Abstract
We set out to deorphanize a subset of putative Drosophila odorant receptors expressed in trichoid sensilla using a transgenic in vivo misexpression approach. We identified farnesol as a potent and specific activator for the orphan odorant receptor Or83c. Farnesol is an intermediate in juvenile hormone biosynthesis, but is also produced by ripe citrus fruit peels. Here, we show that farnesol stimulates robust activation of Or83c-expressing olfactory neurons, even at high dilutions. The CD36 homolog Snmp1 is required for normal farnesol response kinetics. The neurons expressing Or83c are found in a subset of poorly characterized intermediate sensilla. We show that these neurons mediate attraction behavior to low concentrations of farnesol and that Or83c receptor mutants are defective for this behavior. Or83c neurons innervate the DC3 glomerulus in the antennal lobe and projection neurons relaying information from this glomerulus to higher brain centers target a region of the lateral horn previously implicated in pheromone perception. Our findings identify a sensitive, narrowly tuned receptor that mediates attraction behavior to farnesol and demonstrates an effective approach to deorphanizing odorant receptors expressed in neurons located in intermediate and trichoid sensilla that may not function in the classical “empty basiconic neuron” system.
Keywords: behavior, empty neuron, intermediate sensilla, olfaction, orphan receptor, receptor mutant
Introduction
In insects, the antenna is the major substrate for detection of volatile pheromones and food odorants (Ha and Smith, 2009; Ronderos and Smith, 2009). Approximately 500 hair-like sensilla, each housing the dendrites of one to four olfactory neurons, cover its surface (Fig. 1A; Stocker, 1994). The sensilla are classified into four morphological groups, the trichoid, basiconic, coeloconic, and intermediate sensilla (Stocker, 1994; Shanbhag et al., 1999). Most Drosophila olfactory neurons have been characterized according to the sensillum class in which they are located, by the odorant receptor (OR) they express, by which odorants they detect, and by the glomerular targets they innervate in the antennal lobes of the brain (Vosshall et al., 2000; Couto et al., 2005; Benton et al., 2009). However, many of the 62 Drosophila ORs remain orphans with unknown chemical specificity or function.
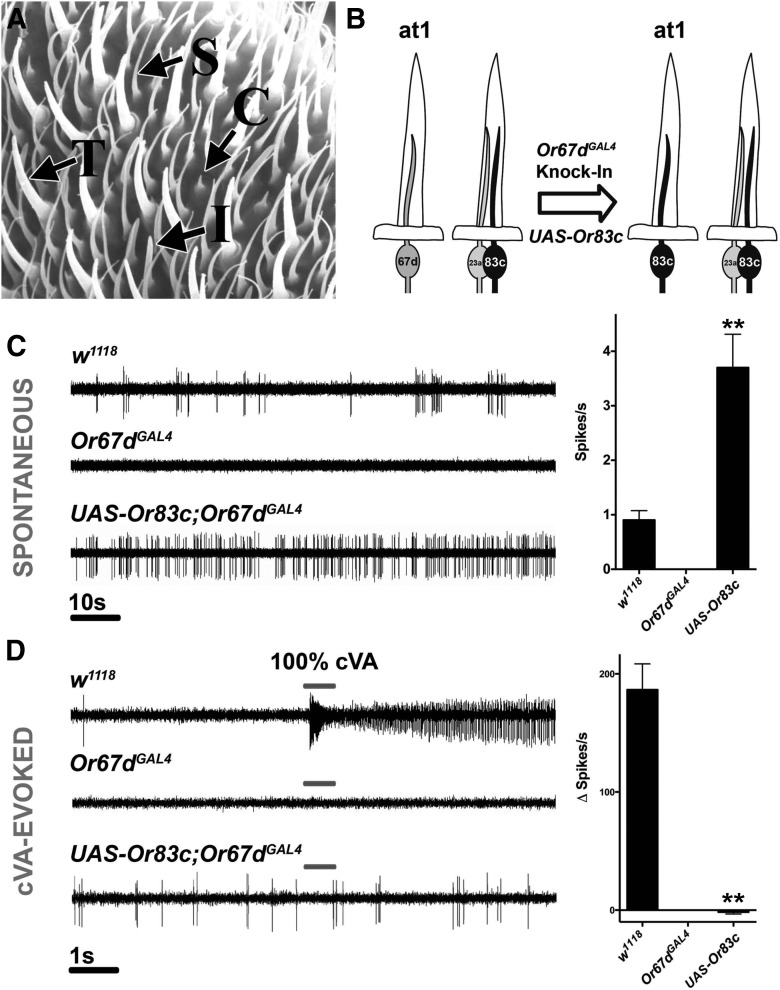
Figure 1.
Misexpression of Or83c in at1 neurons. A, Scanning electron micrograph of antennal surface showing coeloconic (C), trichoid (T), and intermediate (I) sensilla and spinules (S). B, Illustration of the misexpression strategy to characterize Or83c. C, SSR traces showing spontaneous activity of at1 neurons from w1118, Or67dGAL4, or UAS-Or83c;Or67dGAL4 with quantification to the right (**p = 0.0021 between w1118 and Or83c, n = 10–17). D, SSR traces showing cVA-evoked activity of at1 neurons from w1118, Or67dGAL4, and UAS-Or83c;Or67dGAL4 with quantification to the right (**p = 0.0069 between w1118 and Or83c, Student's t test. n = 4–7).
The neurons located in trichoid sensilla are specialized for pheromone detection in many insects. In Drosophila, only one of the nine trichoid receptor neuron classes has been characterized with respect to biological function. The at1 trichoid sensilla contain a single neuron expressing Or67d that is exquisitely tuned to the male-specific pheromone 11-cis-vaccenyl acetate (cVA; Clyne et al., 1997; Ha and Smith, 2006; Kurtovic et al., 2007). Intermediate sensilla, so named because they appear intermediate in morphology between basiconic and trichoid sensilla (Shanbhag et al., 1999), contain olfactory neurons with unknown receptor expression and function.
The Drosophila basiconic receptors were characterized using the “empty neuron” system, in which individual receptors were misexpressed in a defined basiconic neuron that lacked an endogenous tuning receptor (Dobritsa et al., 2003; Hallem and Carlson, 2004, 2006). The unique odorant response profiles of receptors misexpressed in the empty neuron system matched those of endogenous basiconic neuron classes, allowing odorant receptor expression to be correlated with specific functional classes of olfactory neurons. However, expression of trichoid neuron receptors in this basiconic context failed to reveal activating ligands (Hallem and Carlson, 2006). This suggests that trichoid receptors may be narrowly tuned to odorants not tested or may require signaling or processing factors absent in basiconic sensilla. In support of the latter idea, several components essential for the detection of cVA pheromone by at1 trichoid neurons are not broadly expressed in basiconic sensilla, including the odorant-binding protein LUSH and the CD36 homolog Snmp1 (Rogers et al., 1997; Xu et al., 2005; Benton et al., 2007; Jin et al., 2008; Laughlin et al., 2008). Efforts to characterize these ORs in their endogenous sensilla are complicated by the fact that up to four neurons, each expressing up to three receptors, may be colocalized within a sensillum. Therefore, it may not be possible to correlate odor-evoked activity with individual neurons or ORs. To circumvent these problems, we expressed orphan receptors in at1 trichoid neurons lacking the endogenous Or67d tuning receptor. Using this approach, we identified a citrus fruit ligand that activates orphan receptor Or83c and confirmed that this receptor does not function in the basiconic context. We suggest that Or83c functions as a narrowly tuned receptor that participates in localizing citrus fruit substrates.
