Abstract
A system has been engineered for temporally controlled delivery of siRNA from biodegradable tissue regenerative scaffolds. Therapeutic application of this approach to silence prolyl hydroxylase domain 2 promoted expression of pro-angiogenic genes controlled by HIF1α and enhanced scaffold vascularization in vivo. This technology provides a new standard for efficient and controllable gene silencing to modulate host response within regenerative biomaterials.
Keywords: Gene Knockdown, Controlled Release, RAFT, Endosomolytic Nanoparticle, Injectable Scaffold
Clinical translation of siRNA-based therapies has been hampered by delivery barriers, including siRNA susceptibility to nuclease degradation, cell and endosomal membrane impermeability, and inability to achieve sufficient and sustained bioactivity at the target site.[1, 2] Numerous nanotechnological and medicinal chemistry strategies have been tested to enhance the pharmaceutical properties of siRNA,[3] and most of the recent focus has been on delivery of siRNA for cancer and liver targets, with the latter motivated by the fact that many intravenously-administered nanoparticles nonspecifically biodistribute to the liver. Tremendous progress has been made toward systemic delivery applications, and promising clinical data has begun to appear.[4, 5] However, there is a significant, unmet need for clinically-translatable platform technologies that enable controlled and efficient in vivo delivery of small interfering RNA (siRNA) to therapeutically silence expression of disease-related genes.[6] The use of siRNA-based strategies in regenerative medicine and tissue engineering is a relatively understudied but promising application of RNA interference (RNAi). Topical delivery has been pursued clinically: for example, delivery to the eye for macular degeneration,[7] to the lung for RSV,[8] and to the skin for pachyonychia congenita.[9] A primary limitation to topical delivery for regenerative applications is that siRNA has a relatively short half-life, especially in rapidly dividing cells (i.e., representative of regenerating tissue), where the maximum silencing effect has been noted to be at two days post-transfection,[10] with gene silencing bioactivity being exhausted by one week.[11] In one successful approach, agarose hydrogels containing siRNA packaged with Lipofectamine 2000 was found to produce potent siRNA silencing in vivo. However, this commercial transfection reagent is optimized for in vitro use, and rapid diffusion out of the hydrogel or loss of activity of the lipoplexes necessitates multiple applications.[12–14] Other natural biomaterials such as alginate, collagen, and agarose have also been applied as depots for local delivery of siRNA.[15, 16] Other hydrogel and microparticle depots have been developed to achieve sustained, local delivery of siRNA intratumorally and at sites of inflammation,[17–20] though none of these applications provided controlled siRNA delivery from a biomaterial scaffold that promoted host cell infiltration and tissue regeneration.
More recently, biodegradable, synthetic scaffolds developed toward applications in regenerative medicine have demonstrated controlled and sustained siRNA delivery in vitro (i.e., ranging 20–50 days of release in vitro), including prefabricated ε-caprolactone and ethyl ethylene phosphate copolymer (PCLEEP) nanofibers,[21] poly(lactic-co-glycolic acid) (PLGA) nanofibers,[22] and our porous polyester urethane (PEUR) scaffold design.[23] These classes of porous tissue scaffolds have the advantages of being easily tunable and of being adaptable for filling critically-sized defects with biodegradable templates that promote new tissue in-growth. PEUR scaffolds have been shown to promote regeneration in both excisional cutaneous wounds and bone defects and have desirable properties, including the potential for injectable delivery of components that form a porous scaffold in situ, degradability into biocompatible products at rates dictated by the composition of the polyester triol and the isocyanate, and controlled release of growth factors and other therapeutic agents.[24–29] We recently adopted PEUR scaffolds for delivery of siRNA-loaded polymeric nanoparticles,[23] and the current report showcases the ability of this platform to achieve a high level of gene silencing efficiency and tunability in vivo, along with a proof of concept application of this delivery platform for enhancement of angiogenesis within tissue defects.
