Abstract
Hydrogen peroxide (H2O2) is a “green chemical” that has various cleaning and disinfectant uses, including as an anti-bacterial agent for hygienic and medical treatments. However, its efficacy is limited against biofilm-producing bacteria, because of poor penetration of the protective, organic matrix. Here we show new applications for ferromagnetic nanoparticles (Fe3O4, MNP) with peroxidase-like activity in potentiating the efficacy of H2O2 in biofilm degradation and prevention. Our data show that MNP enhanced oxidative cleavage of biofilm components (model nucleic acids, proteins, and oligosaccharides) in the presence of H2O2. When challenged with live, biofilm-producing bacteria, the MNP-H2O2 system efficiently broke down existing biofilm and prevented new biofilm from forming, killing both planktonic bacteria and those within biofilm. By enhancing oxidative cleavage of various substrates, the MNP-H2O2 system provides a novel strategy for biofilm elimination, and other applications utilizing oxidative breakdown.
H2O2 has been used as a general anti-bacterial agent for hygienic and medical treatments. H2O2 generates free radicals which oxidize organic chemicals or biomolecules, but the process is slow with low efficiency and bacteria easily develop resistance,1, 2 especially in a formed biofilm. Biofilms represent an enormous medical challenge, responsible for millions of healthcare-associated infections annually world-wide.3–6 Biofilms consist of cells within a matrix of nucleic acids, proteins and polysaccharides, which connect and protect resident bacteria from external damage.7–9 Nanocatalysts that could potentiate the effects of H2O2 might have great utility for a variety of applications,10–14 potentially including biofilm destruction in the sterilization of medical surfaces, surgical instruments, and indwelling medical devices.
Recently, it has been reported that MNP possess an intrinsic peroxidase-like catalytic activity, which can effectively generate free radicals from H2O2 with high efficiency, similar to horseradish peroxidase (HRP).10, 15 MNP are promising nanocatalysts because of their magnetic properties, high catalytic activity, and adjustability through nanoscale modifications10, 16–19, suggesting possible uses in immunoassays10, 13, 20, 21, organic pollutant degradation12, 22–26, glucose detection17, 18, 27–31 and cancer diagnostics.14 Although it has been reported that MNP alone or combined with H2O2 can prevent biofilm formation by inhibiting bacterial growth,32, 33 there are no reports regarding the efficacy of an MNP-H2O2 system on biofilm destruction and killing of bacteria resident within biofilms—a much more difficult medical challenge.
Results and Discussion
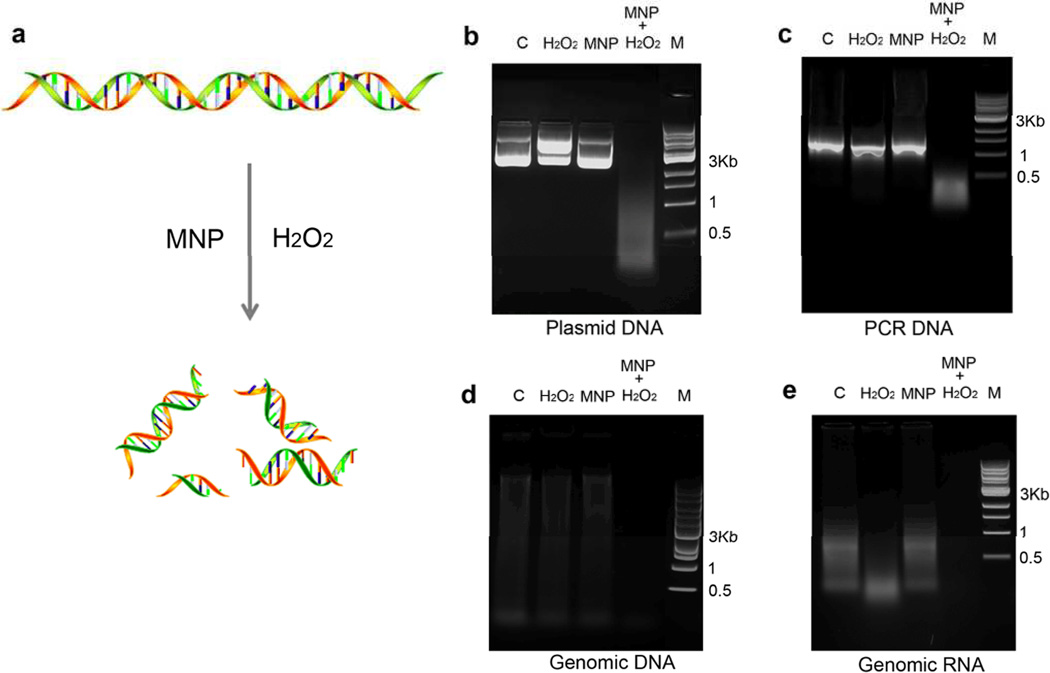
To test the ability of an MNP-H2O2 system to degrade biofilm and kill resident bacteria, we hydrothermally prepared MNP having a bulk morphological diameter of 500 nm, with a rough surface containing 5–10 nm diameter protrusions (Figure S1a). These nanoparticles show very high peroxidase-like activity as demonstrated by the 3,3’,5,5’-tetramethylbenzidine (TMB) colorimetric reaction (Figure S1b). We then used these MNP to test whether the MNP-H2O2 system could degrade each of the 3 major components of biofilms. We used H2O2 at concentrations of 1–3%, consistent with domestic hygiene uses, to investigate first the MNP-H2O2 system’s ability to degrade nucleic acids. Long chain plasmid DNA was completely cleaved into small fragments (Figure 1b). Plasmid DNA in the presence of H2O2 alone showed topological change from supercoiled structure to linearized form but catalysis into fragments was dependent upon the presence of MNP (Figure S2a). Cleavage of plasmid DNA was also dependent upon concentrations of H2O2 and DNA, as well as time and temperature (Figure S2a–d), but was not influenced by pH within the tested range of 4.5–9 (Figure S2e). The only slight difference among the various forms of nucleic acids we tested was that total RNA showed slight cleavage by H2O2 alone (Figure 1e). Our results indicated the MNP-H2O2 system could be used as a universal reagent for cleaving or degrading nucleic acids, suggesting additional applications for this system beyond biofilm degradation.
