Abstract
Self-reported race is generally considered the basis for racial classification in social surveys, including the U.S. census. Drawing on recent advances in human molecular genetics and social science perspectives of socially constructed race, our study takes into account both genetic bio-ancestry and social context in understanding racial classification. This article accomplishes two objectives. First, our research establishes geographic genetic bio-ancestry as a component of racial classification. Second, it shows how social forces trump biology in racial classification and/or how social context interacts with bio-ancestry in shaping racial classification. The findings were replicated in two racially and ethnically diverse data sets: the College Roommate Study (N = 2,065) and the National Longitudinal Study of Adolescent Health (N = 2,281).
Keywords: Race, Racial classification, Genetics, Bio-ancestry
Introduction
For more than 200 years, the measurement of race has been a major component in the United States (U.S.) decennial censuses (Hirschman et al. 2000). Race and ethnicity are standard items in all contemporary population and social surveys. Since the passage of civil rights laws in the 1960s, this information has been used for monitoring racial and ethnic differences in areas such as equal opportunity, affirmative action, the redistributing provisions of the Voting Rights Act, access to health care, exposure to environmental hazards, and medical prevention and treatment strategies. The information is crucial for enforcing policies developed to reduce and eliminate racial and ethnic differences in these areas.
Contemporary surveys and the U.S. censuses since 1960 ask respondents to self-report their race/ethnic category or categories. The U.S. censuses ask household heads to report on other family members’ racial/ethnic category/categories. Farley (1991) interpreted self-report as ethnicity rather than ancestry. Perez and Hirschman (2009) did not consider the census responses on race and ethnicity as measuring ancestry, either, because these responses measure theoretically distinct identities. The consensus is that these measures are without an objective basis beyond self-report (Hirschman et al. 2000:390; Rosenberg et al. 2003:157). As Perlmann and Waters (2002:11) suggested, “the great irony is that the American government gathers data on people’s race through a more or less slippery and subjective procedure of self-identification and then must use these counts as the basis of legal status in an important domain of law and administrative regulation—namely, civil rights.”
The “scientific” racism of the early twentieth century, which held that races were biologically distinct peoples with differential abilities and behaviors, has long been discredited by the scientific community (Gould 1981). However, a socially influenced definition of race need not preclude any logical basis for race/ethnic classifications. Over the past two decades, advances in molecular genetics have yielded a body of evidence showing genetic clustering across geographically separated human populations (Li et al. 2008; Rosenberg et al. 2002). These developments present a prime opportunity to examine the links between bio-ancestry and survey measures of race/ethnicity and to study how bio-ancestry interacts with social factors to shape how individuals respond to survey questions on race/ethnicity.
Our overarching goal is to seek fresh insights into the understanding of racial classification in the contemporary United States by combining a social science perspective with recent advances in human molecular genetics. We aim to (1) establish geographic bio-ancestry as a component of racial classification, and (2) use bio-ancestry measures to examine whether, how much, and how racial self-classification departs from bio-ancestry because of social-contextual influences.
We demonstrate that bio-ancestry (the geographic origin of an individual based on genetic data) and social context interact to influence the classification of race and ethnicity. In other words, the effect of bio-ancestry depends on social, historical, and cultural context. To our knowledge, no social scientist has considered bio-ancestry when studying racial classification, and geneticists do not investigate social context that influences racial classification above and beyond bio-ancestry.
Our contribution is threefold. First, we replicate the match between genetic bio-ancestry and self-reported race across a number of independent data sources (two U.S. and two worldwide sources). We estimate bio-ancestry using saliva DNA in two racially and ethnically diverse data sets from the United States: the College Roommate Study (ROOM, N = 2,065) and the National Longitudinal Study of Adolescent Health (Add Health, N = 2,281).
A general match between genetic bio-ancestry and race has been shown using worldwide populations (Cavalli-Sforza et al. 1994; Li et al. 2008; Rosenberg et al. 2002) and clinical convenience samples in the United States (Fyr et al. 2007; Parra et al. 1998; Reiner et al. 2005; Tang et al. 2005; Yaeger et al. 2008). Others have concluded that the physical characteristics distinguishing East Asians were an adaptive response to living in the Mammoth Steppe environment in Central Asia (Guthrie 1996). However, a number of important differences exist between our work and previous research. Earlier studies focused mostly on the study of human migration spanning the past 50,000 to 100,000 years and population admixture in medical genetic association studies. Integrating bio-ancestry into a study of race and ethnicity requires data sources representative of U.S. ethnic and racial minorities and a social science perspective.
Tang et al. (2005) is a case in point. This study used a large data set of 3,636 U.S. patients with high blood pressure, and showed a 99.86 % match between cluster-analysis assignment and self-classification into white, African American, East Asian, or Hispanic. The study did not consider a social science perspective and did not use a diverse and representative sample. The study treated Hispanics as a race along with blacks and whites; however, Hispanics are considered an ethnicity in the current U.S. census and social surveys. Hispanics can be black, white, and/or Asian. The study obtained a “perfect” match, most likely because all Hispanics in the study are from Starr County, Texas. The Hispanic population in the United States, though, is much more heterogeneous than Hispanics from a single county in Texas. Tang and colleagues did not examine multiracial individuals. As mentioned earlier, the individuals in their study were assumed white, African American, East Asian, or Hispanic. Comparatively, our findings using U.S.-based, nationally representative, and racially and ethnically diverse population samples suggest that a substantial proportion of individuals in the United States is multiracial and cannot be readily assigned to a single racial category.
Second, we show in a test of the “one-drop rule” (the century-old U.S. social and legal practice of treating individuals with any amount of African ancestry as black) that the influence of bio-ancestry on racial classification depends on how black and white are historically and socially defined. In the absence of bio-ancestry, the “one drop” cannot be measured, and thus the rule cannot be tested directly and generally.
Third, we examine the fluidity of racial classification, providing evidence that social context influences whether individuals “change” their racial classification above and beyond bio-ancestry. A common finding in previous work is that multiracial individuals are more likely to change their reported race than mono-racial individuals across occasions (Hitlin et al. 2006) and under different social circumstances (Harris and Sim 2002). Adding the control of bio-ancestry enables us to conclude that given the same proportion of African or Caucasian ancestry, social contextual factors—such as the racial composition of youths’ friendship networks and neighborhoods—contributes to the fluidity of racial classification. Without taking bio-ancestry into account, these social influences cannot be isolated from the influences of bio-ancestry.
Why does bio-ancestry match self-classification of race? After all, individuals typically do not have access to their genetic information. An argument can be made that bio-ancestry underlies phenotypic features (e.g., skin tone, hair color, hair texture, and facial features) and family ancestral history (e.g., race of parents, grandparents, and great grandparents), and that genetic bio-ancestry can be more of a summary measure of bio-ancestry than a measure of phenotypic features and family history. Family history and phenotypic features are usually not measured or are crudely measured in social science studies. This reasoning explains why inaccessible bio-ancestry can be highly correlated with self-report of race.
Background
Social Construction of Racial Classification
Race is much more than human phenotypic or biological characteristics. The meanings of race are grounded in historical, cultural, social, and legal processes (Bonilla-Silva 2001; Davis 1991; Lopez 1996; Omi and Winant 1994; Williamson 1980). The role of bio-ancestry in racial classification must be understood in this larger sociohistorical context. In contemporary perspective, race is widely accepted as predominantly a social, rather than a biological, concept.
The One-Drop Rule or the Rule of Hypodescent
The one-drop rule, which originated in the American South, denoted that one drop of African blood or any amount of African ancestry would define an individual as black (Berry and Tischler 1978:97–98; Davis 1991:5; Myrdal 1944:1–2; Williamson 1980:1–2). The rule implied that even a small amount of black ancestry contaminates, thus disqualifying an individual from being classified as white. Historically, the one-drop rule lay at the heart of socially constructed race for African Americans and, together with anti-miscegenation laws, was designed to preserve racial hierarchy. If all progeny of a black-white union were considered black, and thus those black-white (mixed) individuals could only ever bear (by definition) black children, a sharp color or racial line could be maintained. The one-drop rule was practiced widely in the decades following the Civil War. The rule was further entrenched in the first half of the twentieth century with legalized racial segregation under the Jim Crow system in the South and de facto racial segregation and discrimination in other parts of the United States.
Only individuals with African ancestry are subject to the one-drop rule (Davis 1991; Rockquemore and Brunsma 2001). In the United States, those with one-fourth or less American Indian, Mexican, Chinese, or Japanese ancestry are considered assimilating Americans. The one-drop rule does not apply as strictly to these individuals, and their nonwhite racial backgrounds become ethnic legacies. The one-drop rule is uniquely American. Other countries usually conceptualize race and ethnicity differently, resulting in different systems that determine race based not only on physical characteristics but also on social status, class, and other social circumstances (Surratt and Inciardi 1998; Telles 2006).
Traditional racial and ethnic boundaries have been blurred by the enormous gains in civil rights since the mid-century, by interracial marriage, immigration, and social mobility, and by the new options of multiracial categories introduced in the 2000 U.S. census (Hirschman et al. 2000; Perez and Hirschman 2009). Despite these developments, it remains an open question whether and to what extent the one-drop rule is still observed.
Without measures of bio-ancestry, previous empirical studies of the one-drop rule used “multirace” to measure “one drop” (Fairlie 2009; Roth 2005). Roth’s study examined the race-labeling patterns of black-white married parents for their children ages 15 and younger using the 5 % Integrated Pubic Use Microdata Series (IPUMS) of the 2000 U.S. census (2005). The study considered only the special case in which the “one drop” is approximately 50 % African ancestry.
In this study, we investigated whether the one-drop rule is still observed by respondents in social surveys in the contemporary United States and the amount of African ancestry “required” for an individual to self-classify or be classified by interviewers as black. We also examined the amount of European ancestry required to self-classify or be classified by interviewers as white. Bio-ancestral measures allow a quantitative empirical test of the one-drop rule. Our analysis examined various proportions of African ancestry, including those with 50 % African ancestry as a special case.
