Abstract
The mammalian genome harbors thousands of long noncoding RNA (lncRNA) genes. Recent studies have indicated the involvement of several of these lncRNAs in the regulation of gene expression. LncRNAs play crucial roles in various biological processes ranging from epigenetic gene regulation, transcriptional control to post-transcriptional regulation. LncRNAs are localized in various subcellular compartments and major proportion of these are retained in the cell nucleus; and could be broadly classified as nuclear-retained lncRNAs (nrRNAs). Based on the identified functions, members of the nrRNAs execute diverse roles, including providing architectural support to the hierarchical subnuclear organization and influencing the recruitment of chromatin modifier factors to specific chromatin sites. In this review we will summarize the recently described roles of mammalian nrRNAs in controlling gene expression by influencing chromatin organization, transcription, pre-mRNA processing, nuclear organization and their involvement in disease.
Keywords: long noncoding RNA, nuclear-retained ncRNA, pre-mRNA splicing, MALAT1, nuclear domains, cell cycle, chromatin remodelling
Introduction
The human genome contains less than 2% protein coding sequences, which translate into the requisite number of proteins involved in the myriad of biological functions. The rest of the genomic sequences, till recently were considered as noncoding or “junk”. However, the development of high-throughput sequencing technology has led to the understanding that the majority of the human genome is transcribed to produce non protein coding RNAs (ncRNAs) and constitute the diverse and complex cellular transcriptome (Clark et al., 2011; Kapranov et al., 2007; Wilusz et al., 2009). Interestingly, the complexity of the nuclear transcriptome is fivefold higher compared to the cytosolic transcriptome in terms of transcribed base pairs (Cheng et al., 2005). The ncRNA is classified into broader classes depending upon the length of the RNA sequences. NcRNAs with length > 200 nucleotides are considered as long noncoding RNAs (lncRNAs) whereas the rest of the smaller transcripts are classified as small ncRNAs (Mercer and Mattick, 2013; Wilusz et al., 2009). According to the current lncRNA catalogs, the human genome encodes 10,000–15,000 lncRNAs but they generally display cell type specific expression with only a particular number of them expressed in a specific cell type or specific cell cycle stage. In general, lncRNAs are usually expressed at lower levels, compared to protein coding transcripts (Derrien et al., 2012; Sone et al., 2007; Tripathi et al., 2010).
Due to the complex and diverse nature of the transcripts, lncRNAs are classified into different groups. Depending upon the origin of transcripts with respect to the protein coding genes that are adjacent/overlapping with these transcripts, lncRNAs may be placed in one or more groups, i.e.,sense (noncoding transcript on the same strand) , antisense (noncoding transcript on the opposite strand), bidirectional (transcription of lncRNA gene, and a adjacent protein coding gene on the opposite strand in close genomic proximity), intronic (noncoding transcript generated from the intronic sequences of another transcript) and intergenic (noncoding transcript generated from the genomic region between two protein coding genes) (Carninci et al., 2005; Denoeud et al., 2007; Horiuchi and Aigaki, 2006; Mercer et al., 2009). Retrotransposons and pseudogenes are thought to be involved in the generation and diversification of lncRNAs (Kapusta et al., 2013; Singh and Rath, 2012). LncRNAs are also classified on the basis of their functions, i.e., cis-acting (Xist, Tsix, RepA, Kcnq1ot1, Hottip, ANRIL, Evf2, oct4-ps5) and trans-acting (pRNA, asOct4-ps5), structural scaffold (NEAT1,TUG1), enhancer (eRNAs, Xite, ncRNA-a), co-activator or co-repressor (SINE B2, SRA, Jpx, pRNA) and decoy (Tsix, PTEN-ps) (Khalil et al., 2009; Kim et al., 2010; Koziol and Rinn, 2010; Mohammad et al., 2009; Orom et al., 2010; Spitale et al., 2011; Wang et al., 2011a) lncRNAs.
The majority of the lncRNAs are transcribed by RNA polymerase II (Guttman et al., 2009; White, 2011). Chromatin of a transcriptionally active lncRNA locus displays marks of histone H3 tri-methylated at lysines 4 (H3K4me3) and 36 (H3K36me3) in their regulatory elements and transcription bodies, respectively, imparting the K4-K36 domain-specific chromatin signature. Several of the transcription factors, those that are involved in the transcription of protein-coding genes, are also shown to be responsible for the transcription of lncRNAs (Kim et al., 2005). The cellular expression of lncRNAs is very much cell type- and developmental stage-specific. Several recent studies indicate that the majority of lncRNAs remain associated with distinct subnuclear structures, including specific chromatin regions. Both the cell type specificity and the distinct localization pattern of lncRNAs suggest that their expression is highly regulated, and they control very specific cellular function(s).
The nucleus, one of the important organelles in a eukaryotic cell, harbours the majority of the hereditable genome (Tripathi and Prasanth, 2011). The nucleus is a very complex, dynamic, but highly organized organelle. It is sub-divided into a number of different nuclear zones/subnuclear organelles, characterized by the absence of a membrane boundary around them and also by the presence of a unique set of proteins and/or RNAs within them. Some of the better defined subnuclear domains include nucleoli, perinuclear compartment (PNC), PML (promyelocytic leukemia) bodies, nuclear speckles, paraspeckles and Cajal bodies (Mao et al., 2011). These subnuclear structures are formed and maintained as stochastic self-organization domains, and are actively involved in distinct cellular function(s) (Dundr and Misteli, 2010), including transcription, DNA replication, RNA metabolism and RNA export (Zhao et al., 2009). Apart from proteins, various lncRNAs localized in the subnuclear domains offer another layer of regulatory mechanism in the nucleus (Mattick et al., 2009; Mercer et al., 2009). These so called nuclear-retained regulatory RNAs (nrRNAs) are shown to be involved in key cellular processes associated with gene expression, including but not limited to epigenetic regulation, chromosomal interaction, transcriptional regulation, RNA processing and nuclear domain structure maintenance. In this review, we discuss the recent findings relating to nrRNAs, their involvement in various cellular processes during development and also their association with diseases.
Chromatin remodelling and epigenetic regulation
It has been previously reported that RNAs are involved in the maintenance of higher order chromatin structure, and most of these RNAs are noncoding, derived predominantly from intronic and intergenic regions of the genome (Bernstein and Allis, 2005). This notion is further supported by the recent data from various research laboratories, demonstrating the ability of nrRNAs to specifically interact with proteins that are involved in chromatin maintenance, modification and gene expression regulation (Batista and Chang, 2013; Chu et al., 2011; Guttman and Rinn, 2012; Mondal et al., 2010). Moreover, by associating with specific chromatin modifying complexes, nrRNAs govern the localization of these modifiers to specific chromatin sites, thereby regulating chromatin modifications and gene expression in an allele- or cell type-specific manner (See Figure 1)(Lai et al., 2013).