Figure 1. MNP enhanced oxidative cleavage of nucleic acids.
a, Schematic of MNP-H2O2 cleaving nucleic acids. b, Plasmid DNA cleavage with MNP-H2O2. Plasmid DNA (4 µg, ≈6500bp) was incubated with 3% H2O2 alone, MNP alone, or MNP-H2O2. “C” denotes control with DNA only. “M” denotes DNA marker (1 kb DNA Ladder, NEB) c, Cleavage of a PCR DNA product (1 µg), d, mouse genomic DNA (1 µg), and e, mouse total RNA (from testis) were all performed under similar conditions but with 1% H2O2. All experiments were repeated in triplicate with representative images shown.
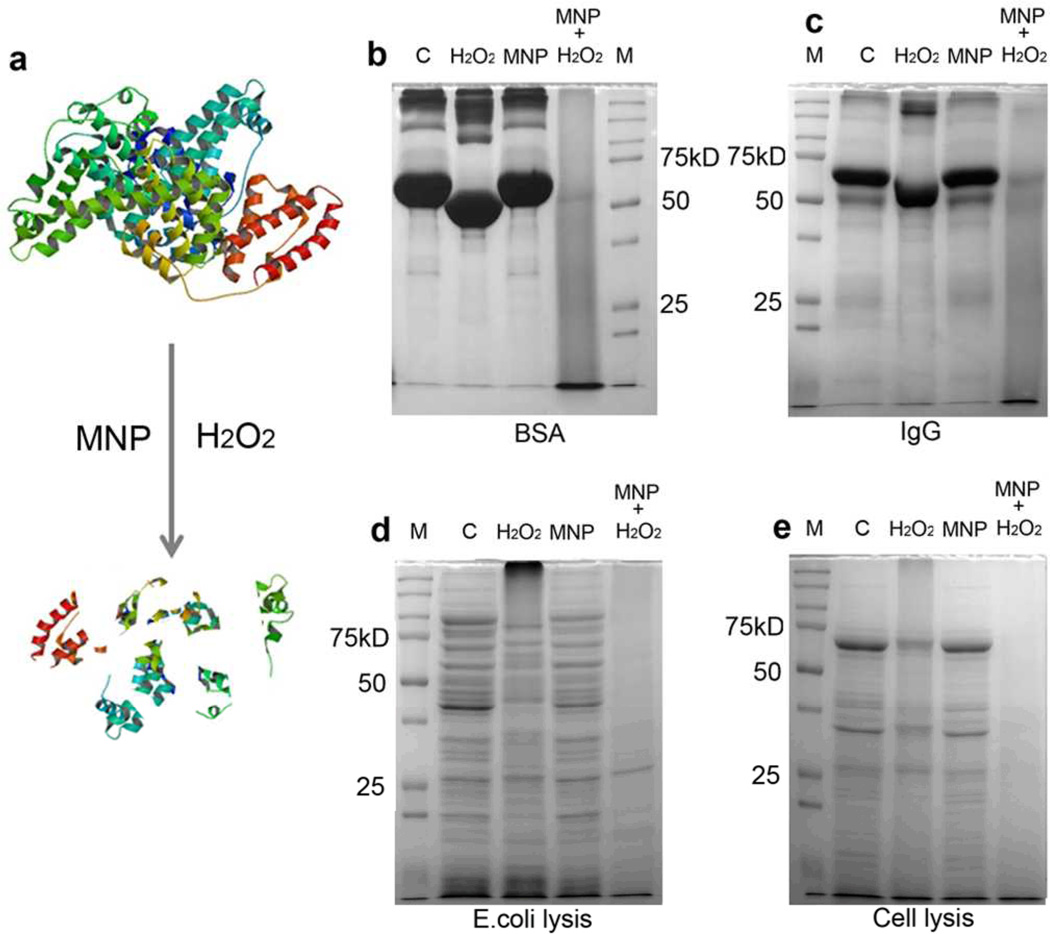
We next investigated the cleavage of proteins by the MNP-H2O2 system using similar conditions as above (Figure 2a). In our first trial, we found that BSA (80µg, MW=66KD), was cleaved into small fragments as seen in a SDS-PAGE gel stained with coomassie blue-R250. In contrast, BSA treated with 3% H2O2 alone showed very limited cleavage (Figure 2b). We next varied the concentrations of H2O2 or BSA while keeping the reaction buffer constant (Figure S3). Although higher concentrations of H2O2 led to some cleavage, complete cleavage was dependent upon the presence of MNP (Figure S3a). Lowering the amount of protein, or increasing the time of reaction or temperature from room temperature to 37°C resulted in more cleavage as expected (Figure S3b–d). However, unlike the effect on DNA cleavage, protein cleavage by the MNP-H2O2 system showed obvious pH dependence with increased cleavage at lower pH (Figure S3e). We assessed the versatility of the cleavage using more complex proteins, including IgG and lysates from whole bacteria and mammalian cells. We found that cleavage was not protein specific; mixtures of the wide variety of proteins found in cell lysates were completely cleaved under the same reaction conditions, demonstrating the generality of the cleavage mediated by the MNP-H2O2 system.