It is important to consider external classification when examining the one-drop rule (Penner and Saperstein 2008). Our analysis included an external interviewer-classification of race/ethnicity. We also examined self-reports because they illuminate the historical consequences of the one-drop rule as both a process of external racial ascription and self-identification. One’s self-report is not independent of social settings. The classic social psychological concept of the “looking-glass self” is often invoked in the discussion of the fluidity of racial identity. Specifically, the concept states that an individual’s self-perception is shaped by others’ perception, and one learns to see oneself as society does (Cooley 1902). Previous work on racial identity has also considered self-reports (Harris and Sim 2002).
The Fluidity of Racial Classification
The fluidity of racial classification refers to the changeability of racial classification across cultures, historical periods, and everyday social contexts. Even the same individual may assume multiple racial classifications under different social circumstances. Racial fluidity is influenced and constrained by historical and contemporary political, legal, and other societal forces that tend to use racial grouping to maintain and perpetuate social stratification (Bonilla-Silva 2001; Gould 1981).
The fluidity and arbitrariness of racial boundaries have been a central theme in the literature on the social construction of race (Brown 1992; Brunsma 2006; Campbell and Troyer 2007; Hahn et al. Teutsch 1992; Harris and Sim 2002; Herman 2010; Khanna 2004, 2010; Nagel 1994; Penner and Saperstein 2008; Saperstein 2006; Tashiro 2002; Thornton et al. 2000; Waters 1990). A respondent’s self-classification in social surveys may be shaped by the purpose of the survey, the explicit or implicit expectation of the circumstances surrounding the survey, and the characteristics of the interviewer (Harris and Sim 2002; Hill 2002). A number of studies have empirically investigated the fluidity of racial classification in the contemporary United States. For example, Harris and Sim (2002) reported that interview contexts when responding to the race/ethnicity questionnaire were related to whether mixed-race individuals rejected or accepted the one-drop rule. Hitlin et al. (2006) reported that multiracial youths were four times more likely to change their reported race between two interviews about eight years apart.
In this study, we empirically investigated social forces associated with a change in racial classification for youth in the United States between an occasion when they were allowed to mark more than one racial category and an occasion when they were asked to mark only one. The analysis controlled for bio-ancestry.
Race and Genetic Clustering Across Geographically Separated Human Populations
Analyzing data from 17 genetic loci, Lewontin (1972) discovered that 94 % of human genetic variations across individuals occurs within a racial group, while the remaining 6 % occurs among the racial groups of Caucasian, African, Mongoloid, South Asian Aborigines, Amerinds, Oceanians, and Australian Aborigines. He concluded that racial classification was of no genetic or taxonomic significance. Lewontin’s pioneering work on the distribution of genetic variance within a population and between populations was confirmed by work using more recent data and statistical methods (e.g., Rosenberg et al. 2002).
Without contradicting Lewontin’s findings, recent work reported that the main genetic clusters occur among Europeans/West Asians, sub-Saharan Africans, and East Asians/Pacific Islanders/American Indians (Li et al. 2008; Rosenberg et al. 2002). The genetic clustering or the structure of various populations today is largely a result of the history of human migration (Cavalli-Sforza et al. 1994). Starting about 100,000 years ago, humans migrated out of Africa and established themselves in new environments. The migrants possessed only a subset of the alleles of the parent population. The smaller the founder population or migrant group, the larger the genetic disparity from the parent population. Furthermore, the reproductive isolation among populations caused by geographical barriers ensures that any differences arising from genetic drift be maintained. As a result, the genetic differences across geographically separated populations would solidify into structured differences between populations.
Relevant to this body of work is the neutral theory of molecular evolution (Kimura 1968, 1983). The theory states that most mutations at the molecular level are selectively neutral or nearly neutral rather than Darwinian-selective. These selectively neutral mutations do not confer functions that increase or decrease evolutionary fitness. The theory is supported by evidence in molecular genetics, which allows comparative studies of amino acid change rates in evolution across related organisms. Frequently, random genetic mutations did not change the amino acid for which a given codon triplet was coding. The majority of mutant polymorphisms could not be functional polymorphisms; otherwise, the stable change rates in amino acids would be much higher. The recognition of a large number of such neutral polymorphisms led to increased attention to the role of random genetic drift in shaping population structure.
The recent work on human migration and the neutral theory together suggest that a small amount of genetic data, which can be much lower than 6 % of the total genetic differences across individuals, is sufficient to predict the continental origins of a person with reasonable accuracy. These genetic differences, however, are largely due to random drift and unrelated to human phenotypes.
For the recent work on human migration, skepticism in social science circles exists with regard to the representativeness of the analyzed samples (Duster 2005; Rotimi 2003, 2004) and whether the way ancestral informative markers (AIMs) are selected might have predetermined the results (Duster 2005). Our replication using the same set of AIMs across four independent data sets addresses the sample representativeness and the potential problem of predetermined results.
Europeans, Africans, and East Asians are important categories because they represent a majority of the human population and because they are the root categories of a great number of subpopulations (Li et al. 2008). However, these population categories are neither the only set nor the most important set of genetic classifications. Given a proper set of genetic markers, genetic clustering can be deciphered within Africans and African Americans (Tishkoff et al. 2009), Europeans (Novembre et al. 2008), Pacific Islanders (Friedlaender et al. 2008), and American Indians in both North and South America (Wang et al. 2007). Most importantly, genetically, although every individual is unique, we all belong to the same human species. All individuals are, to various extents, admixed or genetically mixed from previously isolated human populations.
Data, Measures, and Methods
Data Sources
Our project tapped a total of four data sources. The main analysis was performed on two U.S. data sets: ROOM and Add Health. The panel of ancestral informative markers was selected from the HapMap project (2005). The estimated bio-ancestry using the U.S. data was compared with that from the worldwide Human Genome Diversity Project (HGDP).
ROOM, carried out in the spring semester of 2008 at a large public university, was designed to investigate joint peer and genetic effects on health behaviors on a college campus. The study consisted of a survey component and a saliva-based DNA component; 2,664 (79.5 %) students in the targeted sample completed a Web-based survey, and 2,080 (78.7 % of the survey completers) provided a saliva sample.
Add Health is a nationally representative longitudinal study of the health-related behaviors of about 20,000 U.S. adolescents in grades 7–12 in 1994–1995 (Harris et al. 2009). Our Add Health analysis sample consisted of 2,281 individuals with valid genotype data from the Illumina 1,536 array, including a panel of 186 AIMs and valid survey data from Wave I. These 2,281 individuals represent 87 % of 2,612 individuals whose saliva DNA was collected in 2002 at Wave III. We also analyzed self-report of race and ethnicity from Waves II and III. The findings are similar and not presented. Table 1 shows that the DNA sample characteristics are similar to those in the full Add Health sample at Wave I, suggesting that the DNA sample is also representative of the U.S. population.
Table 1.
Sample characteristics: ROOM, Add Health Wave I genetic sample, and Add Health Wave I full sample
| ROOM | Add Health Wave I Genetic Sample |
Add Health Wave I Full Sample |
|
|---|---|---|---|
|
|
|||
| Sample | Freshmen, Sophomores, and Juniors in a Large Public University |
U.S. Representative Sample Aged 12–18 |
U.S. Representative Sample Aged 12–18 |
| Time of Survey | Spring 2008 | 1994–1995 | 1994–1995 |
| Age of Respondents | 18–20 | 12–18 | 12–18 |
| Male (%) | 39.81 | 47.32 | 45.18 |
| Southern States (%) | 89.01 | 36.09 | 37.11 |
| Race/Ethnicity (%) | |||
| White | 65.19 | 56.31 | 50.39 |
| Black | 13.39 | 17.35 | 20.88 |
| East Asian | 4.18 | 6.67 | 6.03 |
| South Asian | 2.00 | 0.11 | 0.32 |
| Hispanic | 7.40 | 15.42 | 17.05 |
| American Indian | o.19 | 0.18 | 0.55 |
| Other | 1.02 | 0.78 | 0.91 |
| Multiracial | 6.62 | 3.18 | 3.86 |
| Mother’s Education (%) | |||
| Less than high school | 1.51 | 16.94 | 18.16 |
| High school graduate or GED | 7.58 | 39.57 | 39.12 |
| College | 53.23 | 35.66 | 33.79 |
| More than college | 37.68 | 7.83 | 8.92 |
| European Ancestry (%) | 77.03 | 70.67 | – – |
| African Ancestry (%) | 15.79 | 18.70 | – – |
| Asian Ancestry (%) | 7.18 | 10.63 | – – |
| Sample Size | 2,065 | 2,281 | 20,745 |
To cross-check our estimates of bio-ancestry, we reanalyzed the more than 1,000 individuals from 52 worldwide populations in HGDP and compared the estimates of bio-ancestry in HGDP with our estimates from the U.S. data. The HGDP populations spread over most of the inhabited continents (Cann et al. 2002). The same set of AIMs that was genotyped in HGDP was also genotyped in our U.S. data sets. The HapMap project has yielded genotype data for 90 Caucasian individuals from Utah with ancestry in Northern and Western Europe, 45 Han Chinese from Beijing, 44 Japanese from Tokyo, and 90 Yoruban individuals from Ibadan, Nigeria on >6 million single nucleotide polymorphisms (SNPs) located across the genome.
Measures
Genotype
In ROOM, DNA was extracted according to the manufacturer’s instructions from 2ml of saliva (containing buccal epithelial and white blood cells) collected from participants in an Oragene DNA collection kit (DNA Genotek; Ottawa, Ontario, Canada). DNA was plated for Illumina genotyping at 30 μl at >50 ng/μl. Our median DNA yield was 27.33 μg, with a minimum of 0 μg (six individuals) and a maximum of 71.32 μg.
For ROOM, we designed an Illumina GoldenGate assay for 384 candidate SNPs, including 186 ancestral informative markers. Hardy-Weinberg equilibrium tests were performed on each SNP within each race and ethnicity. Less than 1 % of the SNPs yielded a p value smaller than .001. The genetic analysis was based on the 162 of 186 AIMs that were successfully genotyped.
In Add Health, genomic DNA was isolated from buccal cells at the Institute of Behavior Genetics at the University of Colorado, Boulder. The average yield of DNA was 58 ± 1 μg. We designed and genotyped an Illumina GoldenGate assay for 1,536 candidate SNPs, including the same 186 AIMs genotyped in ROOM. In Add Health, 121 of 186 AIMs were successfully genotyped. The literature (briefly described herein) on AIMs suggests that 121 are still likely sufficient for differentiating the continental groups, given our sample sizes.