Materials and Methods
Drosophila stocks.
Wild-type control flies were w1118. The Or67dGAL4 mutants were described previously (Kurtovic et al., 2007). We injected receptor cDNAs regulated by UAS directly into the Or67dGAL4 mutant background to generate transgenic UAS-Or83c flies. Or83cMB11142 is a null mutation generated by a Minos element insertion (Metaxakis et al., 2005) into the first coding exon that was confirmed by sequencing of genomic DNA prepared from mutant animals. Transgenic Snmp1-GAL4 flies were generated using a construct containing a 6.5 kb region 5′ to the start codon of the Snmp1 gene cloned into pCasper4-GAL4 (Pirrotta, 1988). Orco-GAL4 flies were obtained from the Bloomington Stock Center of Indiana University.
Single sensillum electrophysiology.
Extracellular electrophysiological recordings were performed according to Ha and Smith (2006). Briefly, 2- to 7-d-old flies were placed under a constant stream of charcoal-filtered air (36 ml/s; 22–25°C) to prevent any potential environmental odors from affecting activity. Action potentials were recorded by inserting a glass electrode in the base of the sensillum. Signals were amplified 100× (USB-IDAC System; Syntech) and fed into a computer via a 16-bit analog-digital converter and analyzed offline with AUTOSPIKE software (USB-IDAC System; Syntech). The low cutoff filter setting was 200 Hz and the high cutoff setting was 3 kHz. Stimulus consisted of a 300 ms air pulse passed over odorant sources. All recordings were performed from separate sensilla, with a maximum of two sensilla recorded from any single fly. Sensilla were identified based on anatomical location, sensillum morphology, spontaneous activities, and odorant sensitivities. Graphical summaries represent mean ± SEM.
Electroantennography.
Electroantennographs (EAGs) were recorded with capillary glass electrodes (1.5 mm outer diameter) containing Drosophila saline as described previously (Rollmann et al., 2005). The reference electrode was placed in the head capsule close to the base of the antenna. A polished, large diameter (∼40–50 μm) recording electrode was capped onto the anterior distal region of the Drosophila third antennal segment. Control odorant stimulations (1% and 10% cVA) were used to verify that the recording electrode was properly sealed onto the distal antenna. Electrical signals were acquired with an Intelligent Data Acquisition Controller (IDAC-4-USB; Syntech) and quantified by measuring the millivolt value at the greatest deflection in the EAG trace.
Odorant preparation.
Odorants used for electrophysiology stimulus were of the highest purity available and were prepared by placing 30 μl of diluted or undiluted odorant onto a small piece of filter paper inserted into a glass pipette. The odorant dilution value reflects the dilution of odorant applied to the Whatman paper and is much lower at the site of delivery. A similar delivery approach was used with fruit, fruit peel, or live flies. These odorant emitters were placed into a glass pipette with filter paper at the ends. Odorants used to generate the tuning curves in Figure 2, A, B, and E, were undiluted to maximize the probability of a response.
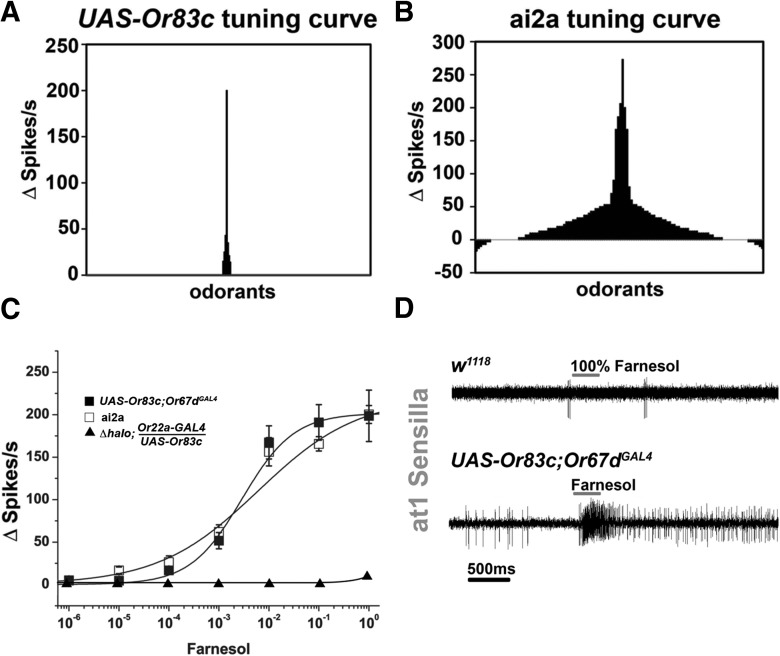
Figure 2.
Or83c detects farnesol and is expressed in ai2 intermediate sensilla. A, Tuning curve for Or83c misexpressed in at1 neurons was determined by plotting each odorant on the x-axis with the responses on the y-axis. The odorants were arranged along the x-axis according to number of spikes elicited, as described in Hallem and Carlson (2006). B, Odorant tuning curve for the endogenous ai2a neuron (n = 5). C, Farnesol-induced dose–response curves from at1 sensilla neurons expressing Or83c (UAS-Or83c;Or67dGAL4), wild-type ai2a intermediate sensilla neurons (ai2), and Or83c expressed in the ab3a empty neuron system (Δhalo;Or22a-GAL4/UAS-Or83c; n = 7). D, SSR traces showing farnesol responses of at1 neurons with and without Or83c.
Projection neuron analyses.
Projection neurons identified in mosaic analysis with a repressible cell marker experiments were traced and registered into a common reference brain and visualized using Amira (Visualization Sciences Group) as described previously (Jefferis et al., 2007; Grosjean et al., 2011). Clustering of projection neuron classes was based on the similarity of axon innervation patterns in the lateral horn, as described previously (Grosjean et al., 2011), with the exception that McQuitty's similarity analysis method was used for the clustering algorithm.
Four-field behavioral assay.
