR, E-616542, Parnate, Forskolin, DZNep | MEFs | none | Hou et al., 2013 |
| VPA | Primary human fibroblast | OS | Huangfu et al., 2008 |
| CHIR, Parnate | Human keratinocytes | OK | Li et al., 2009 |
| NaB, A83-01, PS48, PD | Neonatal human epidermal keratinocytes | O | Zhu et al., 2010 |
Manipulation of other signaling pathways has also been beneficial to cell reprogramming. Wnt signaling is important for maintaining pluripotency of ESCs and self-renewal of adult stem cells.38 CHIR99021 (CHIR), a glycogen synthase kinase 3β (GSK3B) inhibitor, activates Wnt signaling and significantly improve the efficiency of reprogramming of MEFs in the absence of Sox2 and cMyc. 39 Notably, combining CHIR with Parnate, an inhibitor of lysine-specific demethylase 1, they converted human keratinocytes to iPSCs upon ectopic expression of Oct4 and Klf4 (Table 1 and 2). In another study, kenpaullone, a GSK3B inhibitor, functionally replaced Klf4 in the reprogramming of MEFs (Table 1 and 2).40
Using cocktails of functionally diverse small molecules to synergistically improve cell reprogramming has been highly fruitful. Zhu et al. established an optimized combination of small molecules to accomplish the reprogramming of human adult keratinocytes transduced with Oct4 only. 41 PD0325901 and A83-01 induce pluripotency in neonatal human keratinocytes transduced with Oct4 and Klf4. Further screening identified two compounds, sodium butyrate (NaB) and PS48, that allow Klf4 to be omitted (Table 1 and 2). NaB turned out to be superior to VPA as an HDAC inhibitor. Interestingly, mechanistic characterization of PS48 in reprogramming revealed the metabolic switching from mitochondrial oxidation to glycolysis as a fundamental process during reprogramming. Finally, Parnate and CHIR were included in the cocktail to achieve the O-mediated reprogramming of adult human epidermal keratinocytes.41
In one recent report, TFs were completely replaced with chemicals.42 Based on their previous findings that four small molecules (VPA, CHIR, E-616542, Parnate) enabled the reprogramming of MEF under Oct4 only conditions,43 the authors sought to replace that last TF with chemical compounds. They found that the combination of the compounds with Forskolin enabled the dedifferentiation of MEFs during the early phase of reprogramming, as indicated by the increased expression of E-cadherin, as well as the pluripotency-related genes Sall4 and Sox2. At the late stage, 3-Deazaneplanocin A, a global histone methylation inhibitor,44 was added to activate Oct4 expression and furnish the fully reprogrammed cells. In this study, 0.2% of the starting MEFs were converted into iPSCs (Table 1 and 2).
Facilitating hPSC Maintenance
hPSCs can potentially supply an unlimited number of cardiomyocytes for cell-based therapy.45 Ever since hESCs were first established in 1998,2 tremendous efforts have been dedicated to improving the culture conditions for these delicate cells. 46 In the early developed culture conditions, sophisticated media containing mouse feeder cells and/or xenogeneic components were used.2 Subsequently, modified conditions still involved ill-defined, expensive human feeders and/or serum derived-components.47 Better-defined, feeder free conditions were later developed.48, 49 hPSCs are intrinsically prone to apoptosis upon cell dissociation, representing a major hurdle for their preparation and manipulation. hESCs could be maintained in a healthier state by treatment with Rho-associated protein kinase (ROCK) inhibitors, such as Y-7632 and Fasudil.50 Vitamin A promotes self-renewal of human ESCs in feeder-free conditions.51 Studies from this lab further unveiled an adhesion signaling pathway regulating hPSC survival and pluripotency. 52 By high-throughput phenotypic screening, two novel small molecules were identified that promoted hESC survival after trypsin dissociation, namely, Thiazovivin (Tzv) and Pyrintegin (Ptn). Target identification revealed that Tzv inhibits ROCK and thus stabilizes E-cadherin and enhances cell-cell interaction. ROCK inhibitors have been incorporated into simplified, chemically defined conditions for the culture of hPSCs.53, 54
CARDIAC DIFFERENTIATION AND GRAFTS
Improving Cardiac Differentiation
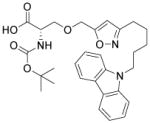
The bona fide cardiomyogenic differentiation potential of human PSCs has long been recognized.55, 56 Human PSCs differentiate into cardiomyocytes only when guided by appropriate extrinsic influences. Substantial efforts have been made to develop conditions that induce cardiac differentiation from human PSCs in an efficient, reproducible and simple manner. Approaches using small molecules will be discussed in the following sections. Phenotypic cell-based screening has been applied to discover small molecules that promote cardiac differentiation of mouse or human PSCs. Ascorbic acid was among the earliest chemicals identified to increase cardiogenic differentiation of ESC (Table 3).57 It also rescues cell line–dependent cardiogenic deficiency of iPSCs.58 The applications of high-throughput screening also led to discoveries of other cardiogenic small molecules, including cardiogenols,59 isoxazolyl-serine-based agonists of peroxisome proliferator-activated receptors (PPARs), 60 verapamil, 61 SB203580, 62 sulfonylhydrazones,63 and cinchona alkaloid derivatives (Table 3).64 While active, cardiogenic small molecules discovered in phenotypic screening assays will continue to serve as immensely useful tools, efforts to elucidate their cellular targets, although challenging, will shed more light on the biology underlying cardiac differentiation.