Race, Ethnicity, and Other Sample Characteristics
ROOM has two sets of self-reported race and ethnicity: one from the housing application form submitted by students when requesting a dorm room to the university housing department before their freshman year, and the second from an online survey. The university housing form allowed students to self-classify as only one of six racial/ethnic groups: white, black, Hispanic, Asian and Pacific Islander, Native Indian, and Other; comparatively, the online questionnaire allowed respondents to mark one or more races.
At Wave I, Add Health’s main race/ethnicity questions predate the format followed in the 2000 U.S. census, allowing identification of more than one racial group. When a respondent selected more than one race during the home interview, the respondent was asked to indicate a single race category that would best describe him or her. Importantly, interviewers were instructed to record the single-best race of the respondent from their observations—not from what the respondent reported. The categories available for interviewers included only single-race categories of white, black, American Indian or Alaska Native, and Asian or Pacific Islander; Hispanic was not an option for interviewers.
The single-race responses in ROOM were recorded from housing application forms submitted to the university’s housing department before the freshman year. In ROOM, the race questionnaire allowing multirace categories was filled out in the spring of 2008. In Add Health, the single-race responses and the multirace responses were recorded in the same survey almost immediately one after the other.
In Add Health, “Southern States” was coded as 1 for individuals who lived in one of the following states at Wave I: Maryland, Virginia, Delaware, Tennessee, Arkansas, Louisiana, Missouri, North Carolina, South Carolina, Mississippi, Alabama, Georgia, Florida, Texas, Oklahoma, West Virginia, and Kentucky. In ROOM, “Southern States” was coded as 1 for those whose permanent address on the housing application form is one of the aforementioned states. The much higher percentage (89 %) of Southern States in ROOM than in Add Health (36 %) is due to the location of the study university (Table 1).
Analytical Strategies
Bio-Ancestry
Our estimation of bio-ancestry relies on a panel of AIMs (rather than one or two distinguishing genetic variants) to estimate bio-ancestry or detect genetic differentiation across human populations. AIMs are sets of genetic polymorphisms whose allele frequencies differ significantly across populations (Frudakis et al. 2003; Parra et al. 1998; Shriver et al. 1997). Our panel of AIMs consists of 186 SNPs and was developed to detect and correct population stratification for genetic association studies (Enoch et al. 2006). The AIMs were selected according to four criteria:
Each AIM differed in allele frequency by a range of 0.7–10 times between at least a pair of continental populations of Europeans, sub-Saharan Africans, and East Asians.
The absolute value of log (RAF1/RAF2) was >1, where RAF1 and RAF2 are the reference allele frequency in continental populations 1 and 2, respectively.1
Each AIM was a genetically independent HapMap SNP with a minimum distance from any other AIM of at least 100 kilo-base pair (kb) to ensure that the AIMs were not in linkage disequilibrium.
The AIMs were evenly distributed throughout the genome for the three continental populations.
The AIM selection was based on the observed reference allele frequencies of the European, African, and Chinese/Japanese populations of the HapMap Project (HapMap data release #16c.1, June 2005). The AIMs were specifically designed for detecting continental populations. As such, these AIMs are much less effective in detecting substructures within a continental population of Europeans, Africans, or East Asians.
Factors such as the minimum number of markers and sample size also affect an AIM panel’s accuracy and informativeness. Bamshad et al. (2004) found that African American populations had roughly 4,700 SNPs that were potentially private to the population (and thus potential AIMs), while Europeans had 580 such SNPs. Rosenberg et al. (2002) found that 100– 160 SNPs were sufficient when the sample size was roughly 1,000; other studies have generally used 150–200, with samples of at least 400 (Halder et al. 2008; Smith et al. 2001; Yang et al. 2005).
We used the AIM panel to estimate biogeographical ancestry via three statistical procedures: PLINK-based cluster analysis (Purcell et al. 2007), STRUCTURE-based cluster analysis (Pritchard et al. 2000), and principal components analysis implemented in the software EIGENSTRAT (Price et al. 2006). All three procedures estimated ancestral population membership without using information from self-report of race.
Cluster analysis has been used to infer population structures and to assign individuals to clusters or groups according to the degree of similarity of genetic data between individuals. Individuals within each cluster share more genetic variants than those in different clusters. However, the traditional cluster analysis assumes that each individual comes from only one population. Pritchard et al. (2000) proposed a method that allows each individual’s ancestral composition to represent a mixture of multiple unobserved populations. This method has been implemented in the software package STRUCTURE.
The particular PLINK procedure we used sets a fixed cluster size or the fixed number of ancestral populations. It assigns individuals into one and only one ancestral population, and the individuals assigned to the same ancestral population are relatively homogeneous with respect to AIM frequencies. To estimate the precision of our PLINK estimates, 95 % bootstrapping confidence intervals (Efron and Tbshirani 1993) were calculated.
The STRUCTURE analysis considers each individual’s genome having potentially arisen from an admixture of multiple populations; it also estimates relative contributions to each individual from multiple ancestral populations. The STRUCTURE analysis assumes a K value that represents the hypothesized number of ancestral populations. It then uses the differences in allele frequencies in the AIMs to predict how much each ancestral population contributed to the genetic ancestry of a given individual. The K contributions from K ancestry populations for each individual sum to 1.
Each STRUCTURE run used a burn-in period of 10,000 iterations, followed by 20,000 iterations from which estimates of bio-ancestry were obtained. To take into account precision of estimates, we performed 20 replicate STRUCTURE runs. All pairwise symmetric similar coefficients (SSC) are greater than 0.995. A SSC measures the similarity of two sets of population structure estimates. Our final figures for bio-ancestry were averaged over the results of the 20 sets of estimates. Our approach is similar to that used in studies of genetic structure among American Indians (Wang et al. 2007) and Pacific Islanders (Friedlaender et al. 2008).
Both the PLINK and STRUCTURE procedures assume that the individuals in the analysis have originated from K populations. K is was chosen for each analysis run, but it can be varied across different runs. Because our panel of AIMs was designed to differentiate continental populations of Europeans, Africans, and East Asians, we set K = 3. However, to test the robustness of our results to choice of K, we performed analyses assuming K = 3, 4, 5, 6, and 7.
The third method, implemented in the software EIGENSTRAT (Price et al. 2006), identifies bio-ancestry through principal components (PCs). Principal component analysis is one of the most widely used techniques to reduce the dimensionality while retaining most of the variation in a data set. In other words, the technique summarizes a large number of variables by a small number of new linearly independent variables. Principal component analysis ranks the relative importance of those components in a descending way, so that the first component contains the largest variation of the original variables. A large number of AIMs provide rich and detailed ancestry-related information for each individual. However, such high-dimensional data make it difficult to visualize the patterns of genetic distances between individuals. When we plot the first and second principal components, genetic distances between individuals (thus genetic clusters) are displayed. The first two principal components represent a significant portion of ancestral information contained in the set of AIMs.
Social Construction of Race
To examine the practice of the one-drop rule, we calculated the percentage of the sample with a proportion of African ancestry that reports itself as black and the percentage’s 95 % bootstrapping confidence interval. We expect that the higher the proportion of African ancestry, the more likely it is that individuals will self-classify or be classified by an interviewer as black. However, the important question is, at what proportions of African ancestry do substantial percentages of individuals begin to self-classify or be classified by an interviewer as black? We also calculated percentage of the sample with a proportion of European ancestry that reports itself as white as well as the percentage’s 95 % bootstrapping confidence interval. Comparing black and white calculations would reveal the likely asymmetry between these two groups: that is, does it take a much higher proportion of European ancestry to self-classify or be classified as white than the proportion of African ancestry needed to self-classify or be classified as black?
Our analysis also takes into consideration three factors expected to affect the practice of the one-drop rule: an individual’s ancestral composition, whether a race questionnaire contains a multiracial option, and/or whether an individual self-classifies or is classified by an interviewer. Our main analysis sample on the one-drop rule included only individuals who self-reported as non-Hispanic black, white, or black-white. A separate analysis using only Hispanics was performed so that Hispanics and non-Hispanics were compared.
For ROOM, we calculated two sets of percentages and their confidence intervals: one set using the self-reported race on the college housing application form that did not have multirace categories; and the second set using the online survey responses, which did allow selection of multiple race categories. For Add Health, we analyzed two samples: the first sample included non-Hispanics, and the second included only Hispanics. Using the first sample, we calculated three sets of percentages and their confidence intervals: the first used self-reported single race, the second used single race recorded by interviewers, and the third used the 2000 U.S. census self-reported questionnaire that allowed the selection of multiple races.
To examine the fluidity of racial classification, we restricted our analysis sample to individuals who were classified by our PLINK analysis as blacks and whites; Hispanics were excluded. First, we investigated the extent to which these individuals “switch” to a multirace category when presented with this option; second, we explored which social circumstances might make individuals more likely to switch racial classification than others. In all the analysis, we controlled for bio-ancestry.
Results
Bio-Ancestry
Table 2 presents results from PLINK cluster analysis, showing both the percentage and case distribution of self-reported race by PLINK-estimated genetic cluster or bio-ancestry. These PLINK estimates (as well as other estimates based on genetic data) are placed in quotation marks to differentiate them from self-reports. The samples were assumed to have derived from three ancestral populations (K = 3). We repeated the analysis, assuming K = 3, 4, 5, 6 and 7, and using a fuller range of self-reported racial classification groupings. The findings from these additional analyses are substantively identical to those in Table 2 and are also available upon request.
Table 2.