A modified four-quadrant olfactometer (Vet et al., 1983; Semmelhack and Wang, 2009) was used to track the olfactory responses of multiple flies at 30 frames/s. Central air passed through a carbon filter before being split into multiple channels, each regulated by a high-resolution flow meter (Cole-Parmer). Electronically controlled three-way solenoid valves (Automate Scientific) regulated whether clean air leaving the flow meters was expelled into the room or entered custom-made odor chambers (Lundström et al., 2010). Teflon tubing was used for odor delivery. The Teflon fly arena was 19.5 cm × 19.5 cm with a thickness of 0.7 cm. Glass plates were secured onto the arena using clamps. The airflow of each quadrant was maintained at a rate of 100 ml/min and verified by an electronic flow meter before each experiment. Farnesol dilutions in 1000 μl of paraffin oil were used in the odor chamber and paraffin oil alone used in the three nonodor control chambers. Forty to 50 flies with an isogenized genetic background (w1118) were used. At the time of the assay, flies were 4- to 6-d-old and had been starved in vials containing 1% agarose for 22–26 h to increase locomotor activity. The dark arena was illuminated by two infrared LED arrays (Advanced Illumination), monitored by an infrared camera (XC-EI50; Sony), and flies tracked by software described previously (Katsov and Clandinin, 2008). Fly traces were recorded for 5 min and data were analyzed by custom MATLAB scripts (written by Junjie Luo). Attraction index (AI) is defined as (Ot5 − Cavgt5)/(Ot5 + Cavgt5), in which Ot5 is the number of tracked flies in the odor quadrant and Cavgt5 is the average number of tracked flies in nonodor quadrants over a 5 min testing period. On average, each fly generates ∼1800 tracked positions per minute. An AI of 1 indicates that all flies were tracked to the odor quadrant and an AI of 0 indicates that flies were distributed equally to all four quadrants.
Olfactory trap assay.
Olfactory attraction behavior was measured using a two-choice trap assay (Potter et al., 2010). Two to 3-d-old mixed populations of male and female flies (1:1) were starved for 40–42 h in vials containing 1% agarose gel. Approximately 100 flies per assay were cold anesthetized and transferred to a 85 mm × 170 mm, 1000 ml glass jar (02-912-305; Fisher) covered by a 150 × 15 mm Petri dish (351058; Falcon) with two nylon mesh screened holes inserted for ventilation. Odor traps were constructed from 40 ml glass vials (B7999-6; National Scientific) with a custom-built polyethylene top containing a cut pipette tip. Traps contained a cotton foam plug to which either 0.5 ml of different dilutions of farnesol dissolved in paraffin oil or paraffin oil alone was added. Experiments with apple cider vinegar used water as the control odorant. The behavioral tests were conducted for 23–24 h in the dark at room temperature.
Results
Or83c detects farnesol
We generated transgenic flies that misexpress orphan receptor Or83c cDNA in at1 neurons lacking the endogenous receptor Or67d (GenBank submission, Accession no. NM_079520.3; Ha and Smith, 2006; Fig. 1B). Or67dGAL4 is a knock-in mutant in which the Or67d coding sequence was replaced by the GAL4 yeast transcription factor gene (Kurtovic et al., 2007). Or83c is one orphan receptor of interest because it is thought to be the sole tuning factor for one of the trichoid sensilla neurons in at2 sensilla (Couto et al., 2005) and its ligand specificity is unknown.
Using single-sensillum electrophysiology (SSR) to record action potentials from the olfactory neurons within a single sensillum, wild-type at1 neurons were shown to have a typical spontaneous firing rate of ∼1 spike/s in the absence of pheromone and were strongly activated by cVA (Clyne et al., 1997; Xu et al., 2005; Fig. 1C,D). Transgenic flies homozygous for the Or67dGAL4 knock-in on the third chromosome (Kurtovic et al., 2007) lack Or67d tuning receptors and have no spontaneous firing (Fig. 1C). Flies homozygous for both Or67dGAL4 on the third chromosome and UAS-Or83c on the second chromosome express the Or83c receptor in at1 neurons in lieu of Or67d (Fig. 1B–D). Driving Or83c in at1 neurons lacking Or67d restores spontaneous activity, but to a distinct higher average firing rate (Fig. 1C). As expected for an at1 neuron expressing Or83c but not Or67d, these neurons are insensitive to cVA in these transgenic flies (Fig. 1D).
We screened a large odorant panel of ∼150 commercially available volatile chemicals to identify ligands that activate Or83c. We found that a single odorant, farnesol, elicited robust activity, peaking at almost 200 spikes/s. Indeed, no other odorant in the panel produced >50 spikes/s even in undiluted form (Fig. 2A). Importantly, farnesol does not activate wild-type at1 neurons, showing that farnesol sensitivity is dependent on the expression of Or83c (Fig. 2D). Farnesol is a known intermediate in the juvenile hormone biosynthesis pathway (Bellés et al., 2005). Therefore, we tested structurally similar compounds and other intermediates in the juvenile hormone pathway, including farnesal, farnesene, methyl farnesoate, farnesoic acid, farnesol methyl ether, geraniol, geranylgeraniol, and juvenile hormone III (Bellés et al., 2005). Like all other odorants in the panel, none of these compounds elicited activity >50 spikes/s, even in undiluted form (Fig. 2A).
We measured the farnesol dose–response relationship for Or83c expressed in at1 neurons. Or83c reliably detects farnesol dilutions down to 1:10,000 when spotted on filter paper and mixed with air (see Materials and Methods for details; Fig. 2C). Saturating firing rates were achieved for dilutions at or above 1:100 dilution. The narrow tuning curve for this receptor combined with high sensitivity to farnesol leads us to conclude that Or83c is selectively activated by farnesol.
Or83c is expressed in intermediate sensilla neurons
Previous in situ hybridization studies reported that Or83c was expressed in at2 trichoid sensilla neurons (Couto et al., 2005). We surveyed wild-type trichoid sensilla to confirm the presence of an endogenous farnesol-activated neuron. Surprisingly, extensive probing failed to identify any trichoid neurons that were activated by farnesol. We therefore tested intermediate sensilla and immediately identified an intermediate sensillum class containing two neurons (Shanbhag et al., 1999), one of which was strongly activated by farnesol. We call this sensillum class “ai2” (antennal intermediate 2) and the neurons within it “ai2a” and “ai2b” (in descending order of spike amplitude), where the “a” neuron is farnesol sensitive (Fig. 3A,F, Table 1). We also identified a second intermediate sensillum class referred to as “ai3” because it contains three olfactory neurons (Shanbhag et al., 1999). None of the ai3 neurons was sensitive to farnesol.
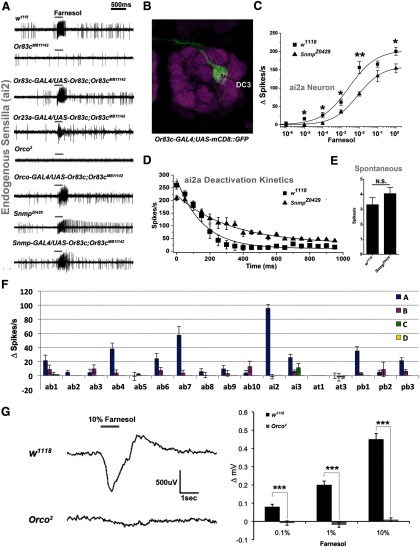
Figure 3.