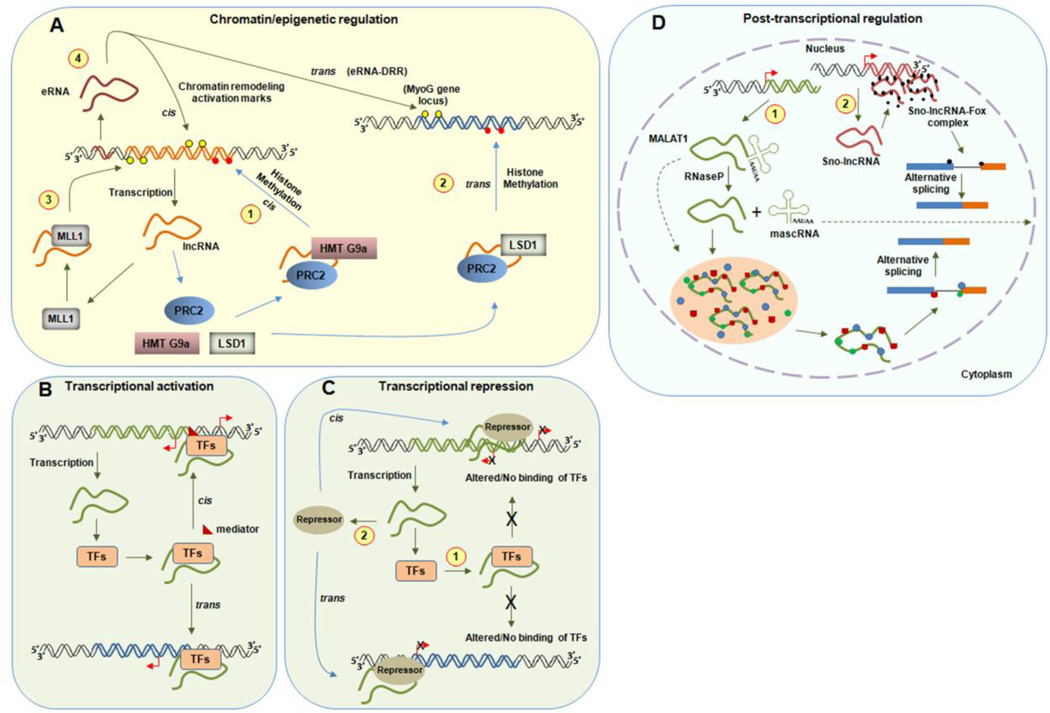
Figure 1. Functions of nuclear retained lncRNAs.
(A) Model showing chromatin/epigenetic regulation by lncRNA. LncRNA interacts with different chromatin modifying complexes and recruits them to specific chromatin target site. (1) shows the deposition of repressive transcription marks (red circles) by histone methylation in cis (Air, Kcnq1ot1, ANRASSF1) and (2) in trans (HOTAIR) leading to transcriptional inactivation. (3) represents the activation of transcription (yellow circle) by deposition of active chromatin marks through the recruitment of specific histone modifiers (Mistral and HOTTIP). (4) eRNAs also enhances the transcription both in cis and trans by chromatin modifications.(B) LncRNAs activate transcription both in cis and trans by recruiting transcription factors (TFs) and mediators. (C) LncRNA represses transcription (shown by cross or ‘x’ at the transcription start site) by (1) inhibiting/altering the binding of transcription factors at the initiation complex and also by (2) recruiting repressors at the site of transcription. (D) Post-transcriptional regulation by lncRNAs. (1) MALAT1 is cleaved by RNaseP to generate a longer MALAT1 lncRNA and a 61 nucleotide mascRNA, which is transported to the cytoplasm; dotted arrow). MALAT1 binds to pre mRNA splicing factors (shown in red, blue and green) in the nuclear speckles and regulates their distribution and splicing activity. (2) Sno-lncRNA interacts with FOX proteins (black circles) and could participate in the FOX-mediated alternative splicing.
X-chromosome inactivation (XCI) in mammals is an example that signifies the distinct role of lncRNAs/nrRNAs in chromatin remodelling, epigenetic modifications and gene repression (Lee, 2011). Among several lncRNAs present in the X-inactivation center (Xic), Xist (X-inactive-specific transcript) was the first-identified and is one of the best-studied nuclear-restricted lncRNAs in mammals [Please see (Augui et al., 2011; Lee and Bartolomei, 2013; Lessing et al., 2013) for an extensive review on X-chromosome inactivation]. Xist is a 17–20 kb long nrRNA expressed from the future inactive X-chromosome (Xi) that spreads all over the surface of Xi in cis (Figure 2A) (Lessing et al., 2013). Repeat A (RepA), another lncRNA, is a conserved direct sequence repeat present at the 5’ end of Xist gene that interacts with the PRC2 (Polycomb repressive complex 2) (Zhao et al., 2008). PRC2 is held responsible for the tri-methylation of histone H3 at Lys27 (H3K27me3), a transcriptionally repressive mark commonly associated with several of the gene loci, the expression of which are regulated in a cell type- or developmental-specific fashion. The initial binding of Xist with PRC2 is mediated by a 1.6 kb long RepA lncRNA showing homology with the 5’ end of the Xist, and further the allele-specific binding of Xist RNA to Xi is facilitated by the YY1 transcription factor (Jeon and Lee, 2011; Zhao et al., 2008). Other lncRNAs from the Xic, including Jpx and Tsix regulate XCI by influencing Xist expression (Lee et al., 1999; Tian et al., 2010). Jpx is transcribed from the Xi chromosome and acts as a positive regulator to Xist RNA. A recent study revealed that Jpx by binding to CTCF, a factor that is known to repress Xist transcription, evicts the binding of CTCF from the promoter of one of the Xist gene alleles, thereby positively influencing Xist gene expression (Sun et al., 2013). On the other hand, Tsix is an antisense lncRNA to Xist RNA, transcribed from the future active X-chromosome and it negatively regulates the expression of Xist RNA. Tsix represses Xist transcription on one of the X-alleles, and thus helps in determining the X-chromosome allele that needs to be activated (Xa). Essentially, Tsix regulates Xist transcription at multiple levels; it controls X-chromosome pairing to provide asymmetry in the epigenetic modifications between the two X-chromosomes. Tsix recruits DNA methyltransferase (Dnmt3a) to inhibit Xist transcription. Finally, Tsix directly binds PRC2 and also duplexes with Xist-RepA RNA, and thus blocks the PRC2 recruitment to Xist by RepA (Do et al., 2008; Lee, 2012).