Figure 2. MNP enhanced oxidative cleavage of proteins.
a, Schematic of MNP-H2O2 cleaving protein. b, Cleavage of BSA with MNP-H2O2. 80µg BSA was incubated with 3% H2O2 alone, MNP alone, or MNP-H2O2. “C” denotes control with BSA only. “M” denotes protein molecular weight marker (Precision Plus Protein™ Dual Color Standards, BIO-RAD) c, Cleavage of rabbit polyclonal IgG (10µg), d, E. coli lysate (40µg), and e, HEK293 mammalian cell lysate (20µg), were all performed under similar conditions except with 1% H2O2. All experiments were repeated in triplicate with representative images shown.
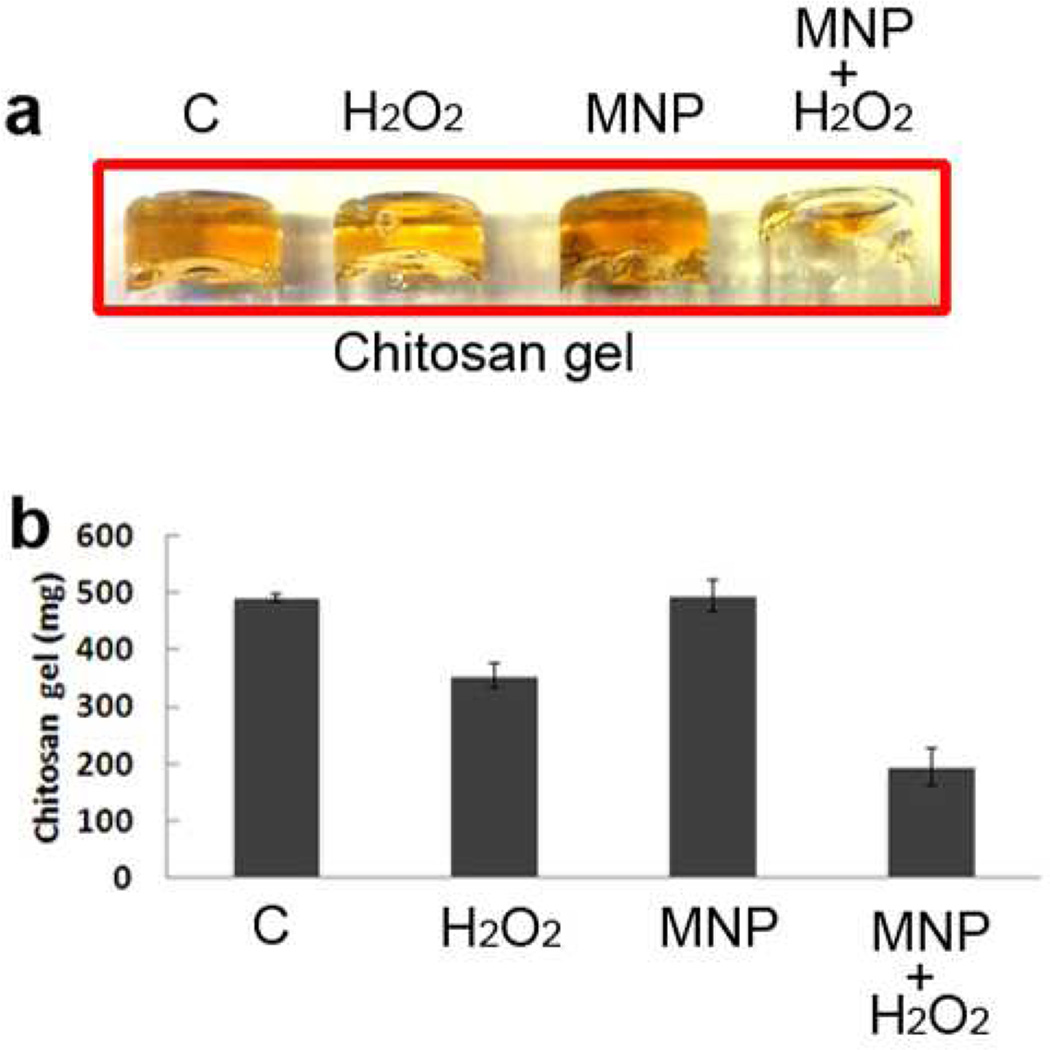
Polysaccharides are the third major component in biofilm matrix.34 We chose chitosan as a model polysaccharide to test the effect of MNP-H2O2 versus H2O2 alone. We prepared a chitosan hydrogel by glutaraldehyde crosslinking in a glass vial. The chitosan gel had a yellow color with high adherence and viscosity, enabling it to stay suspended upside down in a tube (Figure 3a). Although H2O2 alone cleaved the crosslinked chitosan to a degree, MNP-H2O2 had a greater effect at reducing the mass that remained as a gel (Figure 3). We also found that chitosan cleavage was independent of pH, with cleavage occurring under both acidic (pH 4.5) and neutral (pH 7.4) conditions (data not shown).
Figure 3. MNP enhanced cleavage of a polysaccharide (chitosan).
Chitosan gel was prepared at 0.5% with glutaraldehyde crosslinking. Chitosan gels were mixed with 1% H2O2 alone, MNP alone, or MNP-H2O2 to test for cleavage. “C” denotes control with chitosan only. All experiments were repeated in triplicate with representative images shown. When individually compared against the control, H2O2 and MNP-H2O2 were found to be significantly different (p<0.05, n=3, Student’s T test). When compared against each other, the amount of biofilm remaining after treatment with MNP-H2O2 was significantly less than when treated with H2O2 alone (p<0.05, n=3, Student’s T test). Error bars denote standard deviation.