Farnesol responses require Or83c, Orco, and Snmp1. A, SSR traces from the indicated genotypes in response to undiluted farnesol. Wild-type ai2a neurons respond strongly to farnesol, but these responses are absent in Or83c mutants. Farnesol sensitivity of Or83cMB11142 ai2a neurons is rescued by UAS-Or83c driven by Or83c-GAL4, Orco-GAL4, or Snmp1-GAL4, and may be conferred to the ai2b neuron using Or23a-Gal4. Farnesol responses are completely absent in Orco2 flies and display a deactivation defect in Snmp1Z0429 mutant flies. B, Or83c-GAL4 drives reporter expression exclusively in neurons that innervate the DC3 glomerulus of the antennal lobe. C, Dose–response curves for wild-type (n = 5) and Snmp1Z0429 (n = 7) ai2a neurons to farnesol. Snmp1Z0429 mutants show a significant reduction in sensitivity (*p < 0.05, **p < 0.01, Student's t test; actual p-values starting from the lowest concentration: p = 0.126708, *0.015129, *0.039771, *0.029717, **0.001232, 0.108864, and *0.012554, Student's t test). D, Deactivation kinetics of ai2a neurons from Snmp1Z0429 stimulated by undiluted farnesol is significantly prolonged. Deactivation time constants (τ) were calculated for w1118 control and Snmp1Z0429 mutant flies (τ = 151.9 ± 21.8 for w1118, τ = 313.8 ± 67.1 for Snmp1Z0429, n = 7 each, **p < 0.01, Student's t test). E, Spontaneous activity of Snmp1Z0429 mutants (n = 8) is not significantly affected in ai2a neurons (p = 0.145980) compared with w1118 (n = 12). F, Summary of sensillum survey using SSR to measure responses to 1% farnesol (n = 3–6 each). G, Representative EAG traces of w1118 and Orco2 mutant flies stimulated for 1 s with 10% farnesol. The relative change in maximal EAG deflection amplititudes are shown for w1118 and Orco2 mutant flies stimulated by different farnesol concentrations (n = 10–12 each, ***p = 0.0001, 2.8 × 10−8, and 9 × 10−11 from left to right).
Table 1.
Current structural organization of intermediate and trichoid sensilla
| Class | Antennal intermediate | Antennal trichoid | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sensillum | ai2 (formerly at2) | ai3 (formerly at3) | at1 | at3 (formerly at4) | |||||
| Neuron | ai2a | ai2b | ai3a | ai3b* | ai3c* | at1 | at3a* | at3b* | at3c* |
| Receptor | Or83c | Or23a | Or19a-b | Or2a | Or43a | Or67d | Or47b | Or65a-c | Or88a |
*The last letter in the name of the olfactory neuron is assigned based on spike amplitude when measured by SSR, which is not yet correlated with receptor expression for these neurons.
Does the farnesol-sensitive ai2a neuron express Or83c? The odorant tuning curve of ai2a resembles that of Or83c expressed in at1 neurons, although with a wider base reflecting weak responses to a large number of undiluted odorants (compare Fig. 2A,B). The ai2a neuron also had farnesol sensitivity properties nearly identical to misexpressed Or83c (Fig. 2C). To establish conclusively that Or83c is expressed in ai2a neurons, we tested the farnesol sensitivity of Or83cMB11142 mutants (Bellen et al., 2004; Metaxakis et al., 2005). These mutants carry a tranposable element in a coding exon of Or83c that is predicted to eliminate expression (Bellen et al., 2004). Indeed, SSR analysis reveals that the ai2a neurons from Or83cMB11142 mutants are completely insensitive to farnesol (Fig. 3A).
To confirm that the farnesol sensitivity defects in Or83cMB11142 mutants result exclusively from loss of Or83c, we expressed the Or83c cDNA under control of the Or83c-GAL4 driver in the Or83cMB11142 mutant background (Fig. 3A). This driver specifically labels Or83c neurons, which innervate the DC3 glomerulus (Grosjean et al., 2011; Fig. 3B). This transgenic rescue restored farnesol sensitivity in ai2a neurons of Or83cMB11142 flies (Fig. 3A). The Or23a-GAL4 driver (Couto et al., 2005) drives gene expression in the other olfactory neuron that shares the ai2 sensilla, ai2b. Driving UAS-Or83c with Or23a-Gal4 in the Or83cMB11142 mutant background confers farnesol sensitivity to ai2b neurons (Fig. 3A). We conclude that Or83c is a farnesol receptor normally expressed in ai2a intermediate sensilla neurons and that this receptor is necessary and sufficient to confer farnesol sensitivity to both trichoid and intermediate sensilla neurons.
Orco and Snmp1 are required for normal farnesol responses in ai2a neurons
Drosophila odorant receptors are ligand-gated ion channels that require the coreceptor Orco both for dendritic trafficking (Larsson et al., 2004) and odorant responses (Nakagawa et al., 2005; Benton et al., 2006; Sato et al., 2008; Wicher et al., 2008). To confirm that farnesol detection by ai2a neurons requires Orco as a coreceptor, we tested the farnesol sensitivity of these neurons in Orco2 mutant flies (Larsson et al., 2004). ai2a neurons from Orco2 mutants are insensitive to farnesol (Fig. 3A). ai2a neurons normally express Orco, since expression of Or83c using Orco-GAL4 rescues the Or83cMB11142 mutant farnesol insensitivity (Fig. 3A).
Snmp1 is a member of the CD36 protein family and functions as an obligate coreceptor for cVA pheromone sensitivity (Benton et al., 2007; Jin et al., 2008), but is also expressed in nine classes of olfactory sensory neurons, including the Or83c-expressing neurons (Benton et al., 2007) and a single basiconic neuron that is sensitive to the repellant geosmin (Stensmyr et al., 2012). We investigated whether Snmp1 function was similarly required in ai2a neurons for farnesol detection. Interestingly, although they were still activated by farnesol, ai2a neurons from Snmp1Z0429 mutants have defective response kinetics (Fig. 3A,C,D). The time to peak onset, the maximal firing rate, and deactivation kinetics are all impaired in Snmp1Z0429. Deactivation is ∼2 times slower in Snmp1Z0429 mutants compared with w1118 controls (τ = 151.9 ± 21.8 for w1118, and τ = 313.8 ± 67.1 for Snmp1Z0429, n = 6; Fig. 3D). Unlike at1 neurons, spontaneous activity in ai2a neurons is not significantly altered in Snmp1Z0429 (Fig. 3E). We confirmed that Snmp1 is expressed in the ai2a neurons by driving Or83c with the Snmp1 promoter in Or83cMB11142 mutant background, which restored farnesol sensitivity (Fig. 3A). We conclude that Or83c, Orco, and Snmp1 proteins are all normally expressed in ai2a neurons and are required for normal farnesol-induced responses.