Polymeric nanoparticles with pH-dependent endosomal escape behavior have been shown to enhance siRNA intracellular bioavailability.[30–32] To leverage this efficient delivery approach, siRNA loaded nanoparticles were made from the diblock copolymer poly[DMAEMA71-b-(BMA103-co-PAA68-co-DMAEMA57)] (Figure 1A, Mn=43kDA, PDI = 1.41), which was synthesized using reversible addition-fragmentation chain transfer (RAFT). RAFT is a controlled radical polymerization technique amenable to biomedical applications because it enables synthesis of monodisperse and well-defined polymers with block or other architectures and telechelic end chemistries that provide opportunities for site-specific bioconjugation.[33–36] Poly[DMAEMA71-b-(BMA103-co-PAA68-co-DMAEMA57)] was self-assembled into siRNA loaded micellar NPs (si-NPs, Dh = 39.6±12.6 nm, ζ-potential = +20.2 mV) that had been optimized for pH-dependent membrane disruption tuned for endolysosomal escape[30, 37] (Supplemental Figure S1). Trehalose (0, 1.25, 2.5, 5 wt% of PEUR denoted as 0T, 1.25T, 2.5T, and 5T respectively) was added to samples of si-NPs to optimize the stability through lyophilization[38] and to act as a porogen in the cured PEUR scaffolds. Lyophilized si-NPs samples with varied quantities of trehalose were resuspended into polyester triol prepolymers (Figure 1B) and fabricated into scaffolds through a reactive foaming process with lysine triisocyanate (LTI) or hexamethylene diisocyanate trimer (HDIt) (Figure 1C).
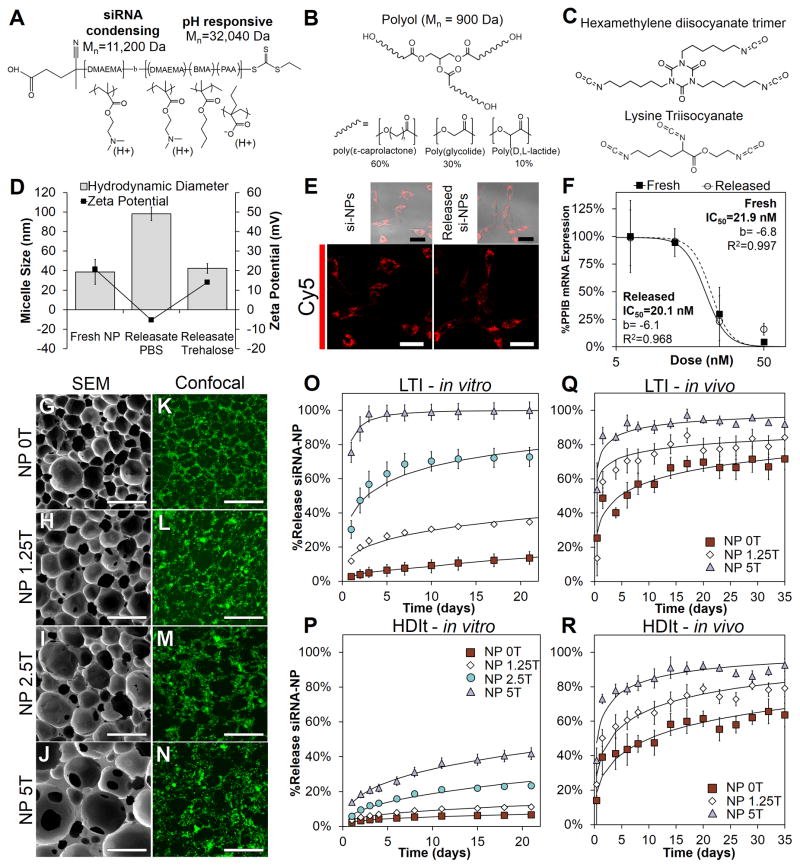
Figure 1.