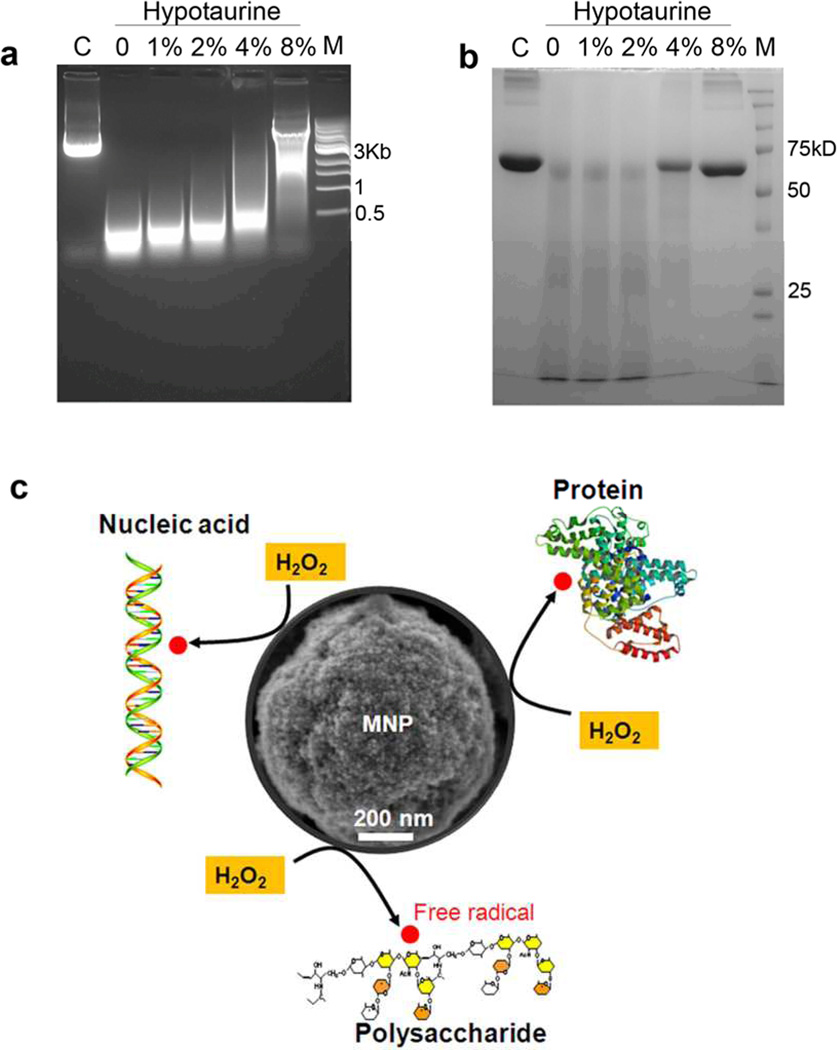
We next sought to verify the mechanism by which the MNP-H2O2 system was able to enhance cleavage of nucleic acids, proteins, and polysaccharides, hypothesizing that the benefits were obtained through production of additional free radical oxidants generated by MNP catalysis of H2O2. To test the hypothesis, we investigated the impacts on cleavage of an anti-oxidant reagent, hypotaurine, which specifically scavenges hydroxyl radicals. Figure 4a shows plasmid DNA cleavage in the presence of varied concentrations of hypotaurine (1%–8%). We observed that cleavage was retarded with increasing concentrations of hypotaurine until it was almost stopped with 8% hypotaurine. Figure 4b shows a similar reduction in cleavage of BSA. These data suggested that enhanced cleavage was dependent on free hydroxyl radicals produced by MNP with peroxidase-like activity (Figure 4c).
Figure 4. Mechanism of MNP-enhanced oxidative cleavage.
The anti-oxidant, hypotaurine, inhibited cleavage of 2µg plasmid DNA (a) or 20µg BSA (b) by MNP-H2O2 in a dose-dependent fashion “M” denotes DNA or protein marker, respectively. “C” denotes control with DNA or BSA only. All experiments were repeated in triplicate with representative images shown. c, Schematic of MNP-enhanced cleavage of nucleic acid, protein and polysaccharide. MNP catalyze H2O2 with high efficiency, making free radicals that attack these biomolecules.
Therefore, it should be possible to control the kinetics or extent of cleavage by adjusting the generation of these free radical oxidants. A simple way to do this would be to change the amount of the catalyst, MNP. We varied the amounts of MNP (0–20µg) while keeping the other components constant in the cleavage reaction and found that 10–20µg of MNP cleaved 2µg of plasmid DNA into small pieces below 100bp, but 5µg of MNP resulted in fragments between 500bp-1000bp (Figure S4). The effects on cleavage of BSA, were similar in that 5µg of MNP only moderately reduced the intensity of the full-length band, with increasing cleavage by 10 or 20 µg MNP (Figure S5).