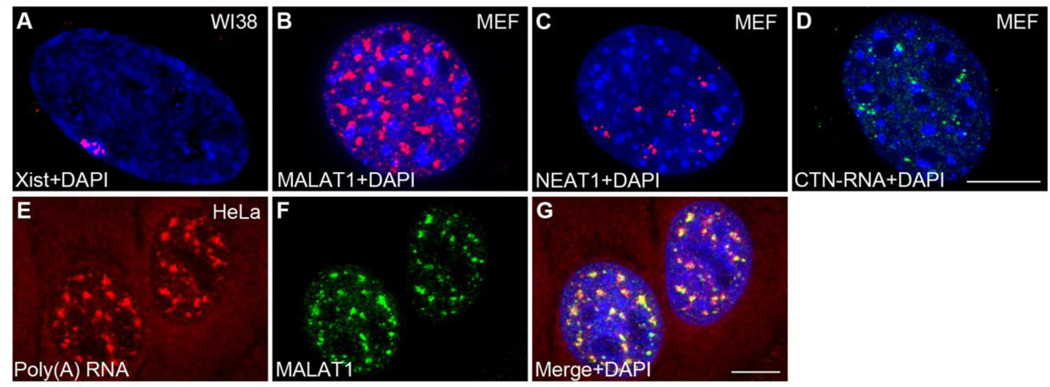
Figure 2. Nuclear-retained regulatory RNAs.
(A) RNA-FISH using probe against the XIST RNA (red) reveal the localization of XIST RNA in the inactive X-chromosome. (B) RNA-FISH analysis reveal nuclear speckle localization of MALAT1 RNA (red) and (C) Paraspeckle localization of NEAT1 RNA (red) and (D) CTN-RNA (green). (E–G) Dual RNA-FISH analyses demonstrate the enrichment of poly adenylated RNA (E, red) and MALAT1 RNA (F, green) in the nuclear speckles. Total Poly (A) RNA is detected using fluorescently-labeled oligo (dT) probes. DNA is counterstained with DAPI (Blue). Bar 5µm.
Other examples of nrRNAs involved in the allele specific epigenetic regulation of gene expression in cis include Air and Kcnq1ot1 (Pandey et al., 2008; Sleutels et al., 2002). Air lncRNA is imprinted; expressed from the paternal allele, and is transcribed from the second intron of mouse insulin-like growth factor 2 receptor (Igf2r) gene. Air transcripts interact with the histone methyltransferases (HMT) G9a and recruit them to the promoters of specific genes (Slc22a3, Slc22a2 and Igf2r) that need to be silenced (Nagano et al., 2008). Kcnq1ot1 is a 90 kb long lncRNA, transcribed from intron 10 of the imprinted gene Kcnq1 from paternal allele (Kanduri, 2011). Kcnq1ot1 recruits PRC2 and HMT G9a to the gene-cluster present in the Kcnq1 domain in an allele-specific manner and creates repressive histone marks i.e., histone H3 tri-methylated at lysine 9 (H3K9me3) and H3K27me3, leading to the silencing of the genes in cis (Terranova et al., 2008).
Very recently, another intronic cis-acting lncRNA ANRASSF1 has been characterized, which is transcribed from the opposite strand of the RASSF1 tumor suppressor gene locus (Beckedorff et al., 2013). ANRASSF1 specifically localizes within the nucleus and has a shorter life-span (half-life of ~50 min) compared to other well characterized cis-acting stable lncRNAs, including Kcnq1ot1 and Air lncRNA. It has been observed that the ANRASSF1 specifically silences the expression of RASSF1A by recruiting PRC2 and repressive histone modifying complexes, but has no effect on the RASSF1C and other neighbouring genes.These studies indicate that several of the lncRNAs have the potential to influence the epigenetic environment of genes in a location-specific manner (Beckedorff et al., 2013).
Besides acting in cis manner, many nrRNAs also dictate epigenetic modifications and control gene expression in trans. For instance, HOTAIR (HOX transcript antisense RNA) antisense lncRNA is transcribed from the HOX-C cluster on chromosome 12. Following transcription, HOTAIR interacts with the PRC2 and LSD1 chromatin modifier complexes utilizing its 5’ and 3’ end, respectively, and recruits them to the Hox-D locus located on chromosome 2 (Figure 1A). This causes H3K27me3 and demethylation of H3K4me2/3 at the Hox-D locus, leading to their silencing (Rinn et al., 2007; Tsai et al., 2010). Besides the Hox-D locus, HOTAIR also targets PRC2 to many other genomic loci for gene silencing by creating repressive histone marks (Chu et al., 2011). Targeted deletion of Hotair in mouse leads to derepression of a large number of genes, including genes from the Hox-D cluster and imprinted clusters, and homeotic transformation of spine and malformation of metacarpal-carpal bones (Li et al., 2013a). In addition, aberrant expression of HOTAIR is implicated in cancer progression and metastasis as elevated expression of HOTAIR is reported in primary and metastatic breast cancer, and its depletion leads to decrease in invasiveness (Gupta et al., 2010).
NrRNAs are also known to promote gene activation by recruiting chromatin modifiers to specific chromatin sites. HOXA transcript at the distal tip (HOTTIP) lncRNA is transcribed from the 5’end of the HOXA locus (Wang et al., 2011b) and coordinates the transcription of several of the 5’ HOXA genes in cis. HOTTIP recruits WDR5/MLL complex to its target genes at the HOX cluster, facilitating H3K4me3 and transcription. HOTTIP is brought to the proximity of its target genes by chromosome looping (Figure 1A). Similar to HOTTIP, Mistral (Mira) lncRNA is also known to positively influence transcription of genes in the HOX clusters in mouse embryonic stem cells (mES) (Bertani et al., 2011). Mistral gene is located in the spacer region separating Hoxa6 and Hoxa7 and in mES cells it is specifically activated upon retinoic acid treatment. Mistral recruits MLL1 to chromatin, induces changes in chromatin structure that results in the activation of Hoxa6 and Hoxa7 genes (Figure 1A) (Bertani et al., 2011).
In addition to establishing histone tail modifications, lncRNAs also influence other epigenetic modifications, including DNA methylation. For example, an lncRNA transcribed from the CEBPA gene locus regulates local DNA methylation (Di Ruscio et al., 2013). The so-called ecCEBPA (extracoding CEBPA) lncRNA is nuclear enriched and non-polyadenylated. It is transcribed from the upstream region of the CEBPA gene and overlaps the entire CEBPA mRNA sequence in the same sense-orientation. Interestingly, ecCEBPA binds to the maintenance DNA methyltransferase 1 (DNMT1) and prevents DNMT1-mediated DNA methylation at the CEBPA gene locus. Finally, in addition to ecCEBPA lncRNA, the authors discoverd several RNA species associated with DNMT1 and confirmed their involvement in regulating DNA methylation and thereby modulating gene expression (Di Ruscio et al., 2013).