Material synthesis and characterization of the PEUR scaffold si-NP delivery platform. A) The structure of the diblock copolymer developed previously[30] contains an siRNA condensing block composed of DMAEMA and a pH-responsive block composed of a copolymer of DMAEMA, BMA, and PAA. B) The polyester alcohol (polyol or triol) that was used in the synthesis of polyurethanes were composed of copolymers of poly(ε-caprolactone), poly(glycolide), and poly(D,L-lactide). C) Isocyanate-containing crosslinking components used for PEUR formation included hexamethylene diisocyanate trimer (HDIt) and lysine triisocyanate (LTI). D) The excipient trehalose stabilized the size and ζ-potential of released si-NPs compared to si-NPs prepared in PBS. E) PEUR scaffold-released si-NPs deliver siRNA into the cytoplasm of cells in vitro similar to freshly prepared si-NPs (scale = 30μm). F) Gene silencing activity was similar in PEUR scaffold-released si-NPs compared to freshly-made si-NPs as revealed by RT-PCR IC50 analysis of target gene expression (p=NS). G–J) SEM images of PEUR scaffolds (LTI-based materials shown) containing varying weight% of trehalose (5% by weight is 5T) demonstrate the porous scaffold architecture (Scale = 300μm). K–N) Maximum intensity projections from confocal microscopy showed homogenous loading of si-NPs into the scaffold (note dark areas correspond to pores, scale = 300μm). O–P) Temporal release profile of si-NPs from PEUR scaffolds in vitro demonstrated diffusion controlled release (characterized by Weibull model) that could be modulated through varying the concentration of trehalose or alteration of the isocyanate chemistry. Q–R) The rate of release of si-NPs in vivo was increased relative to the release in vitro but was also tunable based on varying the concentration of the excipient trehalose.
The scaffolds were then incubated in PBS to trigger diffusion of si-NPs from the PEURs in order to assess the physicochemical properties and bioactivity of the released si-NPs. Analysis of the supernatants revealed that si-NPs released from the PEUR scaffolds were similar to freshly-made si-NP samples in terms of size and ζ-potential, suggesting that no aggregation or destabilization occurred during scaffold formation (Figure 1D). Confocal microscopy of mouse embryo fibroblasts (NIH3T3s) treated with scaffold-released si-NPs with cy5.5-labeled siRNA demonstrated that there was a similar level of uptake and intracellular staining pattern relative to NIH3T3s treated with an equivalent concentration of freshly prepared si-NPs (Figure 1E). Likewise, the gene silencing dose response from scaffold-released si-NPs loaded with siRNA against the model gene peptidylpropyl isomerase B (PPIB) was statistically equivalent to that achieved with freshly made si-NPs (Figure 1F). Addition of the excipient trehalose and preparation/lyophilization of si-NPs in dH2O rather than salt-containing PBS prior to incorporation into PEUR scaffolds improved stability of the si-NPs during scaffold fabrication. Stabilization of si-NP physicochemical properties was also functionally significant in terms of bioactivity and resulted in improved bioactivity relative to our previous in vitro studies where the released si-NPs (lyophilized in salt-containing PBS) were larger in size, had lower ζ-potential, and suffered from a 33% reduction in gene silencing relative to freshly made si-NPs (Figure 1D).[23]
PEUR scaffolds were next cured as cylinders containing si-NPs loaded with FAM-labeled 23-mer double stranded DNAs (a model for siRNA) and 0, 1.25, 2.5, and 5 wt% trehalose relative to the mass of the polyurethane precursors. SEM imaging of the scaffolds demonstrated interconnected pores necessary for si-NP release and cell infiltration (Figure 1G–J), and confocal microscopy revealed homogenous loading of the si-NPs throughout the scaffolds (Figure 1K–N). The kinetics of si-NP release from the scaffolds were monitored using fluorescence. The rate of si-NP release was dependent on the quantity of trehalose in the scaffold (Figure 1O). Trehalose can act as a stabilizer and a porogen,[39] and with this system, the release rate of the si-NPs correlated to the quantity of trehalose present. Trehalose is hydrophilic and microdomains of trehalose rapidly dissolve upon exposure to water creating microchannels that accelerate NP diffusion through the scaffold. The effects of the isocyanate chemistry were also assessed, and the si-NP release rate was measured with PEUR scaffolds made from both LTI and HDIt, the latter being more hydrophobic and is known to degrade more slowly[24, 40]. For in vitro tests, the diffusivity of the si-NPs was lower in the PEUR scaffolds made with HDIt than LTI scaffolds. This property increased the versatility and provided an additional level of control for this system for in vitro siRNA delivery (Figure 1P). Modeling with the Weibull function[41] showed that the release mechanism could be characterized as diffusion-controlled in all cases (Supplemental Table S2 and S3).