Percentage distribution (number of individuals) of self-reported race by genetic markers-based ancestral population membership (three ancestral populations are assumed)
| Ancestral-Informative-Marker-Based Genetic Cluster |
||||
|---|---|---|---|---|
| Self-report | “White” | “Black” | “Non–South Asian” |
Total |
| ROOM | ||||
| White | 99.5(1,399) | 0.28(4) | 0.21(3) | 100(1,406) |
| Black | 0.71(2) | 99.3(279) | 0.0(0) | 100(281) |
| South Asian | 100.00(41) | 0.0(0) | 0.0(0) | 100(41) |
| East Asian | 2.33(2) | 0.0(0) | 97.7(84) | 100(86) |
| American Indian | 75.0(3) | 0.0(0) | 25.0(1) | 100(4) |
| Others | 80.7(50) | 12.9(8) | 6.45(4) | 100(62) |
| Multiracial | 52.9(91) | 36.1(62) | 11.1(19) | 100(172) |
| Total | 1,586 | 353 | 111 | 2,052 |
| Add Health Wave I | ||||
| White | 99.4(1,429) | 0.42(6) | 0.14(2) | 100(1,437) |
| Black | 0.00(0) | 100.0(381) | 0.00(0) | 100(381) |
| East Asian | 6.29(10) | 0(0) | 93.7(149) | 100(159) |
| American Indian | 100.0(19) | 0(0) | 0 (0) | 100(19) |
| Others | 91.1(163) | 5.03(9) | 3.31(7) | 100(179) |
| Multiracial | 72.0(67) | 26.9(25) | 1.08(1) | 100(93) |
| Total | 1,699 | 429 | 160 | 2,268 |
Notes: For ROOM, the bootstrapping 95 % confidence interval for “white,” “black,” and “non–South Asian” are, respectively, [99.0, 99.9], [94.7, 100], and [89.5, 100]. In Add Health, the three confidence intervals are, respectively, [97.7, 99.9], [96.9, 100], and [88.1, 97.5].
In ROOM, of those who self-reported as white, 99.5 % were assigned into the “white” category by the cluster analysis. Of those who self-reported as black, 99.3 % were classified as “black.” We separated South Asians from non–South Asians; previous work suggests that South Asians share substantial bio-ancestry with Europeans (e.g., Rosenberg et al. 2002). Of those self-classifying as non–South Asians (including Chinese, Japanese, Koreans, Filipinos, and Vietnamese), 97.7 % were assigned as “non–South Asians.” Three of the four self-reported American Indians were classified as “white.” The bootstrapping 95% confidence intervals for the three key groups of whites, blacks, and non–South Asians were [99.0, 99.9], [94.7, 100], and [89.5, 100], respectively, indicating that the correspondence between bio-ancestry and self-reports for the three main racial groups is estimated with precision.
The results from Add Health are comparable. Of individuals who self-classified as white, black, or non–South Asian, 99.4 %, 100.0 % and 93.7 %, respectively, were assigned by cluster analysis into the “white,” “black,” and “non–South Asian” categories. The only two self-reported South Asians in Add Health were excluded from the analysis. All self-reported American Indians were classified as “white.” The three confidence intervals for Add Health were [97.7, 99.9], [96.9, 100], and [88.1, 97.5], respectively.
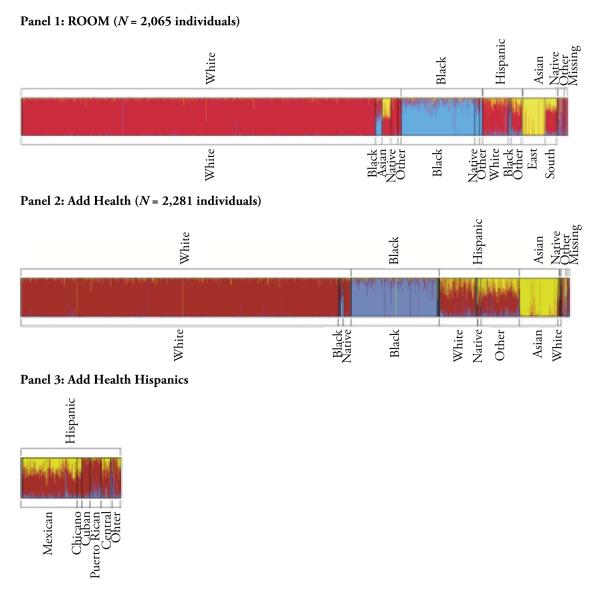
Assuming three ancestral populations, we performed a STRUCTURE analysis (Pritchard et al. 2000) on data from ROOM and Add Health (Fig. 1). This analysis allows each individual to have memberships in as many as three ancestral populations. The horizontal bar graph shows ancestral proportional composition for each individual. Each individual is represented by a vertical line partitioned into as many as three segments; the length of each segment is the measure of each ancestral contribution to an individual’s genome from three ancestral groups. The three continental ancestries are European (red in Fig. 1), black (blue in the figure), and Asian (yellow in the figure). The labels of self-reported race/ethnicity were used to order the individuals or vertical lines in the graph and were added only after each individual’s ancestry had been estimated. There are two sets of labels for white, black, Hispanic, Asians, and so on, with one set above the graph and the other below. The two sets of labels indicate the self-reported single-race and mixed-race individuals.
Fig. 1.
The proportional composition in bio-ancestry for each individual based on the STRUCTURE analysis. Each individual is represented by a vertical line partitioned into as many as three segments, with their lengths corresponding to ancestral contribution to an individual’s genome from up to three ancestral populations of Europeans (red), Africans (blue), and East Asians (yellow). The labels of self-reported race/ethnicity were used to order the individuals or vertical lines in the graphs and were added only after each individual’s ancestry had been estimated. There are two sets of labels of self-reports. The set above a graph is based on responses to a question that instructs a respondent to identify with a single race; the set below is based on a question that allows a respondent to identify with more than a single race.
The results from the STRUCTURE analysis not only confirm the findings described in Table 2 but also demonstrate a close match between the estimated bio-ancestry and self-reported race of multiracial individuals. For example, the bar graph for ROOM shows that the vertical lines for individuals who self-reported as black-white are mostly composed of blue and red colors; the lines for those who self-reported as East Asian-white are largely composed of yellow and red colors. In Add Health, there are fewer respondents who are black-white; the lines of these individuals are composed of red and blue colors. Panel 3 of Fig. 1 magnifies the section of Hispanics in Panel 2, showing that Cubans in Add Health contain a high percentage of European ancestry, that Puerto Ricans contain a significant portion of African ancestry, and that Chicanos are similar in ancestral composition to Mexicans.
Table 3 gives the distribution of average ancestry for each self-reported race/ethnicity assuming three ancestral populations. The results in Table 3 were averaged over the estimates presented in Fig. 1. The results across ROOM and Add Health are consistent. For example, in the two studies, respectively, the average percentage of Caucasian ancestry among self-reported whites is 98.1 % and 98.3 %; the percentage of African ancestry among self-reported blacks is 89.7 % and 93.2 %; and the percentage of East Asian ancestry among self-reported East Asians is 95.5 % and 92.7 %. The ancestry distribution for subgroups within Hispanics in Add Health is also presented.
Table 3.
Distribution of average ancestry for each self-reported race/ethnicity, assuming three ancestral populations
| ROOM |
Add Health Wave I |
|||||||
|---|---|---|---|---|---|---|---|---|
| Self-report | European | African | Asian | N | European | African | Asian | N |
| White | 98.1 | 0.8 | 1.1 | 1,338 | 98.3 | 0.7 | 1.0 | 1,303 |
| White, Black | 42.4 | 54.7 | 2.9 | 25 | 51.1 | 46.0 | 2.9 | 14 |
| White, Asian | 57.3 | 0.8 | 41.9 | 34 | 52.8 | 0.9 | 46.3 | 12 |
| White, Indian | 95.2 | 2 | 2.8 | 27 | 94.3 | 2.0 | 3.7 | 36 |
| White, Other | 97.2 | 1.7 | 1.1 | 13 | 86.6 | 3.8 | 9.6 | 11 |
| Black | 8.7 | 89.7 | 1.7 | 279 | 5.92 | 93.15 | 0.93 | 378 |
| Black, Indian | 17.3 | 81.3 | 1.4 | 17 | 8.9 | 82.2 | 8.9 | 5 |
| Black, Other | 8.8 | 88.3 | 2.9 | 12 | 0.8 | 95.3 | 3.9 | 2 |
| Hispanic White | 86.3 | 6 | 7.7 | 101 | 75.4 | 7.0 | 17.6 | 133 |
| Hispanic Black | 30.7 | 61.4 | 7.9 | 8 | 28.0 | 63.3 | 8.7 | 3 |
| Hispanic Other | 70.8 | 10.2 | 18.9 | 41 | 63.2 | 9.4 | 27.4 | 157 |
| East Asian | 4.0 | 0.5 | 95.5 | 90 | 6.4 | 0.9 | 92.7 | 159 |
| South Asian | 68.4 | 5.1 | 26.5 | 41 | 39.6 | 1.8 | 58.6 | 2 |
| American Indian | 66.5 | 17.5 | 16.0 | 5 | 62.9 | 5.2 | 31.9 | 19 |
| Other | 66.7 | 25.7 | 7.6 | 21 | 61.2 | 10.2 | 28.6 | 34 |
| Missing | 81.5 | 17.4 | 1.1 | 13 | 53.2 | 38.6 | 8.2 | 13 |
| Total | 2,065 | 2,281 | ||||||
| Non-Hispanic | 70.21 | 20.14 | 9.65 | 1,946 | ||||
| Hispanic | 67.73 | 8.53 | 23.74 | 329 | ||||
| Mexican | 63.54 | 5.56 | 30.91 | 197 | ||||
| Chicano | 59.92 | 5.96 | 34.12 | 16 | ||||
| Cuban | 90.36 | 7.07 | 2.57 | 30 | ||||
| Puerto Rican | 75.52 | 20.22 | 4.26 | 39 | ||||
| Central/South American |
66.61 | 11.69 | 21.70 | 33 | ||||
| Other | 64.42 | 14.53 | 21.05 | 34 | ||||
| Total | 2,272 | |||||||
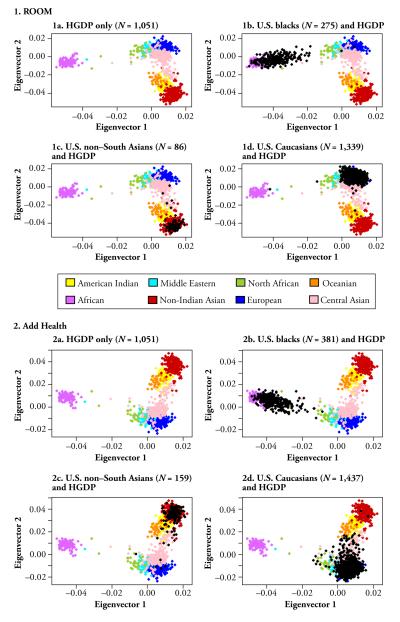
Figure 2 displays the genetic distances among the individuals in ROOM (Panels 1a–1d) and Add Health (Panels 2a–2d) in the context of 52 world populations consisting of more than 1,000 individuals from HGDP. We analyzed the U.S. participants and reanalyze the HGDP study participants in order to compare the two sets of results. Each panel plots the two largest principal components obtained from analyzing the same set of AIMs, and the resulting figure reveals patterns of genetic distances among individuals. Panel 1a plots bio-ancestral distances among the HGDP individuals only. Africans and East Asians are the furthest from each other; American Indians and individuals from Oceania are much closer to East Asians than to Europeans and Africans; and Central Asians and Middle Eastern individuals are closer to Europeans than to East Asians.