To this point, our data were consistent with the Fenton reaction, in which iron and H2O2 interact, yielding radical oxidants. We wished to determine whether cleavage was initiated from the nanoparticles, or from the supernatant which might have leached free Fe ions. To investigate this, we pre-incubated the MNP in the NaAc buffer (0.1M, pH 4.5, 1 hr) without adding H2O2 or DNA/BSA. We then collected the MNP by centrifugation, removed the supernatant, and resuspended the MNP in the same buffer. We separately conducted cleavage reactions as above by adding H2O2 and plasmid DNA or BSA to both the supernatant and the resuspended MNP. We observed significantly higher cleavage for both DNA and BSA in the tubes with the MNP (Figure S6a, b), suggesting that the MNP were primarily responsible for cleavage, which is consistent with our previous research and the findings of others under similar experimental conditions.10, 16, 27
Having demonstrated an ability to enhance cleavage of the individual components of biofilm, we next sought to test the effects of the MNP-H2O2 system on live bacteria. When E. coli were plated, 1% H2O2 in 50µl NaAc buffer (0.1M, pH 4.5) killed 98% of the bacteria within 5 min, in the presence or absence of MNP (Figure S7a). However, we observed equivalent killing when incubated with 0.01% H2O2 in the presence of 20µg MNP, confirming enhanced bacterial killing by the MNP-H2O2 (Figure S7b). This result was consistent with prior studies using MNP for anti-bacterial properties including prevention of biofilm formation.32
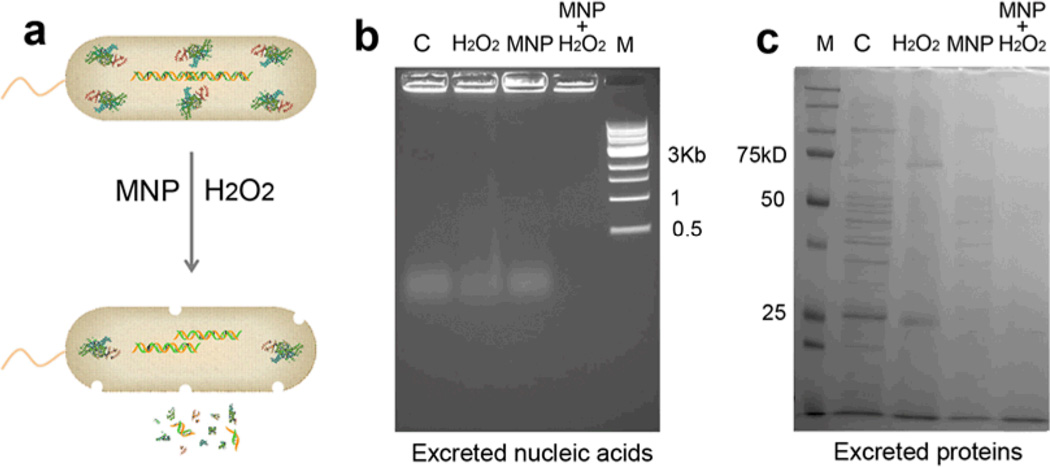
As E. coli die, they release a complex mixture of cellular components including nucleic acids and proteins. The released biomass might accumulate around resistant cells and, therefore, contribute to biofilm formation and protection from disinfectants or antimicrobial treatments. To test whether the MNP-H2O2 system would degrade these complex mixtures, we collected the released nucleic acids and proteins and incubated them with MNP-H2O2 or controls, including either MNP or H2O2. Figure 5 shows that treatment with MNP-H2O2 was successful at degrading these released products, thus not only efficiently killing bacteria, but also degrading the biomass released from the dead cells.
Figure 5. Killing of E. coli and cleavage of released nucleic acids and proteins by the MNP-H2O2 system.
a, Schematic of MNP-H2O2 killing of E. coli and cleavage of released nucleic acids and proteins. b, MNP-H2O2 cleaved nucleic acids released from killed E. coli cells. “C” denotes control with nucleic acids only. “M” denotes DNA marker. c, MNP-H2O2 cleaved proteins released from killed E. coli cells. “M” denotes protein marker. These experiments show that the MNP- H2O2 system was able to degrade the complex mixture of organic components that results from bacterial death. Penetration of organic materials would be an important characteristic of a disinfectant. All experiments were repeated in triplicate with representative images shown.
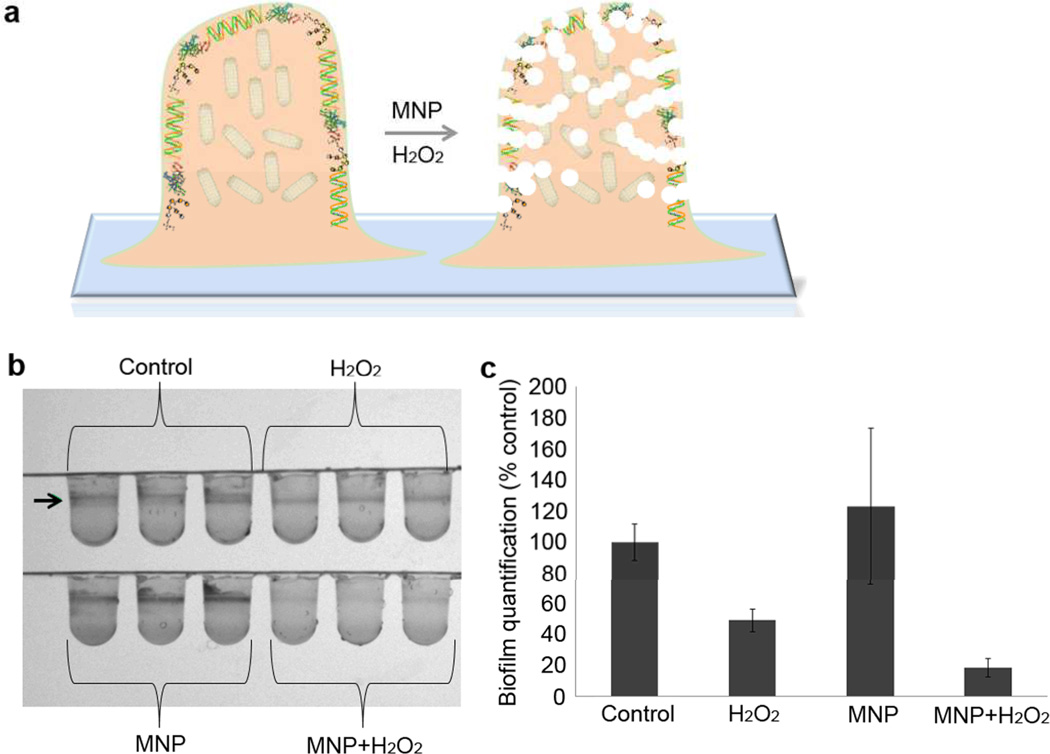
These data led us to study whether the MNP-H2O2 system could penetrate and eliminate biofilm, facilitating killing of resident bacteria as well as planktonic bacteria. We addressed these questions using biofilm from Pseudomonas aeruginosa, a common cause of nosocomial infection and often resistant to treatment due to biofilm. We allowed the bacteria to form biofilm in 96-well plates in advance of treatment with MNP-H2O2, and then observed and quantified remaining biofilm with 0.1 % crystal violet. Treatment with MNP-H2O2 in either NaAc buffer (0.1M, pH 4.5, 1% H2O2, for 2 hr) or biofilm minimal media (M63 medium, pH≈7) both resulted in obvious reduction in remaining biofilm (Figure 6, 18±6.17% of the biofilm remained after treatment). In contrast, wells treated with either H2O2 or MNP alone still showed a clear biofilm band, with 49±7.37% of the biofilm remaining after treatment with H2O2 alone. The difference demonstrated that MNP-H2O2 eliminated formed biofilm with much higher efficiency than H2O2. We next assessed the viability of bacteria resident within the biofilm, quantifying the number of colony forming units (CFU) of bacteria recovered from untreated biofilm (control), versus MNP alone, versus H2O2 alone, versus MNP-H2O2. MNP alone-treated biofilm had almost no change in CFU versus the control. (Figure S8) Incubation with H2O2 alone reduced CFU by approximately 2 orders of magnitude, whereas MNP-H2O2 reduced CFU by an additional factor of 20 (Figure S8).