Table 3.
Representative Small Molecules Enhancing Cardiac Differentiation, Graft Integration, and Heart Regeneration
| Name | Structure | Molecular Function(s) | Ref. |
|---|---|---|---|
| AG1478 |
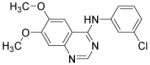
|
EGFR tyrosine kinase inhibitor | Zhang et al., 2011 |
| Ascorbic acid |
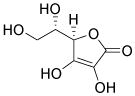
|
Multiple |
Takahashi et al., 2003 Cao et al., 2012 |
| BMS-189453 |
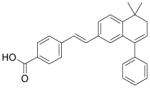
|
Pan-retinoic acid receptor antagonist | Zhang et al., 2011 |
| Cardiogenol C |
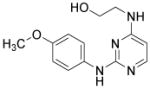
|
Unknown | Wu et al., 2004 |
| Cinchona alkaloid derivative |
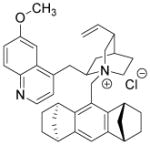
|
Unknown | Berkessel et al., 2010 |
| Diazoxide |
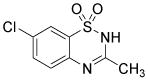
|
ATP-sensitive activator K+ channel | Niagara et al., 2007 |
| 1-EBIO |
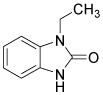
|
Agonist of Ca2+-activated K+ channels | Kleger et al., 2010 |
| Isoxazolyl-serine derivative |
|
PPAR agonist | Wei et al., 2004 |
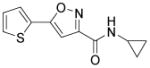
| Isx1 (isoxazole) |
|
Cardiac muscle gene activator. Neuronal reporter genes activator. |
Russell et al., 2013 |
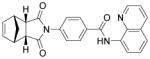
| IWR-1-endo |
|
Wnt inhibitor | Gonzalez et al., 2011 |
| IWP-2 |
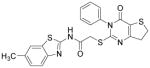
|
Wnt inhibitor | Lian et al., 2012 |
| IWP-4 |
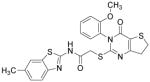
|
Wnt inhibitor | Lian et al., 2012 |
| KY02111 |
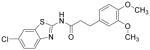
|
Wnt inhibitor | Minami et al., 2012 |
| Pinacidil |
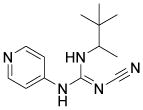
|
ATP-dependent K+ channel opener | Laflamme et al., 2007 |
| Pioglitazone |
|
PPAR-γ activator | Shinmura et al., 2011 |
| Purmorphamine |
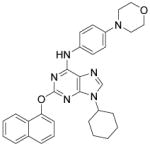
|
Sonic Hedgehog signaling agonist | Gonzalez et al., 2011 |
| RA |
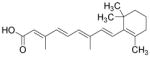
|
Natural ligand of RA receptors | Zhang et al., 2011 |
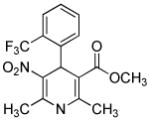
| SB203580 |
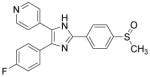
|
p38 MAPK inhibitor | Graichen et al., 2008 |
| Shz-1 (sulfonylhydrazone) |
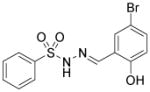
|
Activator of gene Nkx2.5 | Sadek et al., 2008 |
| Verapamil |
|
L-type Ca2+ channel blocker | Sachinidis et al., 2006 |
| ZVAD-fmk |
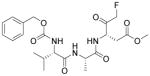
|
Caspase inhibitor | Laflamme et al., 2007 |
Aiming to recapitulate embryonic cardiac development, chemical approaches that systematically target the core signaling pathways involved in each step of cardiogenesis turned out to be extremely successful. Embryonic cardiac development is a well-organized process, involving the sequential formation of mesoderm, cardiac progenitors and cardiomyocytes.3 This stepwise process is finely regulated by multiple signaling pathways. Precise signaling control with appropriate timing using small molecules is critical to the success of chemically guided cardiac differentiation. A number of small molecules that selectively target BMP,65 TGFβ,66, 67 and Wnt, 68, 69, 70 when applied at appropriate time-window, enabled the efficient cardiac differentiation of PSCs, especially those of the human origin. Knowledge gained from these discoveries, in turn, enriches our understandings about the logic of cardiac development.