Fig. 2.
Eigenstrat-generated ancestral distances among U.S. study participants in ROOM (1a–1d) and Add Health (2a–2d) in the context of 51 world populations from the Human Genome Diversity Project (HGDP). The U.S. participants represented by black dots are self-reported blacks (1b and 2b), non–South Asians (1c and 2c), and whites (1d and 2d)
In Panels 1b–1d, the HGDP map of ancestral locations in Panel 1a is used as a backdrop with the U.S. sample (black symbols) imposed onto the HGDP map. The U.S. sample self-classified as African Americans (Panel 1b), East Asians (Panel 1c), and Europeans (Panel 1d). Self-classified East Asians and Europeans in the U.S. sample overlap almost completely with the HGDP East Asians and Europeans, respectively, while self-classified African Americans are located slightly away from the HGDP Africans and closer to the HGDP North Africans and Europeans, which is consistent with the presence of some European ancestry in African Americans. The Add Health results (2a–2d), based on a smaller set of AIMs (121 vs. 162 for ROOM) are similar to those in ROOM. These findings have thus established an agreement among our bio-ancestral results from the PLINK, STRUCTURE, and EIGENSTRAT analyses. We also demonstrate an agreement among the findings based on the U.S. data (ROOM and Add Health), the HGDP, and HapMap.
The One-Drop Rule
Table 4 shows the percentage of a sample with a proportion of African ancestry that reports itself as black for ROOM and Add Health. The related 95 % bootstrapping confidence intervals are given in parentheses. The point estimates are boldfaced to highlight the general patterns across the proportion of African ancestry. We display the information in deciles, but we collapse several deciles where sample sizes are small.
Table 4.
Percentage (95 % bootstrapping confidence interval) of the sample with a proportion of African ancestry reports itself as black: ROOM and Add Health Wave I genetic sample
| The Roommate Study | Add Health Wave I | Add Health Wave I | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Sample -> | Non-Hispanic Black, White, and Black-White by Self-report |
Non-Hispanic Black, White, and Black-White by Self-report |
Hispanic by Self-report | |||||||
|
|
||||||||||
| Proportion of African Ancestry |
Single Race (self-report) |
Multiracial (self-report) |
N | Single Race (self-report) |
Single Race (interviewer) |
Multiracial (self-report) |
N | Single Race (self-report) |
Multiracial (self-report) |
N |
|
| ||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| 0–.02 |
0.00 (0,0) |
0.08 (0.08,0.32) |
1,240 |
0.00 (0, 0) |
0.27 (0.2, 0.9) |
0.00 (0, 0) |
1,193 |
0.00 (0,0) |
0.00 (0,0) |
129 |
| .02–.1 |
1.27 (1.26,5.0) |
0.00 (0,0) |
79 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
136 |
0.00 (0,0) |
0.00 (0,0) |
106 |
| .1–.2 |
16.67 (16.6,50) |
16,67 (16.6,50) |
6 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
8 |
0.00 (0,0) |
0.00 (0,0) |
55 |
| .2–.3 |
0.00 (0,0) |
0.00 (0,0) |
0 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
2 |
4.76 (4.76,14.3) |
0.00 (0,0) |
21 |
| .3–.4 |
100.0 (100,100) |
0.00 (0,0) |
2 |
71.43 (42.9,100) |
57.14 (14.3, 85.7) |
28.57 (14.3, 57.1) |
7 |
0.00 (0,0) |
0.00 (0,0) |
7 |
| .4–.5 |
100.0 (100,100) |
16.67 (16.6,50) |
6 |
66.67 (33.3,100) |
88.89 (66.7,100) |
22.22 (11.1, 55.6) |
9 |
0.00 (0,0) |
0.00 (0,0) |
2 |
| .5–.6 |
100.0 (100,100) |
36.84 (15.7,57.8) |
19 |
90.00 (70,100) |
90.00 (70,100) |
30.00 (10,60) |
10 |
33.33 (33.3,100) |
33.33 (33.3,100) |
3 |
| .6–.7 |
100.0 (100,100) |
50.00 (21.4,71.4) |
14 |
100.0 (100,100) |
100.0 (100,100) |
100.0 (100,100) |
7 |
50 (25,100) |
50 (25,100) |
4 |
| .7–.8 |
100.0 (100,100) |
82.86 (68.5,94.2) |
35 |
100.0 (100,100) |
100.0 (100,100) |
87.10 (74.2,96.8) |
31 |
0.00 (0,0) |
0.00 (0,0) |
0 |
| .8–.9 |
100.0 (100,100) |
85.71 (76.1,93.6) |
63 |
100.0 (100,100) |
100.0 (100,100) |
95.65 (89.1, 100) |
46 |
0.00 (0,0) |
0.00 (0,0) |
0 |
| .9–1 |
100.0 (100,100) |
89.29 (84.6,93.3) |
196 |
100.0 (100,100) |
100.0 (100,100) |
98.67 (96.0,99.3) |
298 |
0.00 (0,0) |
0.00 (0,0) |
1 |
| Total | 1,660 | 1,747 | 328 | |||||||
Note: The point estimates are boldfaced to highlight general patterns across the proportion of African ancestry.
In ROOM, when only a single race was allowed to be self-reported on the housing application, individuals with 30 % to 40 % or more African ancestry always self-classified as black. After the questionnaire in the online survey allowed multiracial categories, the percentages that self-classified as black lowered considerably in comparison with those in the housing form. The lowering or the weakening of the one-drop rule is particularly conspicuous near the 50 % African ancestry mark. Among those with 40 % to 70 % African ancestry (N = 39), when single race was the only choice, 100 % self-identified as black; when offered multiracial options, 24 of the 39 did not self-classify as black in the online survey (column 3 vs. column 2). The 95 % bootstrapping confidence intervals for the online estimates are almost always below those for the housing form (column 3 vs. column 2). On the other hand, the point estimates and confidence intervals in column 3 show that large proportions of individuals with 40 % to 70 % African ancestry still self-classified as black, indicating a cultural influence of the one-drop rule in spite of multiracial options.
The non-Hispanic data from Add Health Wave I displayed a similar pattern as those from ROOM. The large majority of individuals with >30 % African ancestry self-classified as black. The percentages of individuals who self-classified as black also dropped considerably when multiracial categories were an option (column 7). Interviewer-classification did not differ markedly from self-classification. The Hispanic data from Add Health have a small number of persons with >30 % African ancestry—too few to be informative on the one-drop rule (columns 9–11).
Table 5, a mirror image of Table 4, gives the percentage of a sample with a proportion of European ancestry that reports itself as “white” for both ROOM and Add Health. The contrast between Tables 4 and 5 among non-Hispanic individuals is evident. A much larger proportion of individuals with 30 % to 70 % African ancestry self-classified as black (Table 4: 100 % and 38 % in response to a single-race question and a multirace question for ROOM; 82 % and 42 % for Add Health) than the proportion of individuals with 30 % to 70 % European ancestry self-classified as white (Table 5: 3 % and 0 % in response to a single-race question and a multirace question for ROOM; 27 % and 13 % for Add Health). The asymmetry between Tables 4 and 5 is that it takes a higher proportion of European ancestry to self-classify or be classified by an interviewer as white than the proportion of African ancestry needed to self-classify or be classified as black. When multiracial categories come into play, some individuals with a high proportion of European ancestry (columns 3 and 7) switched classification from white to multiracial. Again, interviewer classification does not differ from self-classification noticeably.
Table 5.
Percentage (95 % bootstrapping confidence interval) of the sample with a proportion of European ancestry reports itself as white: ROOM and Add Health Wave I genetic sample
| ROOM | Add Health Wave-I | Add Health Wave-I | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Sample -> | Non-Hispanic Black, White, and Black-White by Self-report |
Non-Hispanic Black, White, and Black-White by Self-report |
Hispanic by Self-report | |||||||
|
|
||||||||||
| Proportion of European Ancestry |
Single Race (self-report) |
Multiracial (self-report) |
N | Single Race (self-report) |
Single Race (interviewer) |
Multiracial (self-report) |
N | Single Race (self-report) |
Multiracial (self-report) |
N |
|
| ||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| 0–.02 |
0.00 (0,0) |
0.65 (0.64,2.5) |
154 |
0.84 (0.4,3.3) |
0.84 (0.4,3.3) |
0.84 (0.4,3.3) |
239 | 0 | 0 | 3 |
| .02–.1 |
0.00 (0,0) |
0.00 (0,0) |
65 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
83 | 0 | 0 | 0 |
| .1–.2 |
0.00 (0,0) |
0.00 (0,0) |
47 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
36 | 0 | 0 | 1 |
| .2–.3 |
0.00 (0,0) |
0.00 (0,0) |
33 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
25 | 0 | 0 | 7 |
| .3–.4 |
0.00 (0,0) |
0.00 (0,0) |
12 |
0.00 (0,0) |
0.00 (0,0) |
0.00 (0,0) |
6 |
18.75 (6.3,37.5) |
18.75 (6.3,37.5) |
16 |
| .4–.5 |
5.26 (5.26,21.0) |
0.00 (0,0) |
19 |
14.29 (14.3,42.9) |
14.29 (14.3,42.9) |
0.00 (0,0) |
7 |
45.71 (28.6,62.9) |
45.71 (6.25,37.5) |
35 |
| .5–.6 |
0.00 (0,0) |
0.00 (0,0) |
5 | 25 (12.5,62.5) |
12.5 (12.5,37.5) |
12.5 (12.5,37.5) |
8 |
29.79 (17.0,42.6) |
29.79 (17.0,42.6) |
47 |
| .6–.7 |
0.00 (0,0) |
0.00 (0,0) |
2 |
55.56 (22.2,88.9) |
55.56 (22.2,88.9) |
33.33 (11.1,66.7) |
9 |
25 (15.0,35.8) |
25 (15.0,35.8) |
68 |
| .7–.8 |
100.00 (100,100) |
100.00 (100,100) |
2 |
100 (100,100) |
100 (100,100) |
100 (100,100) |
10 |
46.55 (29.3,53.4) |
41.38 (34.5,58.6) |
58 |
| .8–.9 |
96.97 (90.9,100) |
78.79 (63.6,90.9) |
33 |
100 (100,100) |
97.87 (93.6,100) |
87.23 (76.6,95.7) |
47 |
55 (35,65) |
50 (40,70) |
40 |
| .9–1 |
100.0 (100,100) |
97.52 (96.6,98.3) |
1288 |
100 (99.5,100) |
99.69 (99.1,99.8) |
97.65 (96.6,98.3) |
1,277 |
83.13 (60.4,84.9) |
73.58 (69.8,90.6) |
53 |
| Total | 1660 | 1,747 | 328 | |||||||
Note: The point estimates are boldfaced to highlight general patterns across the proportion of African ancestry.
