Abstract
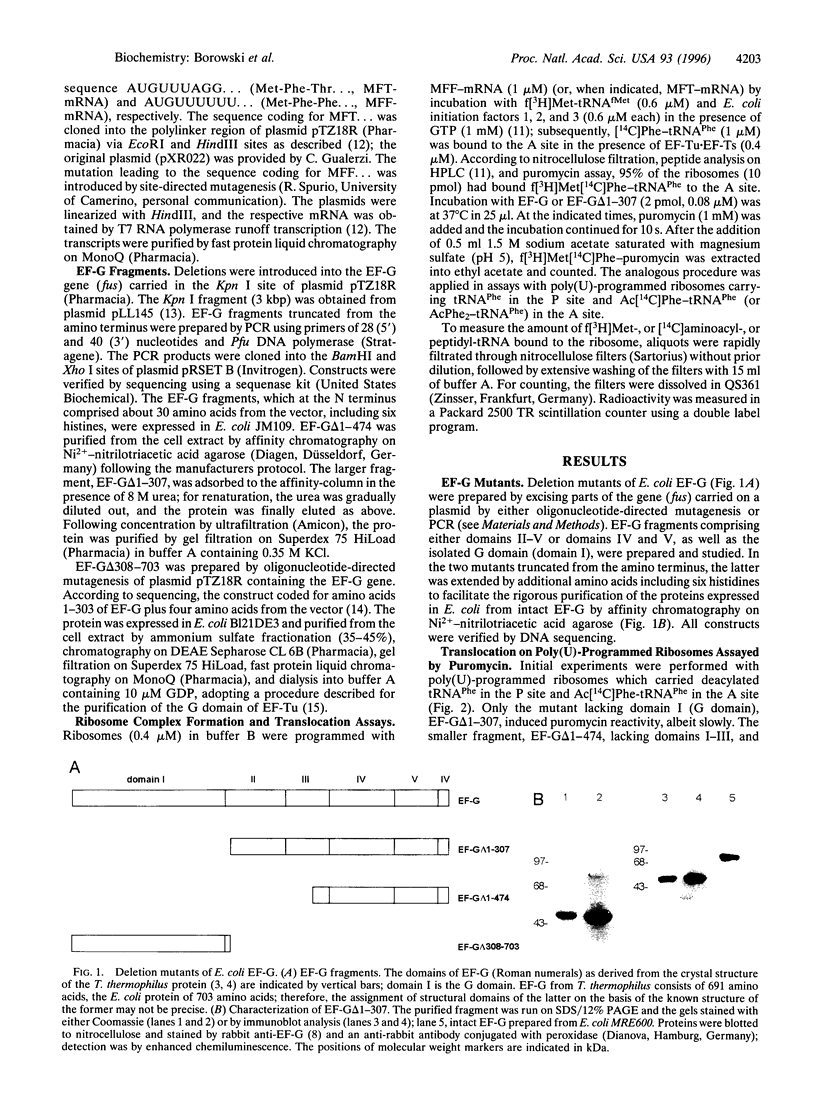
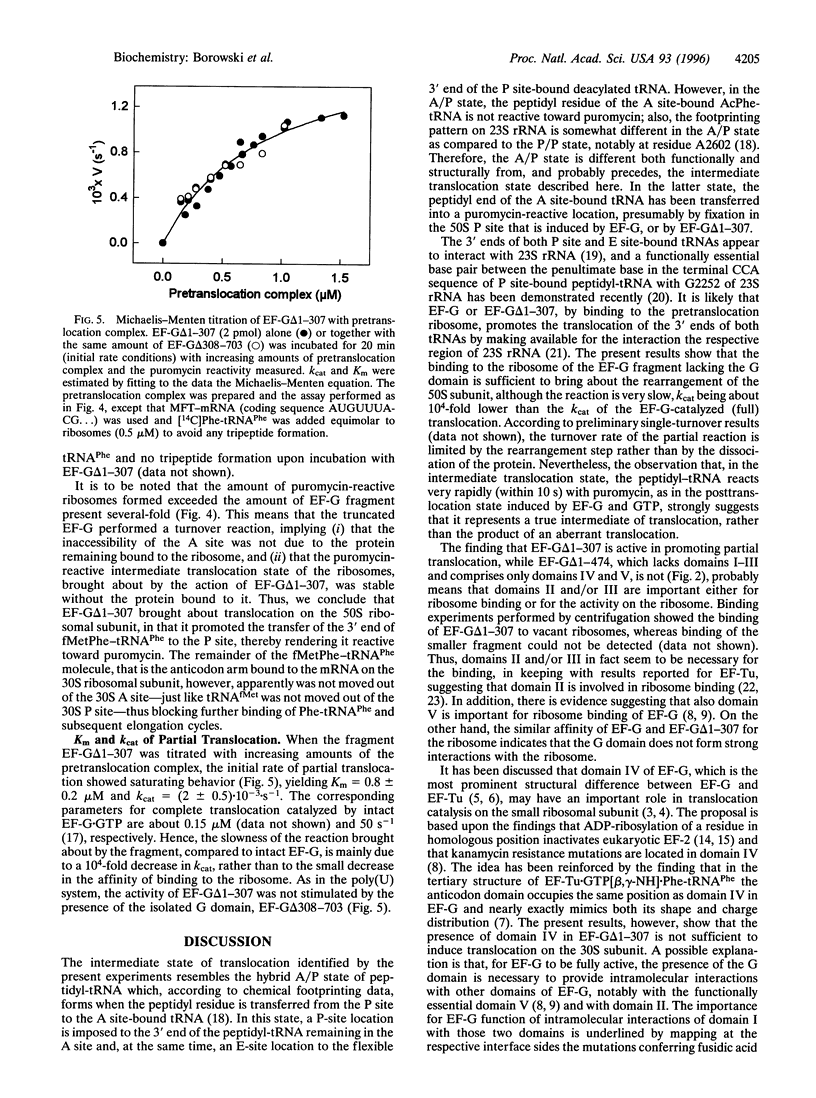
The mechanism by which elongation factor G (EF-G) catalyzes the translocation of tRNAs and mRNA on the ribosome is not known. The reaction requires GTP, which is hydrolyzed to GDP. Here we show that EF-G from Escherichia coli lacking the G domain still catalyzed partial translocation in that it promoted the transfer of the 3' end of peptidyl-tRNA to the P site on the 50S ribosomal subunit into a puromycin-reactive state in a slow-turnover reaction. In contrast, it did not bring about translocation on the 30S subunit, since (i) deacylated tRNA was not released from the P site and (ii) the A site remained blocked for aminoacyl-tRNA binding during and after partial translocation. The reaction probably represents the first EF-G-dependent step of translocation that follows the spontaneous formation of the A/P state that is not puromycin-reactive [Moazed, D. & Noller, H. F. (1989) Nature (London) 342, 142-148]. In the complete system--i.e., with intact EF-G and GTP--the 50S phase of translocation is rapidly followed by the 30S phase during which the tRNAs together with the mRNA are shifted on the small ribosomal subunit, and GTP is hydrolyzed. As to the mechanism of EF-G function, the results show that the G domain has an important role, presumably exerted through interactions with other domains of EF-G, in the promotion of translocation on the small ribosomal subunit. The G domain's intramolecular interactions are likely to be modulated by GTP binding and hydrolysis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AEvarsson A., Brazhnikov E., Garber M., Zheltonosova J., Chirgadze Y., al-Karadaghi S., Svensson L. A., Liljas A. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 1994 Aug 15;13(16):3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova L., Reiser C. O., Schirmer N. K., Sprinzl M., Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993 Sep 9;365(6442):126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Kirsebom L. A., Ehrenberg M., Kurland C. G. Mutations in ribosomal proteins L7/L12 perturb EF-G and EF-Tu functions. Biochimie. 1988 May;70(5):611–618. doi: 10.1016/0300-9084(88)90244-1. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Calogero R. A., Pon C. L., Canonaco M. A., Gualerzi C. O. Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6427–6431. doi: 10.1073/pnas.85.17.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czworkowski J., Wang J., Steitz T. A., Moore P. B. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994 Aug 15;13(16):3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova L. P., Kostiashkina O. E., Koteliansky V. E., Rutkevitch N. M., Spirin A. S. Factor-free ("non-enzymic") and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J Mol Biol. 1976 Mar 15;101(4):537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- Hou Y., Lin Y. P., Sharer J. D., March P. E. In vivo selection of conditional-lethal mutations in the gene encoding elongation factor G of Escherichia coli. J Bacteriol. 