Chemically Defined Conditions for Cardiac Differentiation
The identification of robust cardiogenic chemicals that increase differentiation efficiency and replace complex, ill-defined components (i.e., serum, growth factors, hormones, and extracellular matrix) has allowed for a simple, reliable and cost-effective protocol to chemically induce cardiac differentiation from PSCs. Gonzalez et al. designed a stepwise protocol to generate cardiomyocytes from hESCs (>50% efficiency) in serum and growth factor-free conditions. 71 By systematic screening of 300 known signal transduction modulators, they identified IWR-1-endo (Wnt antagonist), purmorphamine (Sonic Hedgehog signaling agonist), and SB431542 as small molecules that promote differentiation of hESCs into cardiomyocytes (Table 3). Although the system they used was not completely chemically defined (i.e., using MEF conditioned medium and Matrigel), this study demonstrates that exogenous growth factors can be replaced by small molecules for efficient cardiac differentiation. Recently, Lian et al. developed a chemically defined cardiac differentiation system. 72 They showed that timely modulation of Wnt signaling (activated by CHIR during the first 24 hours and then blocked by IWP-2 or IWP-4 during days 3–5) is sufficient and necessary for efficient cardiac induction of hPSCs (up to 98% efficiency) under defined, growth factor–free conditions (Table 3). Similarly, Minami et al. identified a potential Wnt inhibitor KY02111 that, when used in combined with other Wnt modulators, induced robust cardiac differentiation of hPSCs (up to 98% efficiency) in a xeno-free medium devoid of serum, recombinant cytokines or hormones (Table 3).73 While the robustness and reproducibility of these protocols need to be tested on more cell lines and may require further modifications, these significant advancements paved the way to safe, efficient and cost-effective protocols for de novo cardiomyocyte production from hPSCs on a clinically relevant scale.
Reducing Heterogeneity of Cardiomyocytes
Despite the great progress in developing efficient and defined methods for cardiac differentiation of PSCs, methods are still lacking that enrich a specific subtype cardiomyocyte, such as atrial-, ventricular-, or nodal-like cells. Common hPSC differentiation methodologies give rise only to a mixture of all three major subtypes of cardiomyocytes.3 A heterogeneous cellular composition, unfortunately, hampers its utilization in medical research and cell-based therapies. In recent years, several breakthroughs in the field of selective cardiac differentiation conditions were made using the pharmacologic approaches. Kleger et al. found that 1-ethyl-2-benzimidazolinone (1-EBIO), an agonist of Ca2+-activated potassium channels, induces cardiogenesis of murine ESCs and strongly enriches nodal-like cells (from 7.2 to 57.8%) (Table 3).74 By using direct action potential phenotyping, activation of genetic label, and subtype-specific marker expression, Zhu and colleagues demonstrated that NRG-1β/ERBB signaling regulates the ratio of nodal- to ventricular-type cells in hESC-derived cardiomyocytes. Inhibition of NRG-1β/ERBB signaling by its antagonist AG1478 significantly enriched the nodal-like cells (21 to 52%) (Table 3).75 Similarly, Zhang et al. found that retinoic acid (RA) signaling regulates atrial versus ventricular specification during the cardiac differentiation of hESCs. When the RA receptor antagonist BMS-189453 was added to cultures, 83% of the cardiomyocytes showed ventricular-like features, whereas 94% of the cells displayed atrial-like phenotypes when RA was applied (Table 3).76 Overall, these findings highlight the potential of small-molecule-based approaches in directed differentiation of PSCs into specific cardiac subtypes. To efficiently and selectively generate cardiomyocytes of high qualities, it is necessary to further elucidate the mechanisms of cardiac subtype specification and to identify additional chemicals that further improve these methods.
Improving Cardiac Grafts
Besides the production of sufficient amounts of cardiomyocytes of high quality, the integration of transplanted cells into tissues imposes a challenge of no less significance. Several encouraging studies have engrafted and integrated hESC-derived cardiomyocytes into rodent and pig hearts.77, 78 Nevertheless, most cardiomyocytes were lost shortly after transplantation. A large proportion of the cells remaining in the infarcted myocardium also underwent programmed cell death soon afterwards.79, 80 The poor survival of transplanted cells represents a major obstacle for the delivery of long-term value of PSC-derived cardiomyocytes to regenerative therapy.
Attempts with small-molecule approaches to improve graft survival of cardiomyocytes have been conducted. Survival improved in transplanted cells pre-treated with diazoxide, a drug that opens mitochondrial ATP-dependent potassium channels (mitoKATP) in a myocardial infarction (MI) model (Table 3).81 A small molecule pioglitazone, an activator of PPAR-γ signaling, significantly enhanced the viability of transplanted mesenchymal stem cell-derived cardiomyocytes in experimental animals (Table 3).82 Moreover, small-molecule inhibitors of the Rho-associated kinase83 and p38 MAPK84 improve the survival rate of cells before and/or after transplantation.