Figure 6. Biofilm elimination by MNP-enhanced oxidative cleavage.
a, Schematic of MNP-H2O2 eliminating biofilm by cleaving nucleic acids, proteins and polysaccharides in the biofilm. b, Pseudomonas aeruginosa biofilm elimination with MNP-H2O2. The formed biofilm (remaining planktonic cells were removed from wells) was treated with MNP-H2O2 and stained with crystal violet dye (arrow points to the ring of biofilm). This experiment was repeated in triplicate with a representative image shown. c, Quantification of biofilm remaining after MNP-H2O2 treatment. When individually compared against the control, H2O2 and MNP-H2O2 were found to be significantly different (p<0.05, n=3, Student’s T test). When compared against each other, the amount of biofilm remaining after treatment with MNP-H2O2 was significantly less than when treated with H2O2 alone (p<0.05, n=3, Student’s T test). Error bars denote standard deviation.
Conclusions
The combined results of our in vitro assays and biofilm quantification and cell viability assays suggested that the MNP-H2O2 system had marked advantages over the use of H2O2 alone. In particular, the intrinsic peroxidase-like activity of MNP resulted in the enhanced cleavage of nucleic acids, proteins and polysaccharide. The new features could be used for the degradation and penetration of formed biofilm, as well as the killing of planktonic bacteria and prevention of biofilm formation. Enhanced oxidative cleavage by the MNP-H2O2 system therefore provides a promising new approach for cleaning of medical surfaces and instruments that are often resistant to disinfection because of biofilms.
Materials and Methods
Sodium acetate (NaAc), ethanol, Coomassie blue R-250 were purchased from Fisher Scientific. Ethylene glycol was purchased from J.T. Baker. Iron (III) chloride (FeCl3), bovine serum albumin (BSA), hydrogen peroxide (H2O2) (30% stock), chitosan (low molecular weight), ethidium bromide (EB), hypotaurine, HRP, 3,3',5,5'-tetramethylbenzidine (TMB), mouse IgG, and anti-mouse IgG were purchased from Sigma-Aldrich (St. Louis, MO, USA). Plasmid DNA, testis RNA, and lysates from E.coli and HEK293 cells were prepared in our laboratory in the presence of protease inhibitors (Protease Inhibitor Cocktail Tablets, Roche Applied Science) using standard approaches.
Preparation of ferromagnetic nanoparticles with peroxidase-like activity
Ferromagnetic nanoparticles (MNPs) were prepared and used as catalysts in these experiments. The Fe3O4 MNPs were synthesized in one-step in a solvothermal system by combining FeCl3 and NaAc in ethylene glycol. Briefly, 0.82 g of FeCl3 was dissolved in 40 ml of ethylene glycol to form a clear solution. Then, 3.6 g of NaAc was added to the solution with vigorous stirring for 30 min. The mixture was then transferred to a 50 ml teflon-lined stainless-steel autoclave and left to react at 200 °C for 12 h. After the autoclave cooled to room temperature, the black precipitate was collected, rinsed several times using ethanol and then dried at 60°C. The synthesized nanoparticles were characterized using scanning electron microscopy (SEM; Philips XL-30 field, 15 kV). The peroxidase-like activity was tested in a mixture of 500µl NaAc buffer (0.1M, pH 4.5) containing 20µg MNP, 0.3% H2O2 and 100µg TMB. The blue color produced was recorded with a spectrophotometer at 652nm.
Cleavage of nucleic acids and proteins
Unless indicated otherwise, nucleic acid cleavage assays were performed at 37°C for 3 hours in 50µl NaAc buffer (0.1M, pH 4.5) containing 20µg MNP and 1–3% H2O2. In these and later experiments, nucleic acid cleavage products were identified with agarose gel electrophoresis and ethidium bromide staining. Protein cleavage assays were performed at 37°C for 3 hours in 50µl NaAc buffer (0.1M, pH 4.5) containing 20µg MNP and 1–3% H2O2 prior to separation with SDS-PAGE and Coomassie staining.