Transcriptional regulators
Until recently, transcription of RNA was considered to be controlled/regulated by the interplay of the upstream/downstream cis-acting regulatory elements and the various interconnected regulatory proteins/factors (Michalak, 2006). However, with the emerging role of lncRNAs and their widespread distribution in the genome, it is interpreted that lncRNAs may play equally vital roles in the activation or repression of cellular genes. Nuclear-restricted lncRNAs have been reported to regulate gene transcription either by modulating the activities of transcriptional factors or controlling the binding of these factors to the promoter region (Chen and Carmichael, 2010; Khalil et al., 2009; Ng et al., 2013). An example of where lncRNAs induce gene transcription is provided by Evf2, a 3.8 kb long, polyadenylated, alternatively spliced nrRNA, transcribed from the ultraconserved intergenic enhancer region of Dlx-5/6 locus (Feng et al., 2006). Dlx genes are related to the Drosophila Distalless (DII) genes, and are involved in neuronal development (Panganiban and Rubenstein, 2002). Evf-2 was reported to increase the activity of specific enhancer regions (ei and eii) by interacting and recruiting the homeodomain protein Dlx-2 to ei and eii regions, resulting in the induction of Dlx-5/6 gene expression (Figure 1B) (Feng et al., 2006). Thus, Evf-2 functions like a transcriptional coactivator that acts in trans to regulate the expression of homeodomain genes during development. However, in the Evf-2 null mice, the expression of Dlx-5/6 was found to increase despite the absence of Dlx-2 on the ei and eii regions (Bond et al., 2009). In this case, it is not apparent whether the binding of Dlx proteins to some other enhancers compensates for the absence of Dlx-2 on ei, eiiregions or Dlx-2 acts indirectly by preventing the binding of MeCP2, a known repressor of Dlx-5/6, to ei/eii regions.
According to the estimation by the ENCODE project, ~80% of genes in a mammalian genome have an alternative transcriptional start site (Birney et al., 2007; Kawaji et al., 2009). In several instances lncRNAs, transcribed from the upstream region of the protein coding genes have been reported to regulate the transcription of their protein-coding partner by preventing the formation of transcriptional complex at the promoter region (Figure 1C) (Batista and Chang, 2013). For example, the dihydrofolate reductase (DHFR) gene contains both the minor and major promoter regions. In quiescent mammalian cells, an lncRNA transcribed from the upstream region of minor promoter of DHFR gene interacts with the major promoter and general transcriptional factor TFIIB (Martianov et al., 2007). These lncRNAs inhibit transcription of DHFR gene by preventing the association of TFIIB-containing transcriptional complex to the promoter region by forming a stable purine-purine-pyrimidine triple structure with the double stranded DHFR promoter (Martianov et al., 2007).
Linc-HOXA1, located~50 kb away from the Hoxa gene cluster, negatively regulates Hoxa1 expression in mES (Maamar et al., 2013). Linc-HOXA1 represses Hoxa1 in cis by recruiting the transcriptional cofactor PURB at the Hoxa1chromatin site (Figure 1C). Exposure of cells to retinoic acid leads to derepression of the Hoxa1 transcription, possibly due to reduced interaction of linc-HOXA1 with the Hoxa1 transcription site (Maamar et al., 2013). NrRNAs are also shown to activate transcription, by recruiting transcription activators or their co-factors to specific gene promoter regions. In this context, a very recent study revealed the role of nrRNA (lncRNA-JADE) in the activation of a gene (Jade), an integral component of the histone acetyltransferase (HAT) complex (Wan et al., 2013). In human cells, lncRNA-JADE is induced upon double strand DNA break in an Ataxia-telangiectasia mutated (ATM)-dependent manner. LncRNA-JADE transcriptionally activated Jade1 by recruiting Brca1 to the Jade promoter and facilitating the interaction between Brca1 and P300/CBP transcriptional co-factor complex. LncRNA-JADE-mediated transcriptional activation of Jade facilitated the DNA damage-induced hyperacetylation of histone 4. Interestingly, LncRNA-JADE is overexpressed in breast cancer samples and its downregulation inhibits tumor growth in vivo (Wan et al., 2013).
NrRNAs are also implicated in the transcriptional regulation of cell cycle-regulated genes, especially the ones that are activated upon cellular stress. For example, DNA damage induces several lncRNAs from regions located upstream of the CCND1 promoter (Wang et al., 2008). These lncRNAs interact with translocated in liposarcoma (TLS) protein, an RNA binding protein and a known inhibitor of CBP/P300 HAT, and facilitate the association of TLS with CBP/P300. TLS in turn represses the CBP/P300 activity on the CCND1 promoter resulting in the transcriptional inactivation of CCND1. LncRNAs transcribed from regions upstream of CDKN1A gene promoter are reported to regulate gene expression by recruiting RNA binding proteins and transcription factors to specific chromatin sites (Huarte et al., 2010; Hung et al., 2011). LincRNA-p21, located upstream of the CDKN1A (p21) gene, is a p53-responsive gene, and acts as a repressor in the p53-dependent transcriptional responses (Huarte et al., 2010). The tumor suppressor p53 activates the expression of lincRNA-p21 by directly interacting with lincRNA-p21 promoter. LincRNA-p21 in turn inhibits the expression of several of the anti-apoptotic genes that are normally repressed by p53. It has been observed that the interaction between lincRNA-p21 and the heterogeneous nuclear ribonucleoprotein K (hnRNP-K) is required for the genomic association of hnRNP-K with the repressed genes and also for their transcriptional repression (Huarte et al., 2010). PANDA is another p53-induced lncRNA that is also transcribed from the upstream region of CDKN1A gene. PANDA is upregulated upon DNA damage, interacts with NF-YA transcription factor and controls the expression of pro-apoptotic genes. PANDA-depleted fibroblasts show increased sensitivity to DNA damage-induced apoptosis (Hung et al., 2011). A very recent study from Huarte’s laboratory has reported the involvement of another p53-induced lncRNA in gene regulation (Marin-Bejar et al., 2013). Pint (p53-induced noncoding transcript) is an nrRNA in mouse cells that promotes cell proliferation and post DNA-damage cell survival by regulating the expression of genes involved in TGF-beta, MAP kinase and P53 signalling pathways. Similar to several other known lncRNAs, Pint interacts with PRC2 complex and recruits PRC2 to specific chromatin sites for successful H3K27-trimethylation and transcription repression (Marin-Bejar et al., 2013). These studies have highlighted the crucial roles played by lncRNAs in the expression of cell cycle-regulated genes and other genes that are part of the p53-mediated signaling network.