The Hispanics from Add Health in Table 5 show a distinct pattern. Those with 30 % to 60 % European ancestry are more likely than non-Hispanics with African ancestry to self-classify as white (column 9 vs. column 5). For example, about 45 % of Hispanics with 40 % to 50 % European ancestry self-classified as white, compared with about 14 % of non-Hispanics with 40 % to 50 % European ancestry. Hispanics with >60 % European ancestry were less likely to self-classify as white and more likely to self-classify as multiracial (column 9 vs. column 5). For example, only about 50 % of Hispanics with 80 % to 90 % European ancestry self-classified as white, compared with 100 % of non-Hispanics who self-classified as white and who have 80 % to 90 % European ancestry.
Tables 4 and 5 record another asymmetry from both ROOM and Add Health. In the column of the number of individuals by proportion of African ancestry in Table 4, individuals with 10 % to 50 % African ancestry (N = 14 for ROOM and N = 26 for Add Health) are considerably less numerous than individuals with 50 % to 90 % African ancestry (N = 131 for ROOM and N = 94 for Add Health).
The Fluidity of Racial Classification
Table 6 shows the number and percentage of blacks and whites who switched racial classification between the single-race and multirace options. In ROOM, 16.8 % and 2.6 % of blacks and whites, respectively, switched racial classification. The black switchers and nonswitchers scored .76 and .91, respectively, on African ancestry. The white switchers and nonswitchers scored .96 and .98, respectively, on European ancestry. In Add Health, 5.03 % and 2.8 % of blacks and whites, respectively, changed their racial classifications. The changers and nonchangers scored, respectively, .68 and .93 on African ancestry among blacks and .94 and .98 on European ancestry among whites. Among individuals who changed racial classification, more than 70 % of both blacks and whites switched to a multiracial category. Overall, those who changed classification scored higher on bio-ancestry than the nonswitchers within both the African and European samples. The higher probability of classification switching among blacks than whites could be partially attributed to bio-ancestry, suggesting that bio-ancestry needs to be accounted for when examining sociocontextual sources of classification switching.
Table 6.
The number and percentage among blacks and whites who switched racial classification from a survey in which only a single race is allowed to a survey in which a multiracial classification is optional: ROOM and Add Health Wave I genetic sample
| Black Sample | White Sample | |||
|---|---|---|---|---|
|
| ||||
| Self-reported Racial Classification | N or % | Mean African Ancestry |
N or % | Mean European Ancestry |
| ROOM | ||||
| Housing form as black or white | 328 | 1,320 | ||
| Online survey same as housing | 273 | 0.91 | 1,286 | 0.98 |
| Online survey changed from housing | 55 | 0.76 | 34 | 0.96 |
| % changed | 16.77 | 2.58 | ||
| Add Health Wave I | ||||
| Best single race as black or white | 398 | 1,337 | ||
| Multirace optional, black or white | 378 | 0.93 | 1,299 | 0.98 |
| Multirace optional, multiracial | 20 | 0.68 | 38 | 0.94 |
| % changed | 5.03 | 2.84 | ||
Logistic regression was used to examine the sociocontextual sources of classification switching (Table 7). The descriptive statistics of the variables used in the regression models are given in Table 8. The outcome variable was coded as 1 for classification-changers and 0 for nonchangers. In ROOM, Model 1 (which is based on the combined sample of blacks and whites) contains a statistical significance test for the exploratory results described in Table 6, indicating that blacks were about seven times as likely to switch racial classification as whites. This finding is highly statistically significant. However, after primary ancestry—that is, an individual’s most prominent ancestry (African, Caucasian, or Asian bio-ancestry)—is controlled, the odds ratio is reduced from 7.33 to 2.94 (Model 2). Primary ancestry has proved important; an increase of 1 % bio-ancestry reduces the likelihood of classification change by (1 – .94) = 6 %. This result applies to those whose primary ancestry is African and those whose primary ancestry is European. Model 3 shows that students from the South are about 42 % as likely or 58 % less likely to change racial classification as the non-Southern students. Age and gender are not related to classification switching. The findings were obtained after African ancestry was controlled.
Table 7.
Logistic regression of racial classification switching from a survey in which only a single race is allowed to a survey in which a multiracial categorization is optional: ROOM and Add Health Wave-I genetic sample (odds ratios are reported; only non-Hispanics are included)
| ROOM | Add Health Wave I | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Analysis Sample → |
Blacks + Whites |
Blacks + Whites |
Blacks | Whites | Blacks + Whites |
Blacks + Whites |
Blacks | Whites | Blacks | Whites |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| Black | 7.33*** | 2.94*** | 2.07** | 0.56 | ||||||
| Southern State | 0.42* | 1.22 | 1.47 | 0.78 | 2.99 | .58 | ||||
| Racial Composition in Neighborhood |
||||||||||
| 100 % or mostly nonwhite |
– – | – – | – – | – – | – – | |||||
| Half nonwhite | 0.88 | 0.76 | – – | – – | – – | |||||
| Mostly white | 1.19 | o.23* | – – | – – | – – | |||||
| Completely white | 0.85 | 0.25† | – – | – – | – – | |||||
| Black Neighborhood | – – | – – | – – | – – | 0.30* | |||||
| White Neighborhood | – – | – – | – – | – – | 0.27* | |||||
| % Same-Race Friends | ||||||||||
| 0 % to 50 % | – – | – – | – – | – – | – – | |||||
| 51 % to 75 % | 1.20 | 0.49 | – – | – – | – – | |||||
| 76 % to 100 % | 0.81 | 0.30* | – – | – – | – – | |||||
| Racial Heterogeneity of Respondent’s Friend Network |
1.03† | 1.03* | ||||||||
| African Ancestry | 0.94*** | 0.92*** | .91*** | |||||||
| European Ancestry | 0.88*** | 0.89*** | .95*** | |||||||
| Primary Ancestry | – – | 0.94*** | – – | 0.90*** | ||||||
| Age | 1.02 | 0.92 | 0.87 | 0.95 | 0.80 | 1.13 | ||||
| Male | 1.77 | 0.88 | 0.64 | 1.15 | 0.59 | 0.91 | ||||
| −2 Log-Likelihood | 630.65 | 581.5 | 250.87 | 301.52 | 548.59 | 450.5 | 114.22 | 318.99 | 79.3 | 210.7 |
| Sample Size | 1,651 | 1,649 | 328 | 1,311 | 1,743 | 1,743 | 383 | 1,311 | 284 | 946 |
Note: Intercepts are omitted from the table.
p < .10;
p < .05;
p < .01;
p < .001
Table 8.
Descriptive statistics for the data used in the logistic regression analysis of the fluidity of racial classification
| ROOM | Add Health Wave-I | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| African Americans |
European Americans |
African Americans |
African Americans |
European Americans |
European Americans |
|
| Southern States (%) | 90.3 | 90.8 | 72.2 | 77.8 | 29.4 | 30.02 |
| Racial Composition in Neighborhood (%) |
– – | – – | ||||
| Completely or mostly nonwhite |
38.9 | 2.7 | – – | – – | ||
| Half nonwhite | 20.1 | 6.7 | – – | – – | ||
| Mostly white | 35.6 | 62.5 | – – | – – | ||
| Completely white | 5.5 | 28.1 | – – | – – | ||
| Black Neighborhood (%) | – – | – – | 53.1 | 53.5 | ||
| White Neighborhood (%) | – – | – – | 97.0 | 97.15 | ||
| % Same-Race Friends | – – | – – | ||||
| 0 % to 50 % | 31.3 | 6.6 | – – | – – | ||
| 51 % to 75 % | 19.8 | 26.9 | – – | – – | ||
| 76 % to 100 % | 48.9 | 66.5 | – – | – – | ||
| Racial Heterogeneity of Respondent’s Friend Network (%) |
0.27 | 0.21 | ||||
| African Ancestry (%) | 88.0 | 91.2 | 90.8 | |||
| European Ancestry (%) | 98.3 | 96.5 | 97.3 | |||
| Age | 19.4 | 19.5 | 15.9 | 16.0 | 16.0 | 16.0 |
| Male (%) | 29.5 | 41.3 | 47.4 | 44.01 | 47.8 | 48.68 |
| Sample Size | 328 | 1,311 | 383 | 284 | 1,311 | 946 |
For self-reported white participants in ROOM (Model 4), an increase of 1 % European ancestry reduced the likelihood of classification switching by 12 %. Model 4 indicates that in addition to bio-ancestry, social environment also influences classification switching among white students. Those whose neighborhoods were mostly white were 77 % less likely to switch racial classification than those whose neighborhoods were completely or mostly nonwhite. The coefficient estimate for those whose neighborhoods were completely white is similar (.25), but the estimate is statistically significant at the .10 level. White participants whose friends were 76 % to 100 % white were 70 % less likely to change racial classification than those whose friends were 0 % to 50 % white. The neighborhood and friend effects were estimated in the same model.