Cleavage of polysaccharides
Chitosan hydrogel was prepared with glutaraldehyde crosslinking. Briefly, chitosan at 0.5% was dissolved in 500µl NaAc (0.1 M, pH 4.5) and incubated with 0.2% glutaraldehyde at 37°C for 1 hour. The formed hydrogel had yellow color in a glass vial and its weight was recorded as “before cleavage”. 120 µg of MNP, or 120 µg of MNP with 1% H2O2 in 300µl NaAc (0.1 M, pH 4.5) was added to cover the gel in the glass vial and incubated at 37°C for 1 hour. Then the supernatant was discarded and the remaining gel was rinsed 3 times with water. Exposure to filter paper for 1 hour was used to remove water and uncrosslinked chitosan, and the mass of the remaining gel was recorded as “after cleavage”. The volume of gel remaining in the glass vial was visualized after inversion.
Hypotaurine inhibition of cleavage with MNP
To investigate the effect of hypotaurine on cleavage by MNP-H2O2, we incubated either 2µg plasmid DNA or 20µg of BSA at 37°C for 3 hours in 50µl NaAc buffer (0.1M, pH 4.5) containing varying concentrations of hypotaurine (1%–8%), with 20µg MNP and 1% H2O2. To investigate the relative catalytic effects of leached Fe ions versus the MNP, we incubated 20 µg MNP in 50 µl NaAc (0.1M, pH 4.5) for 1 hour at 37°C, and then collected the MNP by centrifugation. The supernatant was mixed with 2µg plasmid DNA or 20 µg BSA and 1% H2O2 and incubated for 1 hour at 37°C. The MNP were resuspended in 50 µl NaAc (0.1M, pH 4.5) containing 2µg plasmid DNA or 20 µg BSA and 1% H2O2 and incubated for 1 hour at 37°C. Efficacy of cleavage was then evaluated by electrophoresis.
Killing of E. coli and cleavage of released cellular components
Killing of E. coli by the MNP-H2O2 system was tested using TOP10 cells (Invitrogen). TOP10 cells were cultured in liquid LB (with Ampicillin, 100 µg/ml) medium at 37°C overnight; the OD600 measured approximately 2.0. Then 20 µl of TOP10 cells were spun down and the pellet was resuspended in 50 µl NaAC (0.1M, pH 4.5), containing 20 µg of MNP and 1% H2O2. After an incubation at 37°C, the mixture was centrifuged, the supernatant removed, and the pellet was resuspended in 1 ml LB (Ampicillin, 100 µg/ml) liquid media. The OD600 was measured after an incubation at 37°C for 6 hours, reflecting the population size.
To investigate the degradation of DNA and protein released from dead bacteria, E. coli cells were boiled at 100°C. The complex mixture of organic cellular components released by and representing the killed cells was incubated in 50µl NaAc buffer (0.1M, pH 4.5) containing 20µg MNP and 1% H2O2. Nucleic acids were identified with agarose gel electrophoresis and ethidium bromide staining. Protein products were identified via SDS-PAGE with Coomassie staining.
Biofilm elimination
Pseudomonas aeruginosa (PA01) was cultured in biofilm minimal media (M63, Amresco) supplemented with 0.2% Glucose, 0.5% Casamino acids (BD), and 1mM MgSO4, in a 96 well plate overnight at 37°C. The wells were rinsed with ddH2O and dried. Wells were then challenged with minimal media only (positive control), minimal media + MNP (20 µg/50 µL), minimal media supplemented with 1% H2O2, or minimal media supplemented with 1% H2O2 + MNP. Challenged plates were then incubated for 2 hours at 37°C with periodic mixing. For the biofilm quantitation assay, the challenged wells were rinsed to remove any remaining planktonic cells and stained with 0.1 % Crystal Violet for 15 minutes. Plates were rinsed to remove the stain and dried. Wells were imaged to visualize biofilm rings that were formed at the air/liquid interface. Subsequently, 10% acetic acid was used to solubilize the dye and then the acetic acid/CV mixture was quantitated using a UV spectrophotometer at 600 nm. The amount of CV present is directly proportional to the number of cells in the biofilm ring35. For the viability assay in the biofilm, challenged wells were rinsed to eliminate any remaining planktonic cells. A sterile cotton swab was used to remove cells resident to the biofilm from the well and was subsequently used to inoculate 500 µL of M63 salts. Dilutions were made and 100 µL aliquots of each dilution were plated onto LB agar plates, and incubated overnight at 37°C. Colonies were counted and the number of CFU was compared for each sample.
Statistics
All experiments were performed in triplicate and data were analyzed using the paired Student’s t-test. When numerical data are presented as bar graphs, lines denote standard deviations. Additional details on methods can be found online in the supplementary material.
Supplementary Material
Acknowledgement
This work was primarily supported by an NIH Pioneer Award (8DP1-EB016541; A.J.T.). This work made use of the Cornell Center for Materials Research Shared Facilities which are supported through the NSF MRSEC program (DMR-1120296) and the Nanobiotechnology Center shared research facilities at Cornell.
Footnotes
Author contributions
L.G., K.M.G., H.S., and A.J.T. conceived of experiments. L.G. and K.M.G. performed the experiments. J.L.N. extracted and purified genomic DNA and RNA. L.G., J.L.N., K.M.G., H.S. and A.J.T. wrote and revised the manuscript.
The authors declare no competing financial interests.