Recent studies have also documented the involvement of lncRNAs in regulating enhancer functions (Shiekhattar, 2013) (Also please see Darrow and Chadwick in this Special issue). Enhancer elements are distal regulatory sequences, which generally promote the transcription of protein coding genes in an orientation independent manner (Bulger and Groudine, 2011). Enhancers are characterized by higher levels of H3K4me1 and H3K27Ac on their chromatin. They also bind to the p300/CBP transcriptional coactivators in a stimulus dependent manner (Bulger and Groudine, 2011; Creyghton et al., 2010; Heintzman et al., 2007). It has been shown that the enhancers are occupied by RNA pol II that leads to the generation of 0.5 to 5.0 kb long lncRNAs referred as enhancer RNAs (eRNAs) and ncRNA-activating (ncRNA-a; ncRNA-a chromatin retains H3K4me3 modifications and contains weak H3K4me1 marks), some of which are polyadenylated (De Santa et al., 2010; Kim et al., 2010; Orom et al., 2010; Wang et al., 2011a). The transcription of eRNAs is regulated in a stage or tissue-specific manner and their activation is generally correlated with the expression of protein-coding genes located in the nearby regions (Figure 1A) (De Santa et al., 2010; Hah et al., 2013; Kim et al., 2010; Li et al., 2013b). The levels of several eRNAs are found to be increased in cells treated with different stimuli, like endotoxins in human macrophage cells and estradiol in MCF-7 cells (Li et al., 2013b). Li and colleagues (2013) reported that 17β-estradiol (E2)-bound estrogen receptor α (ERα)induces a global upregulation of eRNA transcription on enhancers present near the E2-upregulated coding genes in MCF-7 cells (Li et al., 2013b). These eRNAs positively influence ligand-dependent ERα-induced gene activation, in part by facilitating the enhancer-promoter chromatin looping. Similarly, induction of Androgen receptor (AR) signalling in prostate cancer cells also activated specific set of eRNAs located at the AR-regulated gene enhancers (Wang et al., 2011a). These eRNAs promoted enhancer-promoter chromatin and function in combination with lineage specific transcription factors like FOXA1 to create a higher order enhancer chromatin network and regulate transcription in a cell type-specific manner (Wang et al., 2011a). Recently, Lai and colleagues (2013) have shown that activator ncRNA-a interact specifically with Mediator co-activator complex during transcription to regulate chromatin localization as well as the kinase activity (towards H3 serine-10) of the Mediator. Further, by using a heterologous reporter system in mammalian cells, the authors revealed the involvement of Mediator components in the ncRNA-a-mediated activation of genes (Lai et al., 2013). Based on genomic analyses, human cells produce several hundreds of enhancer-associated eRNAs or ncRNA-a and recent studies have started to unravel the roles of these transcripts in diverse cellular activities, including p53-mediated gene activation and muscle differentiation (Lam et al., 2013; Melo et al., 2013; Mousavi et al., 2013; Orom et al., 2010).
Post-transcriptional regulators
Post transcriptional gene regulation works at different levels, including pre-mRNA splicing, RNA editing, transport, stability and degradation of mRNA. All of these processes involve complex networks of various regulatory protein complexes/factors that in turn determine the type and amount of a particular protein to be synthesized at specific time/space to govern a given biological process. Recently, few studies have attempted to understand the involvement of nrRNAs in posttranscriptional gene regulation (Yoon et al., 2012a). Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1; also referred to as NEAT 2) is one such example of a lncRNA that is involved in the post-transcriptional gene regulation via controlling alternate splicing of pre-mRNAs (Zong et al., 2011). MALAT1 is a >7 kb long, abundantly expressed nuclear-restricted lncRNA, originally identified as a lncRNA that is overexpressed in early-stage non-small lung cancers (Hutchinson et al., 2007; Ji et al., 2003; Lin et al., 2007). Recent studies show the involvement of MALAT1 in several important cellular processes, including cell cycle progression, serum stimulation-induced gene activation, neuronal synapse formation and cancer cell migration (Bernard et al., 2010; Tano et al., 2010; Tripathi et al., 2010; Tseng et al., 2009; Yang et al., 2011a). MALAT1 RNA is highly conserved among mammals, and predominantly localizes in the nuclear speckles or SC35 domains, a nuclear structure that contains proteins and RNAs involved in pre-mRNA processing (Figure 2B & 2E-G) (Bernard et al., 2010; Hutchinson et al., 2007; Tripathi et al., 2010). MALAT1, though not shown to be involved in the formation and/or maintenance of nuclear speckles, is found to interact with various pre-mRNA splicing factors, including the serine/arginine family of splicing factors (SR proteins). MALAT1-depleted human cells show alterations in the distribution and activity of SR splicing factors, including aberrant changes in alternative splicing of pre-mRNAs (Figure 1D) (Lin et al., 2011; Tripathi et al., 2010). Recently, we have also demonstrated that MALAT1 modulates the expression of genes involved in the cell cycle progression and is required for G1/S and mitotic progression. MALAT1-depleted human diploid fibroblasts activated p53 and its target genes and showed a robust cell cycle arrest. These cell cycle defects were sensitive to the p53 levels, indicating that p53 is a major downstream mediator of MALAT1 activity (Tripathi et al., 2013). Furthermore, MALAT1-depleted cells display reduced expression of B-MYB (Mybl2), an oncogenic transcription factor involved in G2/M progression. This is due to changes in the association of SR splicing factors on B-MYB pre-mRNA resulting in aberrant alternative splicing. Our studies in human cells indicate that MALAT1 promotes cellular proliferation by modulating the expression and/or pre-mRNA processing of cell cycle-regulated transcription factors (Tripathi et al., 2013). A recent study from Rosenfeld laboratory suggested a role for MALAT1 in cell cycle progression through its involvement in regulating E2F1 transcription factor activity (Yang et al., 2011a). MALAT1-depleted cells showed proliferation defects and an inability to activate E2F target genes upon the addition of serum. The authors described that MALAT1 influenced the interaction of unmethylated polycomb protein, Pc2with E2F1 in serum-activated cells, and such an association facilitates E2F1 SUMOylation, leading to the activation of serum-induced genes (Yang et al., 2011a). Based on all these results it is evident that MALAT1 regulates cell proliferation by interacting with several proteins, including chromatin modifiers and pre-mRNA splicing factors.
Surprisingly, we and others have recently reported that the in vivo MALAT1 knockout (KO) mouse is viable and fertile and MEFs from the knock out (KO) mouse did not show any defects in alternative splicing and SR protein activity (Eissmann et al., 2012; Nakagawa et al., 2012; Zhang et al., 2012). These results implicate that MALAT1 is largely dispensable in mice but plays an important role in particular cell types under specific physiological conditions. The cell type- or organism-specific phenotype observed upon depletion of a particular ncRNA is not specific to MALAT1, as earlier studies had reported similar results for other ncRNAs and protein-coding genes (Berthet et al., 2003; Concepcion et al., 2012; Ortega et al., 2003; Schorderet and Duboule, 2011).