In Add Health, black adolescents were about twice as likely to switch racial classification as white adolescents when bio-ancestry was not controlled. The black and white difference disappeared after bio-ancestry was included in the model (Model 6). Among blacks, “Southern State” as measured in Add Health was not related to classification switching. Among both blacks and whites, living in a census block group in which the mode of racial composition was the same as one’s own race was associated with a 70 % lower likelihood of racial classification switching. We replaced the measure of neighborhood racial composition by a measure of racial heterogeneity in respondent’s friendship networks created from nominated friends in the in-school study at Wave I (Models 9 and 10). The racial heterogeneity ranges from 0 (where all in the networks are of the same race/ethnicity) to .8 (where all five racial/ethnic groups (black, Asian, Hispanic, white, and other) are equally presented). Higher racial heterogeneity is associated with a higher likelihood of classification change for both blacks and whites. The marginal significant result for blacks could be due to the reduction in sample size.
Discussion and Conclusion
Our research demonstrates a close match between estimated bio-ancestry and self-reported race among self-reported blacks, whites, and East Asians in ROOM and Add Health. Our overall analytical strategy for estimating bio-ancestry resembles that used for estimating the links between genetic variations and human traits. That strategy is composed of two essential components. The first is an association between a genetic variant and a human trait, and the second is a replication in one or more independent data sets. This strategy was used in a number of influential publications that identified genetic variants associated with human diseases (e.g., Frayling et al. 2007). In this project, the same panel of AIMs that differentiate European, African, and East Asian populations were first selected in the HapMap data set and then replicated in three independent data sets: the U.S. ROOM, the U.S. Add Health study, and the worldwide HGDP. If either sample representativeness or result predetermination were a serious threat, the replication of these findings across four independent data sources would be unlikely. Our results were also replicated across three different methods (as implemented in PLINK, STRUCTURE, and EIGENSTRAT) that estimate genetic clustering across continental populations.
The extent to which bio-ancestry matches self-classification of race, however, varies across social and cultural contexts. The one-drop rule represents an important case in which social context trumps bio-ancestry. When asked to classify into a single race, most individuals with 30 % to 60 % African ancestry self-report as black; virtually all respondents with >60 % African ancestry self-classify as black. In contrast, a substantially higher proportion of European ancestry is “required” to self-classify or to be classified by an interviewer as white than the proportion of African ancestry necessary to self-classify or be classified as black. However, when given the option of identifying as multiracial, the majority of individuals with 40 % to 60 % African ancestry in both ROOM and Add Health and substantial proportions of individuals with >60 % African ancestry in ROOM stopped self-classifying as only black and primarily chose a multiracial classification.
In summary, although the cultural legacy of the one-drop rule is still evident among the youth in survey responses, the practice has been eroded by recent modifications in survey questions of race and ethnicity. Given the choice of multiracial categories, large proportions of black-white mixed individuals self-classify as multiracial rather than black. This tendency to follow the one-drop rule is observed only among non-Hispanic white, black, and black-white individuals—not among Hispanics. This observation is consistent with the black-nonblack divide discussed recently by Bean et al. (2009) and Lee and Bean (2007). The recent nonwhite racial/ethnic diversity from immigration, the growth of intermarriage, and the rise of multiracial births have not erased the traditional black-white color line. Instead, the United States may simply be redrawing a color line that divides blacks from other racial/ethnic groups.
The fluidity in racial classification represents another major case in which social forces interact with bio-ancestry to shape racial classification. In both ROOM and Add Health, the racial composition of an individual’s social environment is important. In ROOM, white students from a mostly white neighborhood and with mostly white friends are less likely to change racial classification from white to a multiracial category. In Add Health, both black and white students from neighborhoods composed mostly of own-race residents are less likely to change racial classification. Replacing racial composition in neighborhoods by racial composition in one’s friend networks yielded similar results.
After bio-ancestry is adjusted for, blacks are more likely than whites to opt for another racial classification when multiracial categories were an option. This finding was found only in ROOM, not in Add Health. In ROOM, black students from a southern state were less likely than those from other parts of the country to change racial classification. This result may be explained by the observation that the American South is the region where the one-drop rule first originated (Davis 1991) and where racial discrimination and segregation were practiced legally and overtly.
A cautionary note should be made about the comparison between the housing form and the online survey in ROOM, and between ROOM and Add Health. The different responses to the two surveys in ROOM could have resulted from factors other than differences in the questions. Factors such as college education could play a role. Similarly, the differences in the results between ROOM and Add Health could be due to the differences in how responses on racial classification were obtained in the two studies. Students ages 12–18 in Add Health might have treated a race/ethnicity question in a survey less seriously than incoming college freshmen treated a similar question on a housing application form. The information on the housing form would be part of the official university database. Even though the university housing authority did not use race and ethnicity for assigning a dormitory room, students may not have known this. In addition, students may be concerned about whether the expectation created by self-reported race and ethnicity on the housing form would be in agreement with their prospective roommates’ conceptualization of race and ethnicity.
Another case in which self-reports did not match bio-ancestry occurred among those who self-classified as American Indian. Averaging a European ancestry of 67 % and 63 %, respectively, in ROOM and Add Health, and with distal ties to American Indians, these individuals were predominantly of European ancestry. These findings explain the drastic rise in the number of American Indians reported in the U.S. census over the past few decades as a result of ethnic re-identification (Eschbach 1993; Kelly and Nagel 2002; Nagel 1995).
The analysis reveals many fewer individuals with an African ancestry of 10 % to 50 % than individuals with an African ancestry of 50 % to 90 %. This imbalanced distribution is unlikely to result from the fact that there are many more whites than blacks. As long as a mixed union requires a white person and a black person, the marginal distribution in terms of the number of persons (not the proportions) should be balanced. This imbalanced distribution is likely a result of the one-drop rule and/or the minimal miscegenation between African and European Americans since 1865 (Davis 1991: chapters 3–4; Williamson 1980:188). For many decades, mixed-race individuals with one black parent and one white parent were treated as blacks rather than mixed-race individuals. Under such racial exclusion, these mixed-race individuals partnered predominantly with other mixed-race or black individuals rather than whites. These patterns of marriages redistributed the European ancestry in the original mixed-race individuals, “whitening” the general black population and yielding few individuals of more than 50 % European ancestry.
Our findings apply only to the contemporary United States. The dynamics of racial classification in other countries could be quite different. Race is fluid. The racial and ethnic categories as we know them in the contemporary United States are constantly changing. Ongoing immigration, intermarriage, and social mobility are likely to blur contemporary racial and ethnic divisions and boundaries (Perez and Hirschman 2009); therefore, the racial categories we use today may no longer be relevant, or as relevant, in the future.
Our work has a larger theoretical significance on identity studies. Brubaker and Cooper (2000) criticized the overproduction of the word of “identity” in the social analysis of such concepts as race, gender, and sexual orientation in social sciences, cultural studies, ethnic studies, literature, and political philosophy. They argued: “… that the prevailing constructivist stance on identity—the attempt to ‘soften’ the term, to acquit it of the charge of ‘essentialism’ by stipulating that identities are constructed, fluid, and multiple —leaves us without a rationale for talking about ‘identities’ at all and ill-equipped to examine the ‘hard’ dynamics and essentialist claims of contemporary identity politics” (p. 1). For example, they asked, “If [identity] is constructed, how can we understand the sometimes coercive force of external identifications?” (p. 1).
Brubaker and Cooper were not opposed to social construction per se. In the particular case of “race” in the United States, for example, they promoted a detailed analysis of how particular forms of social construction of race “emerge, crystallize, and fade away in particular social and political circumstances” (p. 30). They maintained that construction analysis should not be reduced to an oversimplified and flattened identity account.
Our work demonstrates that in the case of race, social construction could be analyzed and examined against a measurable continental and biological ancestry. Race is, indeed, multiple and fluid, but not all identifications of race are equally constructed. Some deviate more and some less from bio-ancestry. Capitalizing on bio-ancestry, social construction analysis can lay bare whether, how much, and under what social circumstances racial identification departs from bio-ancestry.
Acknowledgements
Two grants to Guang Guo supported the College Roommate Study (the William T. Grant Foundation) and the Illumina 1536 genotyping in Add Health (NSF’s Human and Social Dynamics program BCS-0826913). Data from Add Health were funded by the National Institute of Child Health and Human Development, with cooperative funding from 17 other agencies (www.cpc.unc.edu/addhealth/contract.html) to Kathleen Mullan Harris (P01-HD31921). Special acknowledgment is due Rick Bradley of the Housing Department, Kirk Wilhelmsen of the Genetics Department, Patricia Basta of the Bio-Specimen Process Center, Jason Luo of the Mammalian Genotyping Center, and the Odum Institute at the University of North Carolina, Chapel Hill. We received important assistance in SNP selection and the analysis of HGDP data from David Goldman and his Neurogenetics lab at NIAAA. Many hearty thanks go to Greg Duncan for his important role in the project and his helpful comments on the manuscript.
Footnotes
See Rosenberg et al. (2003) for a technical justification.
Contributor Information
Guang Guo, Department of Sociology and Carolina Population Center, University of North Carolina, CB#3210, Chapel Hill, NC 27599-3210 guang_guo@unc.edu; Carolina Center for Genome Sciences, University of North Carolina, Chapel Hill.
Yilan Fu, Department of Sociology and Carolina Population Center, University of North Carolina, CB#3210, Chapel Hill, NC 27599-3210.
Hedwig Lee, Department of Sociology, University of Washington, Seattle.
Tianji Cai, Department of Sociology, University of Macau, Av. Padre Tomás Pereira, Taipa, Macau.
Kathleen Mullan Harris, Department of Sociology and Carolina Population Center, University of North Carolina, CB#3210, Chapel Hill, NC 27599-3210; Carolina Center for Genome Sciences, University of North Carolina, Chapel Hill.
Yi Li, Department of Sociology and Carolina Population Center, University of North Carolina, CB#3210, Chapel Hill, NC 27599-3210.