Finally, we expressed Or83c in the “empty neuron” basiconic system, in which the ORs are expressed in ab3a basiconic sensilla neurons lacking the endogenous tuning receptor Or22a (Dobritsa et al., 2003). Or83c failed to confer farnesol sensitivity when misexpressed in a basiconic neuron context (Fig. 2C), confirming that basiconic sensilla lack factors necessary for Or83c function and validating the utility of the trichoid misexpression system to study orphan receptors expressed in intermediate and trichoid sensilla.
We screened other olfactory neurons located in the antenna and maxillary palps for farnesol sensitivity using SSR and assayed general antennal farnesol responses using EAG. Most of the basiconic neurons failed to respond or responded weakly to farnesol in SSR recordings (Fig. 3F). The next highest response to farnesol, after ai2a neurons, came from ab7a neurons (Fig. 3F), which are broadly tuned and strongly activated by multiple odorants (Hallem and Carlson, 2006). ab7a responses to farnesol are relatively weak compared with most of the other odorants we tested on this sensilla. To look more globally for farnesol-responsive olfactory neurons, we performed EAG recordings with farensol to measure summed activity from all olfactory neurons in wild-type and Orco2 mutants. EAGs from wild-type showed a significant response to farnesol that was completely abolished in Orco2 mutants, which lack functional OR coreceptors (Benton et al., 2006; Fig. 3G). This finding indicates that farnesol detection is mediated by orco-dependent ORs and rules out ionotropic glutamate receptors (IRs) or antennal gustatory receptors as contributing to farnesol detection (Benton et al., 2009). Together, these experiments indicate that ai2a neurons are the most sensitive and selective farnesol detectors in the Drosophila olfactory system.
Farnesol stimulates attraction behavior in Drosophila
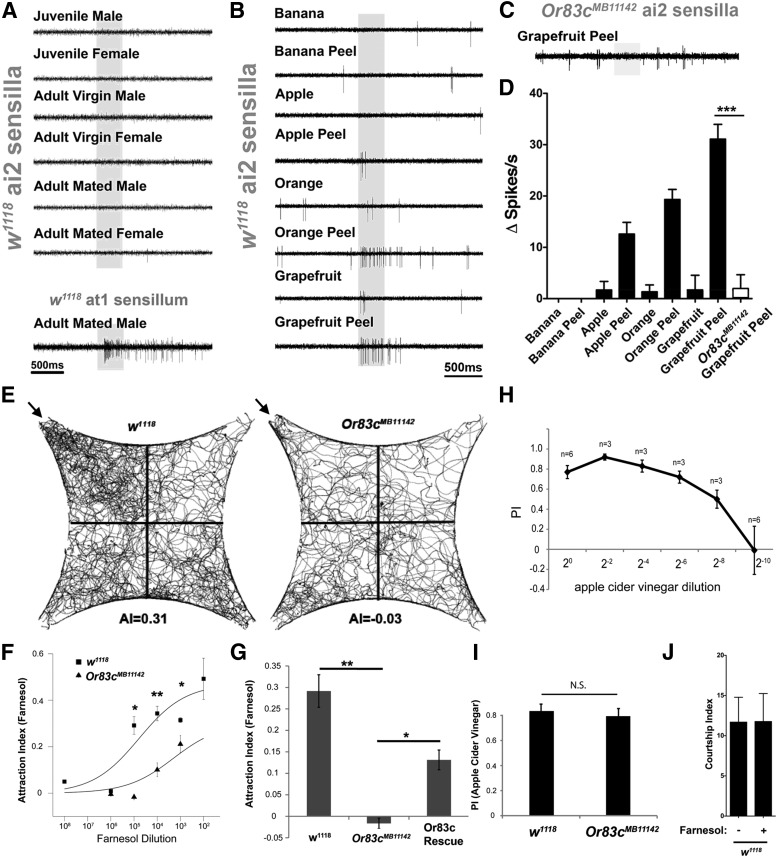
The presence of a narrowly tuned farnesol receptor implies an important ecological role for farnesol detection in Drosophila melanogaster. Farnesol is an intermediate in the juvenile hormone biosynthetic pathway (Bellés et al., 2005) and could potentially function as a volatile pheromone. However, none of the volatiles emitted from live animals at any life stage induced significant activity in the ai2a neurons (Fig. 4A) and farnesol had no effect on mating (Fig. 4J). These data suggest that farnesol may not be a pheromone for flies. Therefore, we considered other sources of farnesol that might have an ecological role. Farnesol is found in the peel of citrus fruits including grapefruit (Njoroge et al., 2005) and oranges (Nigg et al., 1994). We tested the volatiles from several different fruits and fruit peels associated with Drosophila food sources and found that peels known to contain farnesol reliably activated ai2a neurons (Fig. 4B–D). The highest responses were elicited by grapefruit rind and these responses were abolished in Or83cMB11142 mutants (Fig. 4C,D). Interestingly, farnesol is present in the rind but not the flesh of citrus fruits (Choi et al., 2001; Njoroge et al., 2005) and the volatiles from the rind but not the flesh of these fruits activated ai2a neurons (Fig. 4B,D). Although we cannot rule out the possibility that another rind-specific volatile activates Or83c, it is likely that farnesol is the relevant odorant, implicating a role for these neurons in detecting citrus fruit peels.
Figure 4.
Or83c neurons are activated by citrus odors, not fly odors. A, Volatiles from flies fail to activate ai2 sensilla neurons. SSR traces from ai2 sensilla neurons from wild-type flies. Approximately 50 flies of the indicated age and mating status were placed into a glass Pasteur pipette and air was passed through the pipette and introduced into the constant airstream (see Materials and Methods). As a positive control, cVA-secreting male flies produce volatiles that activate at1 neurons. B, Volatiles from the peel of citrus fruits activate ai2 neurons in vivo. SSR traces from wild-type ai2 sensilla in response to air passed over the indicated fruits or peels. C, ai2a sensitivity to citrus peel is Or83c dependent. Shown is an SSR trace from Or83cMB11142 ai2 sensilla stimulated with grapefruit peel. D, Quantification of results from experiments described in B and C. Error bars represent SEM and Student's t test was used to compare w1118 (black bars) and Or83cMB11142 (white bar) responses to grapefruit peel (n = 3–10 for each, ***p = 0.0001). E, Or83c mediates farnesol-dependent attraction behavior. Shown are tracks of 50 flies recorded for 5 min in the 4-quadrant behavior assay. Introduction of farnesol to the top left quadrant (arrows) results in wild-type flies spending more time in this quadrant (10−2 dilution), providing a measure of attraction behavior. F, Dose–response curves for attraction of wild-type or Or83cMB11142 mutant flies to farnesol in the four-quadrant assay. Attraction behavior was significantly reduced in Or83cMB11142 mutant flies at all farnesol concentrations and was abolished at low concentrations (105-103 dilution; *p < 0.05, **p < 0.01, p-values starting from 106 to103 dilution: p = 0.7985, **0.0039, *0.0117, *0.0474, Student's t test). G, Transgenic expression of the Or83c receptor in the Or83c mutant background significantly rescued attraction to 10−5 farnesol (n = 4–6 for each experiment. Or83c rescue: Or83c-GAL4/UAS-Or83c;Or83cMB11142 (error bars indicate SEM, *p < 0.05, **p < 0.01; actual p-values: p = **0.0039 (mutant), *0.0125 (rescue), Student's t test. H, Dose–response curve of starved wild-type (w1118) flies toward apple cider vinegar (ACV) in the two-choice trap assay. PI, preference index. I, Attraction performance index of starved wild-type and Or83cMB11142 mutants toward a 1:16 dilution of ACV were not statistically different, demonstrating that Or83c mutants are capable of attraction behavior. J, Courtship index for wild-type flies is not affected by the presence of farnesol.