An elegant study from Chen and colleagues revealed the role of a novel class of nuclear-retained lncRNAs in splicing regulation (Yin et al., 2012).The so called ‘sno-lincRNAs’ are transcribed from intronic sequences from several genomic sections, including the region in chromosome 15 that has been implicated in Prader-Willi Syndrome (PWS). Sno-lincRNAs are devoid of 5’caps and poly (A) tails and are processed by the snoRNA machinery. The sno-lincRNAs from PWS region accumulate near the sites of their transcription and establish a domain, where the splicing factor Fox2 is accumulated. Functional studies indicate these sno-lincRNAs interact with Fox2, appear to function as a Fox2 ‘sink’, and thereby influence Fox2-mediated pre-mRNA splicing (Figure 1D) (Yin et al., 2012).
Gomafu/RNCR2/MIAT (myocardial infarction associated transcript), a ployadenylated nrRNA with neuron-restricted expression, is another example of an lncRNA that is implicated in posttranscriptional gene regulation (Ishii et al., 2006). Gomafu/MIAT localizes to a novel nuclear compartment, which does not coincide with any of the known nuclear domains (Sone et al., 2007). Gomafu is expressed in differentiating neurons and oligodendrocytes while in progenitor cells its expression is observed following lineage specification (Mercer et al., 2010). Gomafu RNA binds to the branch point-interacting SF1 splicing factor with high affinity utilizing the tandem repeats within the transcript. Gomafu is speculated to influence pre-mRNA splicing efficiency by sequestering the cellular pool of SF1 within the nucleus (Sone et al., 2007; Tsuiji et al., 2011). A general theme emerging from all these studies is that several of the nrRNAs influence pre-mRNA splicing, primarily by titrating the cellular pool of splicing or other pre-mRNA processing factors.
The antisense lncRNAs are known to influence alternative splicing of their sense RNA partners. This has been observed in case of alpha-Thyroid hormone receptor gene (ErbAα), where its antisense transcript RevErbAα dictates the differential synthesis of the alternatively spliced Tra1 and Tra2 mRNAs (Hastings et al., 2000; Hastings et al., 1997) through yet unknown mechanisms.
LncRNAs in nuclear organization
Recent studies indicate that lncRNAs interact and recruit proteins that are involved in the organization and maintenance of specific subnuclear domains (Dundr and Misteli, 2010; Shevtsov and Dundr, 2011). Many of these proteins include RNA processing factors that are involved in RNP metabolism. Proteins present in these discrete domains exchange between the domains with the help of the nuclear domain-resident lncRNAs (Mao et al., 2011). For example, Shevtsov and Dundr (2011) showed that several types of RNAs, both coding and noncoding, can function as structural elements and facilitate in the nucleation of nuclear bodies. They demonstrated transcription as a driving force in the nuclear body formation and RNA acting as a scaffold in the formation of these membraneless structures (Shevtsov and Dundr, 2011). NEAT1 (nuclear enriched abundant transcript 1) or MENε/β (multiple endocrine neoplasia ε/β) or VINC (virus induced noncoding transcript) is a well characterized lncRNA, which acts as an important structural component in the formation and maintenance of paraspeckle domains (Figure 2C) (Fox and Lamond, 2010). The NEAT1 gene encodes two major isoforms i.e., NEAT1_1/MENε (3.7 kb) and NEAT1_2/MENβ (23 kb), both differ at their 3’ end. The long NEAT12 isoform is essential for the formation of paraspeckle while the short NEAT11 though increases the number of paraspeckles when overexpressed, is not found to be sufficient for the maintenance of paraspeckles (Bond and Fox, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009). The paraspeckle contains several RNA binding proteins, including DBHS (Drosophila Behaviour Human Splicing) family proteins i.e., PSPC1, PSF (SFPQ) and p54nrb (NONO) (Fox et al., 2005; Passon et al., 2012). Members of this family, especially NONO, are known to interact with Adenosine-to-Inosine (A-to-I) edited RNAs and thought to influence nuclear retention and paraspeckle localization of A to I edited transcripts (Chen and Carmichael, 2008; Prasanth et al., 2005). Several studies have suggested a role for nuclear paraspeckles and its constituents in the nuclear retention of RNAs (Chen and Carmichael, 2008; Chen and Carmichael, 2009; Prasanth et al., 2005). The mouse-specific CTN-RNA (CAT2 transcribed nuclear RNA) is one such nrRNA that is A-to-I edited at its 3’UTR, interacts with NONO and localizes to paraspeckle (Figure 2D). Upon interferon-γ and lipopolysaccharide treatment, CTN-RNA is post transcriptionally processed at its 3’UTR to produce a smaller transcript that is further transported to cytoplasm for translation (Prasanth et al., 2005). A similar mode of gene regulation was also reported for the synthesis of migration stimulating factor (MSF) upon TGFβ1 treatment (Kay et al., 2005). These examples signify a novel mechanism of gene regulation, where an RNA under normal conditions is sequestered in specific sub-nuclear compartments like paraspeckles and is exported to the cytoplasm upon a specific signal as part of the ‘quick response’ mechanism. Another study from the Carmichael laboratory showed a correlation between the presence of NEAT1, paraspeckle and the nuclear retention of A-to-I edited RNAs in human embryonic stem cells (hES) (Chen and Carmichael, 2009). This study found that hES cells do not express NEAT1 transcripts, therefore, lack paraspeckles and as a result, the hyper A-to-I edited mRNAs transport efficiently to the cytoplasm. However, following differentiation of hES cells to trophoblasts, these cells express NEAT1 and form distinct paraspeckles. Interestingly, differentiated ES cells containing intact paraspeckles showed nuclear retention of A-to-I edited transcripts, further supporting the involvement of NEAT1 and paraspeckle in the nuclear retention of edited RNAs. Future studies will determine the exact role of paraspeckle and A-to-I editing in the nuclear storage and signal-induced cleavage of transcripts.
LncRNAs are also known to sequester proteins in subnuclear domains, as part of the cellular stress response. For example, certain environmental stresses like changes in pH during hypoxia or acidosis result in the nucleolar immobilization of proteins, including VHL (von Hippel-Lindau), a factor that under normal oxygen tension facilitates the degradation of HIFα (hypoxia-inducible factor-α). Similarly, other cellular stresses and oncogenic signals induce the sequestration of HDM2 (MDM2) into the nucleolus (Audas et al., 2012). However, the underlying mechanism responsible for the stress-induced association of factors to nucleolus was not clearly understood. A recent study revealed the role of several lncRNAs transcribed from the intergenic spacer (IGS) region of the rDNA repeats in the stress-induced nuclear detention of proteins (Audas et al., 2012). Unique IGS RNAs are induced upon specific cellular stress and they interact with proteins containing specific peptide codes referred to as nucleolar detention sequence. Interestingly, altering the levels of one type of IGS RNA changes the nuclear detention activity of other IGS RNAs indicating that different members of IGS RNAs function independently and do not influence the function of one another.