References
- Bamshad M, Wooding S, Salisbury BA, Stephens JC. Deconstructing the relationship between genetics and race. Nature Reviews Genetics. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- Bean FD, Feliciano C, Lee J, Van Hook J. The new US immigrants: How do they affect our understanding of the African American experience? Annals of the American Academy of Political and Social Science. 2009;621:202–220. doi: 10.1177/0002716208325256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry B, Tischler HL. Race and ethnic relations. Houghton Mifflin Co.; Boston, MA: 1978. [Google Scholar]
- Bonilla-Silva E. White supremacy and racism in the post-civil rights era. Lynne Rienner Publishers, Inc.; London, UK: 2001. [Google Scholar]
- Brown TN. Predictors of racial label preference in Detroit: Examining trends from 1971 to 1992. Sociological Spectrum. 1992;19:421–442. [Google Scholar]
- Brubaker R, Cooper F. Beyond “identity.”. Theory and Society. 2000;29:1–47. [Google Scholar]
- Brunsma DL. Public categories, private identities: Exploring regional differences in the biracial experience. Social Science Research. 2006;35:555–576. [Google Scholar]
- Campbell ME, Troyer L. The implications of racial misclassification by observers. American Sociological Review. 2007;72:750–765. [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Cavalli-Sforza LL. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Cooley CH. Human nature and the social order. Schocken; New York: 1902. [Google Scholar]
- Davis FJ. Who is black?: One nation’s definition. The Pennsylvania State University Press; University Park, PA: 1991. [Google Scholar]
- Duster T. Medicine. Race and reification in science. Science. 2005;307:1050–1051. doi: 10.1126/science.1110303. [DOI] [PubMed] [Google Scholar]
- Efron B, Tbshirani R. An introduction to the bootstrap. Chapman & Hall; Boca Raton, FL: 1993. [Google Scholar]
- Enoch M, Shen P, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. Journal of Psychopharmacology. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Eschbach K. Changing identification among American Indians and Alaska natives. Demography. 1993;30:635–652. [PubMed] [Google Scholar]
- Fairlie RW. Can the “one-drop rule” tell us anything about racial discrimination? New evidence from the multiple race question on the 2000 census. Labour Economics. 2009;16:451–460. [Google Scholar]
- Farley R. The new census question about ancestry: What did it tell us. Demography. 1991;28:411–429. [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlaender JS, Friedlaender FR, Reed FA, Kidd KK, Kidd JR, Chambers GK, Weber JL. The genetic structure of Pacific Islanders. PLoS Genetics. 2008;4:e19. doi: 10.1371/journal.pgen.0040019. doi:10.1371/journal.pgen.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frudakis T, Venkateswarlu K, Thomas MJ, Gaskin Z, Ginjupalli S, Gunturi S, Nachimuthu PK. A classifier for the SNP-Based inference of ancestry. Journal of Forensic Sciences. 2003;48:771–782. [PubMed] [Google Scholar]
- Fyr CLW, Kanaya AM, Cummings SR, Reich D, Hsueh WC, Reiner AP, Ziv E. Genetic admixture, adipocytokines, and adiposity in black Americans: The health, aging, and body composition study. Human Genetics. 2007;121:615–624. doi: 10.1007/s00439-007-0353-z. [DOI] [PubMed] [Google Scholar]
- Gould SJ. The mismeasure of man. W.W. Norton & Co.; New York: 1981. [Google Scholar]
- Guthrie RD. The mammoth steppe and the origin of mongoloids and their dispersal. In: Akazawa T, Szathmary E, editors. Prehistoric mongoloid dispersals. Oxford University Press; New York: 1996. pp. 172–186. [Google Scholar]
- Hahn RA, Mulinare J, Teutsch SM. Inconsistencies in coding of race and ethnicity between birth and death in US infants. A new look at infant mortality, 1983 through 1985. Journal of the American Medical Association. 1992;267:259–263. [PubMed] [Google Scholar]
- Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: Utility and applications. Human Mutation. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- Harris DR, Sim JJ. Who is multiracial? Assessing the complexity of lived race. American Sociological Review. 2002;67:614–627. [Google Scholar]
- Harris KM, Halpern C, Tabor J, Bearman PS, ones J, Udry JR. The National Longitudinal Study of Adolescent Health: Research design. 2009 [Google Scholar]
- Herman MR. Do you see what I am? How observers’ backgrounds affect their perceptions of multiracial faces. Social Psychology Quarterly. 2010;73:58–78. [Google Scholar]
- Hill M. Skin color and the perception of attractiveness among African Americans: Does gender make a difference? Social Pyschology Quarterly. 2002;65:77–91. [Google Scholar]
- Hirschman C, Alba R, Farley R. The meaning and measurement of race in the US census: Glimpses into the future. Demography. 2000;37:381–393. [PubMed] [Google Scholar]
- Hitlin S, Brown JS, Elder GH. Racial self-categorization in adolescence: Multiracial development and social pathways. Child Development. 2006;77:1298–308. doi: 10.1111/j.1467-8624.2006.00935.x. [DOI] [PubMed] [Google Scholar]
- Kelly ME, Ngel J. Ethnic re-identification: Lithuanian Americans and Native Americans. Journal of Ethnic and Migration Studies. 2002;28:275–289. [Google Scholar]
- Khanna N. The role of reflected appraisals in racial identity: The case of multiracial Asians. Social Psychology Quarterly. 2004;67:115–131. [Google Scholar]
- Khanna N. “If you’re half black, you’re just black”: Reflected appraisals and the persistence of the one-drop rule. Sociological Quarterly. 2010;51:96–121. [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge, UK: 1983. [Google Scholar]
- Lee J, Bean FD. Reinventing the color line immigration and America’s new racial/ethnic divide. Social Forces. 2007;86:561–586. [Google Scholar]
- Lewontin RC. The apportionment of human diversity. Evolutionary Biology. 1972;6:391–398. [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1004. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- López IH. White by law: The legal construction of race. New York University Press; New York: 1996. [Google Scholar]
- Myrdal G, Sterner R, Rose AM. An American dilemma. Harper & Bros; New York: 1944. assisted by. [Google Scholar]
- Nagel J. Constructing ethnicity: Creating and recreating ethnic-identity and culture. Social Problems. 1994;41:152–176. [Google Scholar]
- Nagel J. American Indian ethnic renewal: Politics and the resurgence of identity. American Sociological Review. 1995;60:947–965. [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, IBustamante CD. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi M, Winant H. Racial formations in the United States. Routledge; New York: 1994. [Google Scholar]
- Parra EJ, Marcini A, Akey L, Martinson J, Batzer MA, Cooper R, Shriver MD. Estimating African American admixture proportions by use of population-specific alleles. American Journal of Human Genetics. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner AM, Saperstein A. How social status shapes race. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19628–19630. doi: 10.1073/pnas.0805762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AD, Hirschman C. The changing racial and ethnic composition of the US population: Emerging American identities. Population and Development Review. 2009;35:1–51. doi: 10.1111/j.1728-4457.2009.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann J, Waters MC. Introduction. In: Perlmann J, Waters MC, editors. The new race question: How the census counts multiracial individuals. Russell Sage Foundation; New York: 2002. pp. 1–32. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Ziv E, Lind DL, Nievergelt CM, Schork NJ, Cummings SR, Kwok PY. Population structure, admixture, and aging-related phenotypes in African American adults: The cardiovascular health study. American Journal of Human Genetics. 2005;76:463–477. doi: 10.1086/428654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockquemore KA, Brunsma DL. Beyond black: Biracial identity in America. Sage Publications; Thousand Oaks, CA: 2001. [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. American Journal of Human Genetics. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Roth WD. The end of the one-drop rule? Labeling of multiracial children in black intermarriages. Sociological Forum. 2005;20:35–67. [Google Scholar]
- Rotimi CN. Are medical and nonmedical uses of large-scale genomic markers conflating genetics and “race?”. Nature Genetics. 2004;36:S43–S47. doi: 10.1038/ng1439. [DOI] [PubMed] [Google Scholar]
- Rotimi CN. Genetic ancestry tracing and the African identity: A double-ediged sword? Developing World Bioethics. 2003;3:151–158. doi: 10.1046/j.1471-8731.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- Saperstein A. Double-checking the race box: Examining inconsistency between survey measures of observed and self-reported race. Social Forces. 2006;85:57–74. [Google Scholar]
- Shriver MD, Smith MW, Jin L, Marcini A, Akey JM, Deka R, Ferrell RE. Ethnic-affiliation estimation by use of population-specific DNA markers. American Journal of Human Genetics. 1997;60:957–964. [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O’Brien SJ. Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. American Journal of Human Genetics. 2001;69:1080–1094. doi: 10.1086/323922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surratt HL, Inciardi JA. Unraveling the concept of race in Brazil: Issues for the Rio de Janeiro Cooperative Agreement site. Journal of Psychoactive Drugs. 1998;30:255–260. doi: 10.1080/02791072.1998.10399700. [DOI] [PubMed] [Google Scholar]
- Tang H, Quertermous T, Rodriguez B, Kardia SLR, Zhu XF, Brown A, Risch NJ. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. American Journal of Human Genetics. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro CJ. Considering the significance of ancestry through the prism of mixed-race identity. Advances in Nursing Science. 2002;25(2):1–21. doi: 10.1097/00012272-200212000-00002. [DOI] [PubMed] [Google Scholar]
- Telles EE. Race in another America: The significance of skin color in Brazil. Princeton University Press; Princeton, NJ: 2006. [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton MC, Taylor RJ, Brown TN. Correlates of racial label use among Americans of African descent: Colored, Negro, black, and African American. Race and Society. 2000;2:149–164. [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Ruiz-Linares A. Genetic variation and population structure in Native Americans. PLoS Genetics. 2007;3:2049–2067. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MC. Ethnic options: Choosing identities in America. University of California Press; Los Angeles: 1990. [Google Scholar]
- Williamson J. New people: Miscegenation and mulattoes in the United States. The Free Press; New York: 1980. [Google Scholar]
- Yaeger R, Avila-Bront A, Abdul K, Nolan PC, Grann VR, Birchette MG, Joe AK. Comparing genetic ancestry and self-described race in African Americans born in the United States and in Africa. Cancer Epidemiology Biomarkers & Prevention. 2008;17:1329–1338. doi: 10.1158/1055-9965.EPI-07-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Li HZ, Criswell LA, Gregersen PK, Alarcon-Riquelme ME, Kittles R, Seldin MF. Examination of ancestry and ethnic affiliation using highly informative diallelic DNA markers: application to diverse and admixed populations and implications for clinical epidemiology and forensic medicine. Human Genetics. 2005;118:382–392. doi: 10.1007/s00439-005-0012-1. [DOI] [PubMed] [Google Scholar]