1994 Jan;176(1):123–129. doi: 10.1128/jb.176.1.123-129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Yaskowiak E. S., March P. E. Carboxyl-terminal amino acid residues in elongation factor G essential for ribosome association and translocation. J Bacteriol. 1994 Nov;176(22):7038–7044. doi: 10.1128/jb.176.22.7038-7044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M., Cool R. H., Mortensen K. K., Clark B. F., Parmeggiani A. Structure-function relationships of elongation factor Tu. Isolation and activity of the guanine-nucleotide-binding domain. Eur J Biochem. 1989 Jun 15;182(2):247–255. doi: 10.1111/j.1432-1033.1989.tb14824.x. [DOI] [PubMed] [Google Scholar]
- Johanson U., Hughes D. Fusidic acid-resistant mutants define three regions in elongation factor G of Salmonella typhimurium. Gene. 1994 May 27;143(1):55–59. doi: 10.1016/0378-1119(94)90604-1. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen P., Thirup S., Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993 Sep 15;1(1):35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- Lill R., Robertson J. M., Wintermeyer W. Binding of the 3' terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989 Dec 1;8(12):3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989 Nov 9;342(6246):142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B. F., Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995 Dec 1;270(5241):1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Nygård O., Nilsson L. Reduced ribosomal binding of eukaryotic elongation factor 2 following ADP-ribosylation. Difference in binding selectivity between polyribosomes and reconstituted monoribosomes. Biochim Biophys Acta. 1985 Feb 20;824(2):152–162. doi: 10.1016/0167-4781(85)90092-2. [DOI] [PubMed] [Google Scholar]
- Omura F., Kohno K., Uchida T. The histidine residue of codon 715 is essential for function of elongation factor 2. Eur J Biochem. 1989 Mar 1;180(1):1–8. doi: 10.1111/j.1432-1033.1989.tb14607.x. [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha R. R., Green R., Noller H. F. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995 Sep 28;377(6547):309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- Spirin A. S. Ribosomal translocation: facts and models. Prog Nucleic Acid Res Mol Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- Swart G. W., Parmeggiani A., Kraal B., Bosch L. Effects of the mutation glycine-222----aspartic acid on the functions of elongation factor Tu. Biochemistry. 1987 Apr 7;26(7):2047–2054. doi: 10.1021/bi00381a038. [DOI] [PubMed] [Google Scholar]
- Tubulekas I., Hughes D. A single amino acid substitution in elongation factor Tu disrupts interaction between the ternary complex and the ribosome. J Bacteriol. 1993 Jan;175(1):240–250. doi: 10.1128/jb.175.1.240-250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel J. M., Archer R. H., Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984 Feb 24;12(4):2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]