Besides small molecules, growth factors such as IGF1,85 TGFβ2,86 and erythropoietin87 were also brought into play to protect transplanted cells. Laflamme et al. designed a ‘pro-survival cocktail’ consisting of Matrigel, IGF1, a Bcl XL peptide (to block mitochondrial death pathways), pinacidil (to open mitoKATP), peptide cyclosporin A (to attenuate cyclophilin D–dependent mitochondrial pathways) and the caspase inhibitor ZVAD-fmk (Table 3).77 Employing this combination of multiple pro-survival factors, 7-fold increase in graft size in a rat MI model was achieved.78 Future efforts to further enhance graft survival will most likely involve searching for novel combinations of small molecules and pro-survival factors as well as other strategies, such as pre-conditioning, immunosuppressing, and bioengineering.
IN SITU HEART REPAIR AND REGENERATION
While addressing challenges that cell transplantation therapy is currently faced with, researchers have sought to develop new strategies for in situ heart repair and regeneration. The advantages and drawbacks of a variety of therapeutic strategies for cardiac regenerative medicine are listed in Table 4. Progress in emerging, promising strategies in this direction will be the focus of the following sections. Potential applications of small-molecule approaches in these strategies will be tentatively suggested.
Table 4.
Comparison between PSC-based Cardiac Cell Therapy and in situ Heart Regeneration
| Issues | PSC-based Cardiac Cell Therapy | in situ Heart Regeneration |
|---|---|---|
| Therapeutic mechanisms | Replacing the damaged myocardium through transplantation of in vitro generated cardiac cells into the heart | Modulating the heart’s own regenerative response by simulating or reprogramming endogenous cells |
| Cell sources | Theoretically unlimited amounts. Well-controlled cell type and quality. | Cell type, quality and amounts typically restricted and context-dependent. |
| In vitro bioengineering | Applicable | Not applicable |
| Cellular maturation | Fetal or neonatal cardiomyocytes-like features | Often adult cardiomyocyte-like features |
| Risk of tumor formation | Possible due to residue pluripotent cells | Possible due to modifying host genome by transgenes and uncontrollable transgene expression |
| Risk of immune rejection | Possible but ameliorable with iPS technology | Unlikely |
| Risk of arrhythmias | Possible due to autorhythmicity, immaturity and inorganization of graft cells | Possible due to potentially unpredictable and incomplete reprogramming |
| Graft survival and host-graft integration | Challenging | Not necessary |
| Ease of implementation | Low | Relatively high |
| Cost | Relatively high | Relatively low |
Cardiomyocyte Dedifferentiation and Proliferation
During mammalian embryonic development, the heart grows through the proliferation of cardiomyocytes but switches to hypertrophic growth soon after birth. As cardiomyocytes exit the cell cycle at this point, further increases in cardiac mass are mainly due to the increase in cardiomyocyte size instead of number.88 In contrast to hearts of some lower organism, such as zebrafish, which have a robust regenerative response upon injury mainly through cardiomyocytes dedifferentiation and proliferation, an adult mammalian heart typically has extremely limited renewal capacity and is incapable of restoring the damaged myocardium after injury. 89 Nonetheless, mammalian cardiomyocytes can slowly self-renew and turnover under physiological condition,90, 91 presumably through the division of pre-existing cardiomyocytes instead of differentiation of residue progenitor cells,92 even although this multiplication capacity is clearly not sufficient to repair a damaged heart. Genetic93, 94 and pharmacological95 strategies to enhance the intrinsic renewal capacity have improved cardiac function after infarction. With the rapid progress in stem cell biology and high-content screening platform, systematic screenings have been performed to identify chemicals inducing cardiomyocyte proliferation.96 Future efforts to continually identify novel small molecules that robustly induce dedifferentiation and proliferation of cardiomyocytes will be of great value to fulfill the potential of this regenerative strategy.
Direct Cardiac Conversion of Non-Myocytes
The establishment and advances in iPSC technology have re-galvanized research on direct reprogramming somatic cells from one lineage into another without entering the pluripotent stage, a process conventionally known as transdifferentiation.14 Transdifferentiation of endogenous or explanted fibroblasts represents a fascinating, novel regenerative approach. Fibroblasts account for up to 50% of all cells in an adult human heart. After cardiac injury, fibroblasts are hyper-proliferated and lead to fibrosis and scar formation within the damaged area. 97 Properly reprogrammed into cardiomyocytes, the ample fibroblasts can serve as an attractive cell source to replenish the myocardial muscle. Proof-of-principle demonstration of the successful cardiac reprogramming of fibroblasts has been achieved by ectopic induction of multiple cardiac-enriched transcription factors (Gata4, Mef2c and Tbx5). 98 On the other hand, Efe et al. established the cell-activation and signaling-directed (CASD) cardiac reprogramming, which involves transient expression of the Yamanaka factors in conjunction with cardiogenic signal simulation.99 The success of CASD lineage conversion reveals a common paradigm for both transdifferentiation and reprogramming towards pluripotency. Cardiogenic transdifferentiation was also accomplished by transfection of cardiac fibroblasts with microRNAs (miR-1, miR-133, miR-208 and miR-499).100
Impressively, in vivo delivery of reprogramming factors into infarcted mouse hearts regenerated the post-infarcted, damaged myocardial muscle in situ by converting resident cardiac fibroblasts into cardiomyocytes.100, 101, 102 The induced cardiomyocytes generated in their native environment displayed mature, adult-like phenotype and improved heart function.101, 102 Evidence of electrical coupling to the host myocardial tissue was also observed.101 Despite the encouraging results observed, several safety concerns need to be addressed, including the viral delivery of transgenes and partially reprogrammed cells that potentially disturb the cardiac rhythm. Small molecules that can avoid the usage of transcription factors and/or enhance the in situ transdifferentiation will have tremendous impact on the successful translation of this attractive strategy from bench to beside.