NrRNAs and Disease
Since many of the nrRNAs are involved in the fine tuning of chromatin reprogramming, epigenetic modification, transcriptional and post-transcriptional gene regulation, their aberrant expression in the cell alters gene expression that leads to diseases like cancer. Several studies showed higher expression of some of the lncRNAs in cancerous cells or tissues; and their induced expression in mouse models result in tumor formation and metastasis, while some other lncRNAs act as tumor suppressors (Prensner and Chinnaiyan, 2011). As discussed earlier, HOTAIR interacts with PRC2 and LSD1/CoREST/REST complex leading to epigenetic changes by H3K27 methylation and H3K4 demethylation (Rinn and Chang, 2012). Its higher expression has been reported in several type of cancers like breast cancer, hepatocellular carcinoma, colorectal cancer, pancreatic and gastrointestinal stromal cancer (Gupta et al., 2010; Kim et al., 2013; Kogo et al., 2011; Niinuma et al., 2012; Yang et al., 2011b). It has been observed that the over-expression of HOTAIR in cancer samples leads to poor prognosis while its loss decreases the invasiveness of cancerous tissues. Another lncRNA ANRIL (antisense noncoding RNA in the INK4 locus) transcribed from the p15/CDKN2B-p16/CDKN2A-p14/ARF gene cluster is associated with breast cancer, basal cell carcinoma, gliomas, intracranial aneurysm and coronary artery disease (Burd et al., 2010; Pasmant et al., 2011; Shete et al., 2009; Stacey et al., 2009; Turnbull et al., 2010). ANRIL functions as a repressor of tumor suppressor gene p15INK4B by interacting with SUZ12 and recruit PRC2 to thep15INK4B transcription site (Kotake et al., 2011). Nuclear-localized MALAT1 over expression is observed in lung, liver, breast, pancreas, prostate, colon and cervical cancer samples (Guo et al., 2010; Ji et al., 2003; Lin et al., 2007; Schmidt et al., 2011). A study by Tano and colleagues suggested that MALAT1 regulates the activity of genes (CTHRC1, CCT4, HMMR and ROD1) that are involved in cell motility thereby influencing cancer metastasis (Tano et al., 2010).
Recently, Calin laboratory has described the involvement of a novel nuclear-retained lncRNA (CCAT2) in metastatic progression and chromosomal instability in colon cancer (Ling et al., 2013). CCAT2 transcript maps to the highly conserved 8q24.21 region and is overexpressed in the microsatellite-stable colorectal cancer samples. CCAT2 promotes tumor growth, as subcutaneous transplantation of CCAT2-overexpressing cells resulted in larger tumors in the immuno-compromised mice compared to empty vector-transduced cells. Furthermore, CCAT2 lncRNA induces the expression of MYC by regulating the activity of TCF7L2 transcription factor, an essential protein that is required for the transcriptional activation of genes involved in WNT signaling pathway. These data support a novel role for CCAT2 lncRNA in regulating MYC and WNT signaling pathways (Ling et al., 2013).
PCAT-1 is one among several lncRNAs that show elevated expression in prostate cancer tissue samples (Prensner et al., 2011). In vitro and in vivo studies indicate that PCAT-1 facilitates cancer cell proliferation. Functional studies suggest that PCAT-1 acts as a transcriptional repressor of genes involved in mitosis and cell cycle progression. The same investigators have recently identified another prostate cancer-associated lncRNA; Schlap1 (second chromosome locus associated with prostate-1), that is specifically overexpressed in a subset of prostate cancers (Prensner et al., 2013). Overexpression of Schlap1 in cancer samples predicts poor outcomes, including metastasis and prostate cancer-specific mortality. Functional studies indicate that Schlap1 negatively regulates the genome-wide association and function of SWI/SNF chromatin remodelling complex (Prensner et al., 2013).
Conclusion
Recent studies have highlighted the important roles played by lncRNAs in key biological/cellular processes. Majority of the lncRNAs remain in the nucleus, and this provides a better opportunity for nuclear-retained lncRNA or nrRNAs to regulate several of the nuclear functions in a sequence-specific manner, and also to function as structural elements to maintain proper nuclear territories/domains. In addition, regulatory RNAs are retained within the nucleus and are exported out to the cytoplasm upon specific signals/stimuli (Carrieri et al., 2012; Kay et al., 2005; Prasanth et al., 2005). Antisense ubiquitin carboxy terminal hydrolase L1 (AS Uchl1) is one such nuclear-retained lncRNA that is complementary to the mRNA, which encodes Uchl1. AS Uchl1 interacts with Uchl1 mRNA, enhances translation of Uchl1 mRNA by facilitating the formation of active polysomes on Uchl1 mRNA. Interestingly, when cells are inhibited of mTORC1 activity, AS Uchl1 lncRNA is exported out of the nucleus to the cytoplasm. This in turn increases Uchl1 mRNA translation (Carrieri et al., 2012).
Similar to what is known for proteins, lncRNAs are also shown to execute more than one mutually exclusive function. For example, human translational regulatory lncRNA (treRNA) earlier referred as ncRNA-a7, is known to function as an enhancer-like RNA to regulate Snail transcription in the nucleus (Orom et al., 2010). It is also involved in the translational repression of specific cellular mRNAs in the cytoplasm (Gumireddy et al., 2013; Orom et al., 2010). TreRNA specifically downregulates the expression of epithelial marker E-cadherin by altering the polysomal distribution on E-cadherin mRNA. TreRNA mediates its function through its association with an RNP complex consisting of several RNA-binding proteins (hnRNP K, FXR1 and FXR2), PUF60 and SF3B3 (Gumireddy et al., 2013). Similarly, LincRNA-p21, in addition to regulating the transcription of genes in the p53 signaling pathway, was also recently shown to repress translation of specific mRNAs (Yoon et al., 2012b). The human genome contains several thousands of lncRNAs. Given the complexity provided by several of the characterized lncRNAs, it is highly likely that many nuclear-retained lncRNAs with additional functions in gene regulation will be identified in the near future. Future studies will also determine how these lncRNAs are being regulated and also their involvement in several human diseases.
Acknowledgements
We would like to thank Drs. Stephanie Ceman (UIUC), Ashish Lal (NCI, NIH) and Supriya G. Prasanth (UIUC) for critical reading of the manuscript. We would like to thank Dr. Aparna Anantharaman, Dr. Vidisha Tripathi and Sumanprava Giri for their assistance in RNA-FISH analyses. Research in the KVP laboratory is supported by grants from American Cancer Society (RSG-11-174-01-RMC) and NIH/NIGMS (GM088252).