Electronic Supplementary Information (ESI) available: Magnetic nanoparticle with peroxidase activity, Cleavage details on DNA and BSA, killing of E.coli, Cell viability of Pseudomonas aeruginosa in biofilm. See DOI: 10.1039/c000000x/
Notes and references
- 1.Cochran WL, McFeters GA, Stewart PS. J Appl Microbiol. 2000;88:22–30. doi: 10.1046/j.1365-2672.2000.00825.x. [DOI] [PubMed] [Google Scholar]
- 2.Mah TF, O'Toole GA. Trends in microbiology. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 3.Francolini I, Donelli G. FEMS immunology and medical microbiology. 2010;59:227–238. doi: 10.1111/j.1574-695X.2010.00665.x. [DOI] [PubMed] [Google Scholar]
- 4.Donlan RM. Emerging infectious diseases. 2001;7:277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Costerton JW, Stoodley P. Nature reviews. Microbiology. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. Appl Environ Microbiol. 1999;65:4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flemming HC, Wingender J. Nature reviews. Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 9.Davies D. Nature reviews. Drug discovery. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 10.Gao LZ, Zhuang J, Nie L, Zhang JB, Zhang Y, Gu N, Wang TH, Feng J, Yang DL, Perrett S, Yan X. Nat Nanotechnol. 2007;2:577–583. doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- 11.Natalio F, Andre R, Hartog AF, Stoll B, Jochum KP, Wever R, Tremel W. Nat Nanotechnol. 2012;7:530–535. doi: 10.1038/nnano.2012.91. [DOI] [PubMed] [Google Scholar]
- 12.Zhang JB, Zhuang J, Gao LZ, Zhang Y, Gu N, Feng J, Yang DL, Zhu JD, Yan XY. Chemosphere. 2008;73:1524–1528. doi: 10.1016/j.chemosphere.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 13.Gao LZ, Wu JM, Lyle S, Zehr K, Cao LL, Gao D. J Phys Chem C. 2008;112:17357–17361. [Google Scholar]
- 14.Fan KL, Cao CQ, Pan YX, Lu D, Yang DL, Feng J, Song LN, Liang MM, Yan XY. Nat Nanotechnol. 2012;7:459–464. doi: 10.1038/nnano.2012.90. [DOI] [PubMed] [Google Scholar]
- 15.Wei H, Wang EK. Chemical Society reviews. 2013;42:6060–6093. doi: 10.1039/c3cs35486e. [DOI] [PubMed] [Google Scholar]
- 16.Liu SH, Lu F, Xing RM, Zhu JJ. Chem-Eur J. 2011;17:620–625. doi: 10.1002/chem.201001789. [DOI] [PubMed] [Google Scholar]
- 17.Zhang XQ, Gong SW, Zhang Y, Yang T, Wang CY, Gu N. J Mater Chem. 2010;20:5110–5116. [Google Scholar]
- 18.Yu FQ, Huang YZ, Cole AJ, Yang VC. Biomaterials. 2009;30:4716–4722. doi: 10.1016/j.biomaterials.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Q, Li WX, Yao YX, Liu HY, Su HY, Ma D, Gu XK, Chen LM, Wang Z, Zhang H, Wang B, Bao XH. Science. 2010;328:1141–1144. doi: 10.1126/science.1188267. [DOI] [PubMed] [Google Scholar]
- 20.Park KS, Kim MI, Cho DY, Park HG. Small. 2011;7:1521–1525. doi: 10.1002/smll.201001886. [DOI] [PubMed] [Google Scholar]
- 21.Kim MI, Ye Y, Woo MA, Lee J, Park HG. Advanced healthcare materials. 2013 doi: 10.1002/adhm.201300100. in press. [DOI] [PubMed] [Google Scholar]
- 22.Xu LJ, Wang JL. Environ Sci Technol. 2012;46:10145–10153. doi: 10.1021/es300303f. [DOI] [PubMed] [Google Scholar]
- 23.Niu HY, Dizhang, Meng ZF, Cai YQ. J Hazard Mater. 2012;227:195–203. doi: 10.1016/j.jhazmat.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Huang YM. J Hazard Mater. 2011;191:163–169. doi: 10.1016/j.jhazmat.2011.04.057. [DOI] [PubMed] [Google Scholar]
- 25.Luo W, Zhu LH, Wang N, Tang HQ, Cao MJ, She YB. Environ Sci Technol. 2010;44:1786–1791. doi: 10.1021/es903390g. [DOI] [PubMed] [Google Scholar]
- 26.Zhang SX, Zhao XL, Niu HY, Shi YL, Cai YQ, Jiang GB. J Hazard Mater. 2009;167:560–566. doi: 10.1016/j.jhazmat.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 27.Wei H, Wang E. Anal Chem. 2008;80:2250–2254. doi: 10.1021/ac702203f. [DOI] [PubMed] [Google Scholar]
- 28.Yang LQ, Ren XL, Tang FQ, Zhang L. Biosens Bioelectron. 2009;25:889–895. doi: 10.1016/j.bios.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Shi WB, Zhang XD, He SH, Huang YM. Chem Commun. 2011;47:10785–10787. doi: 10.1039/c1cc14300j. [DOI] [PubMed] [Google Scholar]
- 30.Kim MI, Shim J, Li T, Lee J, Park HG. Chemistry-a European Journal. 2011;17:10700–10707. doi: 10.1002/chem.201101191. [DOI] [PubMed] [Google Scholar]
- 31.Kim MI, Ye Y, Won BY, Shin S, Lee J, Park HG. Advanced Functional Materials. 2011;21:2868–2875. [Google Scholar]
- 32.Taylor EN, Webster TJ. Int J Nanomed. 2009;4:145–152. [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor E, Webster TJ. Int J Nanomed. 2011;6:1463–1473. doi: 10.2147/IJN.S22021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutherland IW. Trends in microbiology. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole GA, Kolter R. Molecular microbiology. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.