Activation of Endogenous Cardiac Progenitor Cells
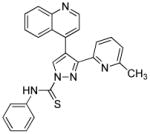
The existence of resident cardiac progenitor cells (CPCs) in the adult rodent and human heart has been well demonstrated over the last decade.103 Although many efforts have been made on the identification, in vitro expansion and subsequent differentiation of these cells, little is known about their roles and behaviors within the naïve heart niche under physiological and pathological conditions. In view of their well-characterized cardiogenesis potential both in vitro and in vivo,103 it is reasonable to envisage a CPC-based therapy that allows the proper mobilization of resident CPCs to replace the lost or damaged cells in situ, avoiding the problems of limited graft survival, restricted integration to the host tissue, and potential immune rejection. The feasibility of this approach has been established in recent years. Using genetic fate mapping, Loffredo et al. demonstrated that exogenously delivered bone marrow–derived cells could stimulate resident CPCs and promote the endogenous cardiomyocyte refreshment.104 Remarkably, Smart et al. described that thymosin β4, a known pro-angiogenic peptide, mobilized an epicardial origin of progenitor population, induced concomitant cardiac differentiation and regeneration of myocardial tissue, and ultimately improved heart function post-infarction.105 Similarly, Zangi et al. found that intramyocardial injection of synthetic modified RNA encoding human VEGF-A resulted in the expansion and directed differentiation of endogenous CPCs, and markedly improved heart function in a mouse MI model.106 An attempt to target CPCs using small molecules has recently been reported.107 Russell et al. found that a 3,5-disubstituted isoxazole, Isx1, could activate cardiac genes expression in residential, multipotent Notch-activated epicardium-derived cells in vivo and induced the generation of CPCs (Table 3).107 Unfortunately, MI abrogated Isx1’s cardiogenic effects and led to fibrosis.107 Nonetheless, the possibilities to develop novel small-molecule tools to achieve safe, robust and cost-effective activation of CPCs for heart regeneration are undoubtedly alluring.
CONCLUSIONS AND OUTLOOK
The discovery and improvement of iPSC technology, as well as the development of efficient cardiac differentiation system offer tremendous hope for novel cell replacement therapies to improve cardiac function in compromised individuals. The eventual success of complete small-molecule-based reprogramming through activation of endogenous expressions of genes enabling pluripotency will greatly propel the realization of the clinical potentials of iPSCs.
Multiple concerns still hinder the applications of cardiac cell transplantation therapy, including insufficient quantities and qualities of cardiomyocytes, ineffective delivery and retention, acute graft death and rejection. The tremendous potentials of small molecules to address these issues have been well recognized. Since a broad spectrum of small molecules have been identified that can replace factors during iPSC generation and facilitate the transition of partially reprogrammed cells into ground state pluripotency,41 it may be possible to eliminate the risk associated with editing of host genome by viral genes, increase the overall efficiency of cardiac reprogramming, and improve the functional integrity of induced cardiomyocytes using pharmacological approaches. The in vitro generated cardiomyocytes might be further engineered pharmacologically and serve as suitable materials for direct transplantation. Methods for temporal- and spatial-controllable in vivo delivery of small molecule must be developed to achieve their therapy values. Hopefully, drugs that facilitate the transplantation can be developed based on small molecules that improve the survivals and functions of transplanted tissues.
In contrast to the significant achievement of small molecules approaches made on modulating cardiac cell fate and function, their potential in in situ heart repair is yet to be explored. The possibilities to convert resident non-cardiomyocytes into myocardium, activate and/or enhance the intrinsic regenerative capacity of cardiac cells by pharmacologic means will provide alternative, fascinating options for regenerative therapy. Towards these ultimate goals, high-throughput screening will continue to serve as a powerful strategy to discover more novel chemicals with desired properties. Applications of combinations of small molecules to garner their synergistic effects have already proven advantageous. Efforts will be continuously made to search for the optimal cocktails for specific therapeutic purposes. Last but not least, better understanding of these cell reprogramming and developmental processes will ultimately benefit stem cell biology as well as regenerative therapy.