Abbreviations
- ANRIL
Antisense nocoding RNA in the INK4 locus
- AR
Androgen receptor
- A-to-I
Adenine to Inosine
- CCAT2
Colon cancer associated transcript 2
- CEBPA
CCAAT/Enhancer Binding Protein (C/EBP), Alpha
- DBHS
Drosophila Behaviour Human Splicing
- DHFR
Dihydrofolatereductase
- ecCEBPA
extracoding CEBPA
- eRNA
enhancer RNA
- ER α
Estrogen receptor α
- H3K4me3
Histone 3 Lysine 4 rimethylation
- H3K27me3
Histone 3 Lysine 27 trimethylation
- HAT
Histone acetyl transferase
- hES
Human embryonic stem cells
- HMT
Histone methyltransferase
- hnRNP-K
heterogeneous nuclear ribonucleoprotein K
- HOTAIR
HOX transcript antisense RNA
- HOTTIP
HOXA transcript at the distal tip
- Igf2r
Insulin-like growth factor 2 receptor
- IGS
Intergenic spacer
- lncRNA
Long noncoding RNA
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- mES
Mouse embryonic stem cells
- MIAT
Myocardial infarction associated transcript
- ncRNAa
Noncoding RNA activator
- nrRNA
Nuclear-retained RNA
- PRC2
Polycomb repressive complex 2
- PWS
Prader-Willi Syndrome
- RepA
Repeat A
- Schlap1
Second chromosome locus associated with prostate-1
- TCF7L2
transcription factor 7-like 2
- treRNA
Translational regulatory lncRNA
- Uchl1
Ubiquitin carboxyterminal hydrolase L1
- VHL
von Hippel-Lindau
- XCI
X-chromosome inactivation
- Xi
Inactive X-chromosome
- Xic
X-chromosome inactivation center
- Xist
X-inactive-specific transcript
References
- Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT, Menck CF, Reis EM, Verjovski-Almeida S. The Intronic Long Noncoding RNA ANRASSF1 Recruits PRC2 to the RASSF1A Promoter, Reducing the Expression of RASSF1A and Increasing Cell Proliferation. PLoS Genet. 2013;9:e1003705. doi: 10.1371/journal.pgen.1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, Forrest AR, Carninci P, Biffo S, Stupka E, Gustincich S. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22:357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–2196. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, Dike S, Wyss C, Henrichsen CN, Holroyd N, Dickson MC, Taylor R, Hance Z, Foissac S, Myers RM, Rogers J, Hubbard T, Harrow J, Guigo R, Gingeras TR, Antonarakis SE, Reymond A. Prominent use of distal 5' transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res. 2007;17:746–759. doi: 10.1101/gr.5660607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D'Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013 doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do JT, Han DW, Gentile L, Sobek-Klocke I, Stehling M, Scholer HR. Enhanced reprogramming of Xist by induced upregulation of Tsix and Dnmt3a. Stem Cells. 2008;26:2821–2831. doi: 10.1634/stemcells.2008-0482. [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, Calin GA, Huang Q. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J. 2013 doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li Y, Liu Y, Wang J, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J Biol Chem. 2000;275:11507–11513. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–4300. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol. 2011;22:343–350. doi: 10.1016/j.semcdb.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaji H, Severin J, Lizio M, Waterhouse A, Katayama S, Irvine KM, Hume DA, Forrest AR, Suzuki H, Carninci P, Hayashizaki Y, Daub CO. The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol. 2009;10:R40. doi: 10.1186/gb-2009-10-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RA, Ellis IR, Jones SJ, Perrier S, Florence MM, Schor AM, Schor SL. The expression of migration stimulating factor, a potent oncofetal cytokine, is uniquely controlled by 3'-untranslated region-dependent nuclear sequestration of its precursor messenger RNA. Cancer Res. 2005;65:10742–10749. doi: 10.1158/0008-5472.CAN-05-2038. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, Lee CY, Watt A, Grossman TR, Rosenfeld MG, Evans RM, Glass CK. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lessing D, Anguera MC, Lee JT. X Chromosome Inactivation and Epigenetic Responses to Cellular Reprogramming. Annu Rev Genomics Hum Genet. 2013 doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY. Targeted Disruption of Hotair Leads to Homeotic Transformation and Gene Derepression. Cell Rep. 2013a doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013b;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, Huang L, Moreno I, Navarro A, Monzo M, Garcia-Foncillas J, Rinn JL, Guo S, Huarte M. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P. RNA world - the dark matter of evolutionary genomics. J Evol Biol. 2006;19:1768–1774. doi: 10.1111/j.1420-9101.2006.01141.x. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs Promote Transcription by Establishing Chromatin Accessibility at Defined Genomic Loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- Passon DM, Lee M, Rackham O, Stanley WA, Sadowska A, Filipovska A, Fox AH, Bond CS. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc Natl Acad Sci U S A. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013 doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Shiekhattar R. Opening the Chromatin by eRNAs. Mol Cell. 2013;51:557–558. doi: 10.1016/j.molcel.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Rath PC. Long interspersed nuclear elements (LINEs) show tissue-specific, mosaic genome and methylation-unrestricted, widespread expression of noncoding RNAs in somatic tissues of the rat. RNA Biol. 2012;9:1380–1396. doi: 10.4161/rna.22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Scherer D, Hemminki K, Rudnai P, Gurzau E, Koppova K, Botella-Estrada R, Soriano V, Juberias P, Saez B, Gilaberte Y, Fuentelsaz V, Corredera C, Grasa M, Hoiom V, Lindblom A, Bonenkamp JJ, van Rossum MM, Aben KK, de Vries E, Santinami M, Di Mauro MG, Maurichi A, Wendt J, Hochleitner P, Pehamberger H, Gudmundsson J, Magnusdottir DN, Gretarsdottir S, Holm H, Steinthorsdottir V, Frigge ML, Blondal T, Saemundsdottir J, Bjarnason H, Kristjansson K, Bjornsdottir G, Okamoto I, Rivoltini L, Rodolfo M, Kiemeney LA, Hansson J, Nagore E, Mayordomo JI, Kumar R, Karagas MR, Nelson HH, Gulcher JR, Rafnar T, Thorsteinsdottir U, Olafsson JH, Kong A, Stefansson K. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–1551. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Prasanth KV. Cell Nucleus. Encyclopedia of Life Sciences. 2011 [Google Scholar]
- Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng JJ, Hsieh YT, Hsu SL, Chou MM. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol Hum Reprod. 2009;15:725–731. doi: 10.1093/molehr/gap071. [DOI] [PubMed] [Google Scholar]
- Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011;16:479–490. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den Ouweland A, Houlston R, Ross G, Langford C, Pharoah PD, Stratton MR, Dunning AM, Rahman N, Easton DF. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Hu X, Liu Y, Han C, Sood AK, Calin GA, Zhang X, Lu X. A novel non-coding RNA lncRNA-JADE connects DNA damage signalling to histone H4 acetylation. EMBO J. 2013 doi: 10.1038/emboj.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011a;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011b;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011a;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011b;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional Gene Regulation by Long Noncoding RNA. J Mol Biol. 2012a doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012b;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]