We next tested whether farnesol alone could drive Or83c-dependent olfactory behaviors. We first determined whether farnesol was behaviorally attractive or repulsive to Drosophila. To test innate olfactory behaviors, we used a four-quadrant olfactometer in which odorant is introduced into one quadrant and clean air into the other three quadrants (Vet et al., 1983; Semmelhack and Wang, 2009; see Materials and Methods). We found that wild-type flies were robustly attracted by farnesol even at dilutions of 1:100,000 in air (Fig. 4E). Farnesol attraction in Or83cMB11142 mutant flies was significantly impaired at all concentrations tested and completely abolished at 105 or greater dilutions (Fig. 4E–G). This defect was rescued by transgenic expression of a single copy of Or83c in the ai2a neuron, thereby supporting the conclusion that the mutant farnesol attraction defect was due to the lack of Or83c (Fig. 4G).
In an independent attraction behavioral paradigm, we performed olfactory two-choice bait trap assays in which starved motile flies in a large arena were allowed to choose between vials containing an odorant or a solvent control (Larsson et al., 2004; Potter et al., 2010; Fig. 5A). Wild-type flies showed significantly higher preference for farnesol compared with paraffin control, with robust farnesol preference maintained even at a 1:10,000 dilution (Fig. 5B). In Or83cMB11142 mutant flies, olfactory attraction behavior to farnesol was completely abolished (Fig. 5B). Transgenic expression of the Or83c receptor in the Or83cMB11142 mutant flies rescued attraction behavior to farnesol (Fig. 5C). As a positive control, we assayed the attraction behavior of starved flies to the potently attractive odor apple cider vinegar (Semmelhack and Wang, 2009; Ai et al., 2010). Wild-type flies were significantly attracted to the vial containing up to a 512-fold dilution of apple cider vinegar (Fig. 4H). Attraction behavior of Or83cMB11142 flies remained intact toward apple cider vinegar (Fig. 4I), indicating that mutant flies could exhibit normal locomotor and attraction activity toward other odors. Together, these data indicate that Or83c mediates farnesol-induced attraction behavior in Drosophila by activating ai2a neurons.
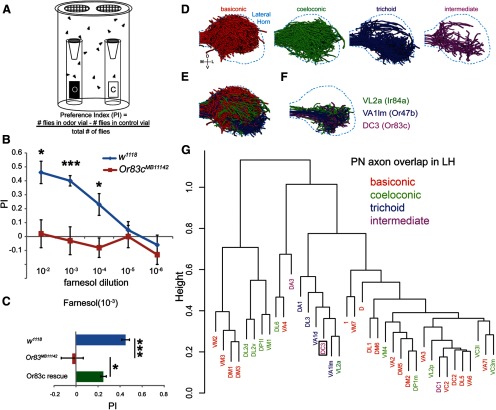
Figure 5.
Or83c mediates farnesol-induced attraction behavior. A, Diagram showing the two-choice behavioral attraction assay. The formula for calculating the attraction preference index (PI) is also shown. B–C, Or83c mediates farnesol-induced attraction behavior. B, Wild-type flies display strong attraction behavior toward farnesol, which is abolished in Or83cMB11142 mutant flies (n = 4–6 for each experiment). C, Bar graphs depicting the attraction performance index of mutant and wild-type flies toward a 1:1000 dilution of farnesol. Attraction behavior in the Or83cMB11142 mutant flies can be significantly rescued by transgenic expression of Or83c in the Or83c neuron (the Or83c rescue genotype is Or83c-GAL4/UAS-Or83c;Or83cMB11142. Error bars indicate SEM (*p < 0.05, **p < 0.01; actual p-values starting from 102 to 106 dilution: p = *0.0141, **0.0012, *0.039771, 0.5976, 0.4309, p value of Or83c rescue = *0.0468, Student's t test.). D–G, DC3 (Or83c) projection neurons target pheromone-processing centers in the brain. D, E, Computer modeling of 37 different classes of PN axons innervating the lateral horn region of the Drosophila brain. PN axons are color coded according to sensillar class. F, DC3 PN axons (purple) significantly overlap with VA1lm (blue) and VL2a (green) axons in brain regions previously hypothesized to be dedicated solely to courtship behaviors (Jefferis et al., 2007; Grosjean et al., 2011). G, Clustering analysis of axon overlap in the lateral horn among 37 classes of projection neurons highlights relationships between sensillar class and innervation of the lateral horn. DC3 (boxed) clusters with trichoid signaling projection neurons and not food-odor signaling (basiconic) projection neurons. The y-axis (height) reflects normalized overlap scores based on McQuitty's method (see Materials and Methods). LH, Lateral horn.
Projection neurons relay Or83c signals to pheromone-processing centers
Because Or83c is likely to mediate attraction behavior, we investigated whether farnesol might activate higher-order brain regions that process attractive olfactory food signals. Most olfactory neurons expressing a single tuning odorant receptor target a single antennal lobe glomerulus, which in turn contacts one type of output projection neuron (PN) to send signals to higher brain centers such as the mushroom body calyx and lateral horn (Fig. 5D–F; Jefferis et al., 2007; Min et al., 2013). PNs from the food sensing neurons innervate a distinct domain of the lateral horn compared with the PNs that relay pheromone information (Jefferis et al., 2007). Or83c olfactory neurons target the DC3 glomerulus (Fig. 3B) and therefore signal primarily to DC3 projection neurons (Jefferis et al., 2007). Surprisingly, DC3 projection neuron axons target the lateral horn in a region that overlaps with PNs relaying pheromone information from glomeruli VA1lm and VL2a (Fig. 5F,G). The VA1lm glomerulus is innervated by trichoid neurons expressing Or47b (Couto et al., 2005; Fishilevich and Vosshall, 2005), which is activated by female odors (Masuyama et al., 2012). The ORNs that target the VL2a glomerulus express Ir84a and are narrowly tuned to the food odor phenylacetic acid, which augments male courtship behaviors and innervates the pheromone-activated domain of the lateral horn (Grosjean et al., 2011). As mentioned previously, we found no changes in courtship index when mating pairs were exposed to farnesol (Fig. 4J). These findings indicate that farnesol odorant information targets a brain region that was previously associated with pheromone-induced courtship behaviors, but which apparently may also be used for chemoattraction.