Acknowledgments
Sheng Ding is supported by funding from National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, and National Eye Institute/National Institute of Health, California Institute for Regenerative Medicine, and the Gladstone Institutes. We thank Gary Howard for critical reading and editing of this manuscript. The authors apologize to all scientists whose research could not be properly discussed and cited in this review owing to space limitations.
KEY WORDS
- Small Molecule
a defined chemical entity, often an organic compound with a molecular weight smaller than 900 Daltons
- PSC
pluripotent stem cell, a stem cell possessing the potential to differentiate into all cell types in the body
- Cell Reprogramming
the artificial conversion of one particular cell state and/or fate into another, often referring to the generation of stem cells from more differentiated cells
- TF
transcription factor, a protein, either on its own or in complex with other proteins, which binds to specific DNA sequences and thereby controls the transcription of genes
- Signaling Pathway
the relaying of signals among a group of molecules which ultimately triggers cellular responses
- Differentiation
a cellular process in which stem cells become more specialized cell types
- Transdifferentiation
the conversion of one type of somatic cells into another cell type without passing through the pluripotent state
- Cardiac Regeneration
a process to replenish lost myocardial tissues and restore cardiac function in post-injured hearts
Footnotes
The authors declare no competing financial interest.
References
- 1.Harvey PA, Leinwand LA. The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol. 2011;194:355–365. doi: 10.1083/jcb.201101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–329. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 3.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka S. Strategies and new developments in the generation of patient specific pluripotent stem cells. Cell Stem Cells. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: from genome association to function. Cell Stem Cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 10.Engle SJ, Puppala D. Integrating human pluripotent stem cells into drug development. Cell Stem Cell. 2013;12:669–677. doi: 10.1016/j.stem.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Hong SG, Dunbar CE, Winkler T. Assessing the Risks of Genotoxicity in the Therapeutic Development of Induced Pluripotent Stem Cells. Mol Ther. 2013;2:272–281. doi: 10.1038/mt.2012.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frye SV. The art of the chemical probe. Nat Chem Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Li K, Wei W, Ding S. Chemical approaches to stem cell biology and therapeutics. Cell Stem Cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nie B, Wang H, Laurent T, Ding S. Cellular reprogramming: a small molecule perspective. Curr Opin Cell Biol. 2012;24:784–792. doi: 10.1016/j.ceb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyssiotis CA, Lairson LL, Boitano AE, Wurdak H, Zhu S, Schultz PG. Chemical control of stem cell fate and developmental potential. Angew Chem Int Ed Engl. 2011;50:200–242. doi: 10.1002/anie.201004284. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 18.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Bio. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 21.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 22.Kretsovali A, Hadjimichael C, Charmpilas N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells Int. 2012;2012:184154. doi: 10.1155/2012/184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nature Biotechnology. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 27.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nature Biotechnology. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 29.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Do JT, Desponts C, Hahm HS, Schöler HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 34.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, Ding S. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of TGF-β signaling replaces Sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maherali N, Hochedlinger K. Tgfβ Signal Inhibition Cooperates in the Induction of iPSCs and Replaces Sox2 and cMyc. Curr Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan X, Wan H, Zhao X, Zhu S, Zhou Q, Ding S. Brief report: Combined chemical treatment enables oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells. 2011;29:549–553. doi: 10.1002/stem.594. [DOI] [PubMed] [Google Scholar]
- 38.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Zhou H, Abujarour R, Zhu S, Young JJ, Lin T, Hao E, Schöler HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, Bollong M, Kunick C, Brinker A, Cho CY, Schultz PG, Jaenisch R. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci USA. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of Human Primary Somatic Cells by OCT4 and Chemical Compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, Wu Y, Li H, Liu K, Wu C, Song Z, Zhao Y, Shi Y, Deng H. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21:196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 46.Abbasalizadeh S, Baharvand H. Technological progress and challenges towards cGMP manufacturing of human pluripotent stem cells based therapeutic products for allogeneic and autologous cell therapies. Biotechnol Adv. 2013;31 doi: 10.1016/j.biotechadv.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Amit M, Margulets V, Segev H, Shariki K, Laevsky I, Coleman R, Itskovitz-Eldor J. Human feeder layers for human embryonic stem cells. Biol Reprod. 2003;68:2150–2156. doi: 10.1095/biolreprod.102.012583. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, Llanas RA, Thomson JA. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 49.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Khillan JS. Promotion of feeder-independent self-renewal of embryonic stem cells by retinol (vitamin A) Stem Cells. 2008;26:1858–1864. doi: 10.1634/stemcells.2008-0050. [DOI] [PubMed] [Google Scholar]
- 52.Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, Gepstein L. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–1523. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, Lee RT. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 58.Cao N, Liu Z, Chen Z, Wang J, Chen T, Zhao X, Ma Y, Qin L, Kang J, Wei B, Wang L, Jin Y, Yang HT. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012;22:219–236. doi: 10.1038/cr.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu X, Ding S, Ding Q, Gray NS, Schultz PG. Small molecules that induce cardiomyogenesis in embryonic stem cells. J Am Chem Soc. 2004;126:590–591. doi: 10.1021/ja038950i. [DOI] [PubMed] [Google Scholar]
- 60.Wei ZL, Petukhov PA, Bizik F, Teixeira JC, Mercola M, Volpe EA, Glazer RI, Willson TM, Kozikowski AP. Isoxazolyl-serine-based agonists of peroxisome proliferator-activated receptor: design, synthesis, and effects on cardiomyocyte differentiation. J Am Chem Soc. 2004;126:16714–16715. doi: 10.1021/ja046386l. [DOI] [PubMed] [Google Scholar]
- 61.Sachinidis A, Schwengberg S, Hippler-Altenburg R, Mariappan D, Kamisetti N, Seelig B, Berkessel A, Hescheler J. Identification of small signalling molecules promoting cardiac-specific differentiation of mouse embryonic stem cells. Cell Physiol Biochem. 2006;18:303–314. doi: 10.1159/000097608. [DOI] [PubMed] [Google Scholar]
- 62.Graichen R, Xu X, Braam SR, Balakrishnan T, Norfiza S, Sieh S, Soo SY, Tham SC, Mummery C, Colman A, Zweigerdt R, Davidson BP. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 63.Sadek H, Hannack B, Choe E, Wang J, Latif S, Garry MG, Garry DJ, Longgood J, Frantz DE, Olson EN, Hsieh J, Schneider JW. Cardiogenic small molecules that enhance myocardial repair by stem cells. Proc Natl Acad Sci USA. 2008;105:6063–6068. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berkessel A, Seelig B, Schwengberg S, Hescheler J, Sachinidis A. Chemically induced cardiomyogenesis of mouse embryonic stem cells. ChemBioChem. 2010;11:208–217. doi: 10.1002/cbic.200900345. [DOI] [PubMed] [Google Scholar]
- 65.Hao J, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu J, Peterson RT, Hatzopoulos AK, Hong CC. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Willems E, Cabral-Teixeira J, Schade D, Cai W, Reeves P, Bushway PJ, Lanier M, Walsh C, Kirchhausen T, Izpisua Belmonte JC, Cashman J, Mercola M. Small Molecule-Mediated TGF-β Type II Receptor Degradation Promotes Cardiomyogenesis in Embryonic Stem Cells. Cell Stem Cell. 2012;11:242–252. doi: 10.1016/j.stem.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci USA. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, Cashman J, Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Hao J, Hong CC. Cardiac induction of embryonic stem cells by a small molecule inhibitor of Wnt/betacatenin signaling. ACS Chem Biol. 2011;6:192–197. doi: 10.1021/cb100323z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gonzalez R, Lee JW, Schultz PG. Stepwise Chemically Induced Cardiomyocyte Specification of Human Embryonic Stem Cells. Angew Chem Int Ed Engl. 2011;50:11181–11185. doi: 10.1002/anie.201103909. [DOI] [PubMed] [Google Scholar]
- 72.Lian X, Hsiao S, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval K, Zhang J, Kamp TJ, Palecek SP. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minami I, Yamada K, Otsuji TG, Yamamoto T, Shen Y, Otsuka S, Kadota S, Morone N, Barve M, Asai Y, Tenkova-Heuser T, Heuser JE, Uesugi M, Aiba K, Nakatsuji N. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012;2:1448–1160. doi: 10.1016/j.celrep.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 74.Kleger A, Seufferlein T, Malan D, Tischendorf M, Storch A, Wolheim A, Latz S, Protze S, Porzner M, Proepper C, Brunner C, Katz SF, Pusapati GV, Bullinger L, Franz WM, Koehntop R, Giehl K, Spyrantis A, Wittekindt O, Lin Q, Zenke M, Fleischmann BK, Wartenberg M, Wobus AM, Boeckers TM, Liebau S. Modulation of calcium-activated potassium channels induces cardiogenesis of pluripotent stem cells and enrichment of pacemaker-like cells. Circulation. 2010;122:1823–1836. doi: 10.1161/CIRCULATIONAHA.110.971721. [DOI] [PubMed] [Google Scholar]