Discussion
Or83c is a farnesol sensor
Basiconic sensilla typically contain neurons expressing broadly tuned odorant receptors that are activated directly by food odors (de Bruyne et al., 2001; Hallem et al., 2004), although a few receptors are narrowly tuned (Suh et al., 2004; Jones et al., 2007; Kwon et al., 2007; Stensmyr et al., 2012). Coeloconic neurons typically express IRs that detect humidity and a variety of volatile compounds including acids, ammonia, and the insect repellant DEET (Yao et al., 2005; Benton et al., 2009; Kain et al., 2013; Min et al., 2013). Trichoid sensilla contain olfactory neurons specialized for pheromone detection in most insects. Or83c-expressing neurons were previously categorized as trichoid neurons based on in situ experiments (Couto et al., 2005), but we show that these neurons are actually located in intermediate sensilla. The intermediate and trichoid sensilla were previously characterized and named based on morphology and the number of neurons contained (Shanbhag et al., 1999). Accordingly, we refer to the intermediate sensillum containing two neurons as “ai2” (previously called “at2” in Couto et al., 2005) and we refer to the intermediate sensillum with three neurons as “ai3” (Table 1). A recent study refers to the intermediate sensillum containing three neurons as “ai2” (Dweck et al., 2013). In the interest of consistency and clarity, we propose the intermediate nomenclature described in Table 1.
We found that Or83c is a narrowly tuned odorant receptor selectively activated by farnesol and expressed in neurons located in intermediate sensilla. Flies are attracted to farnesol and this attraction is defective in flies lacking Or83c expression. We did not find farnesol emitted by Drosophila flies, pupae, or larvae, suggesting that farnesol does not act as a pheromone cue. However, volatiles from citrus fruit peels known to contain farnesol could reliably activate Or83c-expressing neurons. Given the known presence of farnesol in these rinds and the farnesol-selective activation of Or83c, it is very likely that the odorant emitted by the fruit rinds is farnesol. Drosophila encounter farnesol in citrus fruit peels and consume the yeast that grows on them (Begon, 1982; Choi et al., 2001; Njoroge et al., 2005). In addition, it has been shown recently that Drosophila females prefer to lay their eggs in citrus fruit peels (Dweck et al., 2013). This preference is mediated by Or19a neurons expressed in ai3 intermediate sensilla that are tuned to detect valencene and other citrus volatiles. Nonetheless, the citrus volatiles that activate Or19a neurons do not promote olfactory attraction (Dweck et al., 2013). Because farnesol is attractive even at very low concentrations, our results suggests that Drosophila might use farnesol and Or83c activation to guide long-range attraction to citrus fruit peels followed by activation of Or19a neurons by other citrus volatiles to guide egg-laying decisions. If farnesol detection is conserved in other, more destructive insect species, then farnesol may be a useful component of lures and traps to protect citrus crops.
Or83c function requires Snmp1 and other factors not found in basiconic sensilla
Misexpression of Or83c in basiconic neurons fails to confer farnesol sensitivity. However, misexpression in at1 neurons confers farnesol sensitivity, as does misexpression in Or23a-expressing neurons located in intermediate sensilla neurons that are normally insensitive to this odorant. This indicates that there are farnesol sensitivity factors expressed in intermediate and trichoid sensilla that are lacking in basiconic sensilla. We show that Snmp1 is one factor lacking in most basiconic sensilla that is important for normal responses to farnesol in trichoid and intermediate sensilla. However, when Or83c is expressed in intermediate or trichoid sensilla lacking Snmp1 (in Snmp1Z0249) it can still respond to farnesol, but when Or83c is expressed in basiconic sensilla neurons, the responses are completely abolished. This suggests that there are likely additional, as yet unknown factors in intermediate and trichoid sensilla that are required for Or83c function. Interestingly, ab4b is the only basiconic neuron expressing Snmp1 (Benton et al., 2007) and these neurons are narrowly tuned to detect geosmin, a microbial-produced volatile that alerts flies to the presence of potentially toxic molds and bacteria (Stensmyr et al., 2012). The Or19a neurons shown recently to mediate oviposition toward citrus substrates also express Snmp1 (Benton et al., 2007; Dweck et al., 2013), but it is not known whether Snmp1 plays any role in the detection of either valencene or geosmin. However, it is interesting that all of the Snmp1-expressing olfactory neurons characterized to date appear to have relatively specialized functions.
Farnesol-detecting circuits innervate the pheromone zone of the lateral horn
Neurons located in trichoid sensilla typically mediate responses to pheromones (Clyne et al., 1997; Xu et al., 2005) and basiconic neurons respond primarily to food odorants (Hallem and Carlson, 2004, 2006). The Or83c DC3 PNs innervate higher brain centers also targeted by PNs from VA1lm (Or47b) and VL2a (Ir84a) glomeruli (Grosjean et al., 2011). This region of the lateral horn is associated with mating behaviors, but we show here that farnesol signaling leads to food-seeking behaviors despite innervating this region of the lateral horn. What might account for this discrepancy in wiring and function? One possibility is that the farnesol-activated projection neurons synapse with different populations of lateral horn neurons than those associated with mating, thus activating distinct circuits. VA1lm and VL2a are two of three projection neuron populations (the third being DA1) that express the sexually dimorphic fruitless (FruM) transcription factor (Stockinger et al., 2005), which is required for establishing the entire olfactory neural circuit from olfactory neurons to motor neurons that regulate mating behaviors (Ruta et al., 2010). DC3 PNs, which do not express FruM, might not integrate into this courtship circuit, perhaps targeting interneurons that regulate other olfactory behaviors. Another possibility is that organization of the lateral horn might not be a simple pheromone versus food division, as postulated previously, but instead may predominately reflect continuation of a sensillar organization (basiconic vs intermediate vs trichoid vs coeloconic) into higher brain centers. Nonetheless, the surprising overlap of courtship and food attraction in the lateral horn requires future investigation on how these different olfactory signals are further processed to mediate distinct behaviors.
Finally, we show here that Or83c is a selective farnesol receptor that can confer farnesol sensitivity on intermediate and trichoid olfactory neurons in a cell autonomous fashion. Therefore, misexpression of Or83c in other trichoid or intermediate olfactory neurons in the Or83cMB11142 mutant background may provide an odorant-driven strategy to gain insights into the behavioral outputs of these circuits.
Notes
Supplemental material for this article is available at https://docs.google.com/spreadsheet/ccc?key=0ArzPfdWRURgqdG1nT1k5T2RPeWxKRlZaWXJrNjZqVkE&usp=sharing. This material has not been peer reviewed.
Footnotes
This work was supported by the National Institutes of Health (Grants R01 DCD011751 and R01 DCD02539 to D.P.S. and R01 DC01307901 to C.J.P.) and the Whitehall Foundation (Grant 2011-05-04 to C.J.P.). We thank Mariya E. Cameron for help generating UAS-Or83c transgenic fly lines; Samarpita Sengupta for collecting part of the data used to construct Figure 1D; Tal Soo Ha, Olena Riabinina, and Greg Jefferis for assistance with data analysis; Junjie Luo for the use of custom MATLAB scripts; Rob Rawson for juvenile hormone III; Barry Dickson for Or83c-GAL4, Or23a-Gal4 and Or67dGAL4 flies; the Center for Sensory Biology Imaging Facility (funded by NIH Grant P30DC005211) for LSM700 confocal microscopy; and Terry Shelley for custom equipment fabrication.
The authors declare no competing financial interests.
References
- Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GS. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon M. Yeasts and Drosophila. In: Ashburner M, Carson HL, Thompson JN Jr, editors. The genetics and biology of Drosophila. Vol 3A. London: Academic; 1982. pp. 345–384. [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, Hoskins RA, Spradling AC. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellés X, Martín D, Piulachs MD. The mevalonate pathway and the synthesis of juvenile hormone in insects. Annu Rev Entomol. 2005;50:181–199. doi: 10.1146/annurev.ento.50.071803.130356. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Kondo Y, Sawamura M. Characterization of the odor-active volatiles in citrus hyuganatsu (Citrus tamurana Hort. ex Tanaka) J Agric Food Chem. 2001;49:2404–2408. doi: 10.1021/jf001467w. [DOI] [PubMed] [Google Scholar]
- Clyne P, Grant A, O'Connell R, Carlson JR. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/S0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/S0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Dweck HK, Ebrahim SA, Kromann S, Bown D, Hillbur Y, Sachse S, Hansson BS, Stensmyr MC. Olfactory preference for egg laying on citrus substrates in Drosophila. Curr Biol. 2013;23:2472–2480. doi: 10.1016/j.cub.2013.10.047. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Grosjean Y, Rytz R, Farine JP, Abuin L, Cortot J, Jefferis GS, Benton R. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478:236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- Ha TS, Smith DP. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J Neurosci. 2006;26:8727–8733. doi: 10.1523/JNEUROSCI.0876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha TS, Smith DP. Odorant and pheromone receptors in insects. Front Cell Neurosci. 2009;3:10. doi: 10.3389/neuro.03.010.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. The odor coding system of Drosophila. Trends Genet. 2004;20:453–459. doi: 10.1016/j.tig.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Ha TS, Smith DP. Snmp is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci U S A. 2008;105:10996–11001. doi: 10.1073/pnas.0803309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kain P, Boyle SM, Tharadra SK, Guda T, Pham C, Dahanukar A, Ray A. Odour receptors and neurons for DEET and new insect repellents. Nature. 2013;502:507–512. doi: 10.1038/nature12594. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Katsov AY, Clandinin TR. Motion processing streams in Drosophila are behaviorally specialized. Neuron. 2008;59:322–335. doi: 10.1016/j.neuron.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Laughlin JD, Ha TS, Jones DN, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell. 2008;133:1255–1265. doi: 10.1016/j.cell.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström JN, Gordon AR, Alden EC, Boesveldt S, Albrecht J. Methods for building an inexpensive computer-controlled olfactometer for temporally-precise experiments. Int J Psychophysiol. 2010;78:179–189. doi: 10.1016/j.ijpsycho.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26:89–102. doi: 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Ai M, Shin SA, Suh GS. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 2013;110:E1321–E1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- Nigg HN, Mallory LL, Simpson SE, Callaham SB, Toth JP, Fraser S, Klim M, Nagy S, Nation JL, Attaway JA. Caribbean fruit fly, Anastrepha suspensa (Loew), attraction to host fruit and host kairomones. J Chem Ecol. 1994;20:727–743. doi: 10.1007/BF02059609. [DOI] [PubMed] [Google Scholar]
- Njoroge SM, Koaze H, Karanja PN, Sawamura M. Volatile constituents of redblush grapefruit (Citrus paradisi) and pummelo (Citrus grandis) peel essential oils from Kenya. J Agric Food Chem. 2005;53:9790–9794. doi: 10.1021/jf051373s. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez RL, Denhart DT, editors. Vectors: a survey of molecular cloning vectors and their uses. Boston: Butterworths; 1988. pp. 437–456. [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ME, Sun M, Lerner MR, Vogt RG. Snmp-1, a novel membrane protein of olfactory neurons of the silk moth Antherea polyphemus with homology to the CD36 family of membrane proteins. J Biol Chem. 1997;272:14792–14799. doi: 10.1074/jbc.272.23.14792. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Mackay TF, Anholt RR. Pinocchio, a novel protein expressed in the antenna, contributes to olfactory behavior in Drosophila melanogaster. J Neurobiol. 2005;63:146–158. doi: 10.1002/neu.20123. [DOI] [PubMed] [Google Scholar]
- Ronderos DS, Smith DP. Diverse signaling mechanisms mediate volatile odorant detection in Drosophila. Fly (Austin) 2009;3:290–297. doi: 10.4161/fly.9801. [DOI] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Semmelhack JL, Wang JW. Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature. 2009;459:218–223. doi: 10.1038/nature07983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, Müller B, Steinbrecht RA. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. International Journal of Insect Morphology and Embryology. 1999;28:377–397. doi: 10.1016/S0020-7322(99)00039-2. [DOI] [Google Scholar]
- Stensmyr MC, Dweck HK, Farhan A, Ibba I, Strutz A, Mukunda L, Linz J, Grabe V, Steck K, Lavista-Llanos S, Wicher D, Sachse S, Knaden M, Becher PG, Seki Y, Hansson BS. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151:1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Suh GS, Wong AM, Hergarden AC, Wang JW, Simon AF, Benzer S, Axel R, Anderson DJ. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- Vet L, Lenteren J, Heymans M, Meelis E. An airflow olfactometer for measuring olfactory responses of hymenopterous parasitoids and other small insects. Physiological Entomology. 1983;8:97–106. doi: 10.1111/j.1365-3032.1983.tb00338.x. [DOI] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/S0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Xu P, Atkinson R, Jones DN, Smith DP. Drosophila OBP LUSH is required for activity of Pheromone-sensitive neurons. Neuron. 2005;45:193–200. doi: 10.1016/j.neuron.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]