- 75.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res. 2010;107:776–786. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, Xu Y, Cao H, Meng Q, Chen L, Tian T, Wang X, Li P, Hescheler J, Ji G, Ma Y. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 78.Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, Van Biber B, Dardas T, Mignone JL, Izawa A, Hanna R, Viswanathan M, Gold JD, Kotlikoff MI, Sarvazyan N, Kay MW, Murry CE, Laflamme MA. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang M, Methot D, Poppa V, Fujio Y, Walsh K, Murry CE. Cardiomyocyte grafting for cardiac repair: graft cell death and anti-death strategies. J Mol Cell Cardiol. 2001;33:907–921. doi: 10.1006/jmcc.2001.1367. [DOI] [PubMed] [Google Scholar]
- 80.van Laake LW, Passier R, Monshouwer-Kloots J, Verkleij AJ, Lips DJ, Freund C, den Ouden K, Ward-van Oostwaard D, Korving J, Tertoolen LG, van Echteld CJ, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res. 2007;1:9–24. doi: 10.1016/j.scr.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 81.Niagara MI, Haider HK, Jiang S, Ashraf M. Pharmacologically preconditioned skeletal myoblasts are resistant to oxidative stress and promote angiomyogenesis via release of paracrine factors in the infarcted heart. Circ Res. 2007;100:545–555. doi: 10.1161/01.RES.0000258460.41160.ef. [DOI] [PubMed] [Google Scholar]
- 82.Shinmura D, Togashi I, Miyoshi S, Nishiyama N, Hida N, Tsuji H, Tsuruta H, Segawa K, Tsukada Y, Ogawa S, Umezawa A. Pretreatment of human mesenchymal stem cells with pioglitazone improved efficiency of cardiomyogenic transdifferentiation and cardiac function. Stem Cells. 2011;29:357–366. doi: 10.1002/stem.574. [DOI] [PubMed] [Google Scholar]
- 83.Braam SR, Nauw R, Ward-van Oostwaard D, Mummery C, Passier R. Inhibition of ROCK improves survival of human embryonic stem cell-derived cardiomyocytes after dissociation. Ann N Y Acad Sci. 2010;1188:52–67. doi: 10.1111/j.1749-6632.2009.05083.x. [DOI] [PubMed] [Google Scholar]
- 84.Yeghiazarians Y, Gaur M, Zhang Y, Sievers RE, Ritner C, Prasad M, Boyle A, Bernstein HS. Myocardial improvement with human embryonic stem cell-derived cardiomyocytes enriched by p38MAPK inhibition. Cytotherapy. 2012;14:223–231. doi: 10.3109/14653249.2011.623690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kutschka I, Kofidis T, Chen IY, von Degenfeld G, Zwierzchoniewska M, Hoyt G, Arai T, Lebl DR, Hendry SL, Sheikh AY, Cooke DT, Connolly A, Blau HM, Gambhir SS, Robbins RC. Adenoviral human BCL-2 transgene expression attenuates early donor cell death after cardiomyoblast transplantation into ischemic rat hearts. Circulation. 2006;114:I174–I180. doi: 10.1161/CIRCULATIONAHA.105.001370. [DOI] [PubMed] [Google Scholar]
- 86.Singla DK, Singla RD, Lamm S, Glass C. TGF-β2 treatment enhances cytoprotective factors released from embryonic stem cells and inhibits apoptosis in infarcted myocardium. Am J Physiol Heart Circ Physiol. 2011;300:H1442–H1450. doi: 10.1152/ajpheart.00917.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Robey TE, Saiget MK, Reinecke H, Murry CE. Systems approaches to preventing transplanted cell death in cardiac repair. J Mol Cell Cardiol. 2008;45:567–581. doi: 10.1016/j.yjmcc.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat Rev Mol Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, Ogórek B, Ferreira-Martins J, Goichberg P, Rondon-Clavo C, Sanada F, D’Amario D, Rota M, Del Monte F, Orlic D, Tisdale J, Leri A, Anversa P. Cardiomyogenesis in the adult human heart. Circ Res. 2010;107:305–315. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 94.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 95.Engel FB, Hsieh PC, Lee RT, Keating MT. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci USA. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uosaki H, Magadum A, Seo K, Fukushima H, Takeuchi A, Nakagawa Y, Moyes KW, Narazaki G, Kuwahara K, Laflamme M, Matsuoka S, Nakatsuji N, Nakao K, Kwon C, Kass DA, Engel FB, Yamashita JK. Identification of Chemicals Inducing Cardiomyocyte Proliferation in Developmental Stage-Specific Manner with Pluripotent Stem Cells. Circ Cardiovasc Genet. 2013;6:624–633. doi: 10.1161/CIRCGENETICS.113.000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. 2006;291:H1015–H1026. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- 98.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nature Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 100.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sturzu AC, Wu SM. Developmental and regenerative biology of multipotent cardiovascular progenitor cells. Circ Res. 2011;108:353–364. doi: 10.1161/CIRCRESAHA.110.227066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8:389–398. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smart N, Bollini S, Dubé KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Später D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT, Chien KR. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Russell JL, Goetsch SC, Aguilar HR, Frantz DE, Schneider JW. Targeting native adult heart progenitors with cardiogenic small molecules. ACS Chem Biol. 2013;7:1067–1076. doi: 10.1021/cb200525q. [DOI] [PMC free article] [PubMed] [Google Scholar]