To measure release kinetics in vivo, PEUR scaffolds were synthesized containing si-NPs made with cy5-labeled siRNA (description in Supplemental Information). The scaffolds were implanted subcutaneously in balb/c mice, and the temporal release profile was characterized in vivo through fluorescence imaging with an IVIS200®. Release of si-NPs from the PEUR scaffolds was faster in vivo relative to in vitro (Figure 1Q–R, images in Supplemental Figure S2), which may be attributable to both increased mechanical forces and cell-mediated effects on the scaffold (i.e. oxidative degradation).[24] Similar to the in vitro studies, the in vivo release kinetics were tunable based on the quantity of trehalose added, and the release mechanism was found to be diffusion-controlled based on the Weibull model (Figure 1O–R, black line). Table 1 reports the time that it took for each formulation tested to release 50, 60, or 75% of the total payload in vivo. ANOVA analysis showed that isocyanate chemistry and trehalose concentration were significant predictors of release kinetics when this variable was tested across all of the scaffold formulations (p<0.05). This analysis suggests that, through the right combination of isocyanate and trehalose concentration, one can tune the system to achieve optimal release kinetics for a wide variety of clinical applications. Relatively long-term release has been achieved in vivo using biodegradable hydrogel depots[42] but no previous platform has demonstrated locally-sustained siRNA release for several weeks and an ability to finely tune the release kinetics in vivo from a tissue scaffold that promotes cell infiltration and regeneration. Reports on regenerative scaffolds including PCLEEP nanofibers, PLGA nanofibers, and porous PEUR have been applied to achieve sustained release in vitro,[21–23] but tunability and in vivo validation were not achieved. The current platform provides the unique capability to match siRNA delivery to the time course of expression of a target gene and to tune the system so that siRNA release, scaffold degradation, and cell/tissue infiltration can be temporally aligned in order to optimize tissue regeneration.
Table 1.
Days of release required for 50%, 60%, and 75% release to be reached
| Group | Formulation | 50% (days ± SE) | 60% (days ± SE) | 75% (days ± SE) |
|---|---|---|---|---|
| 1 | LTI – 0T | 4.67 (±1.46)4,6 | 10.57 (±2.94) 4,6 | 45.27[a] |
| 2 | LTI – 1.25T | 2.89 (±0.37)4,6 | 5.41 (±4.97)4 | 7.73(±6.86)3 |
| 3 | LTI – 5T | 0.07[a] | 0.21[a] | 1.16(±0.31)2,5,6 |
| 4 | HDIT – 0T | 10.09(±5.17)1,2,5,6 | 20.86(±2.81) 1,2,5,6 | 61.27[a] |
| 5 | HDIT – 1.25T | 2.90(±1.32)4 | 6.16(±2.31) 4 | 18.83(±7.49)3 |
| 6 | HDIT - 5T | 0.52(±0.17)1,2,4 | 1.21(±0.13) 1,4 | 4.30(±1.83)5 |
Extrapolated from Weibull model
Superscripts denote significance (p<.05) to the designated group
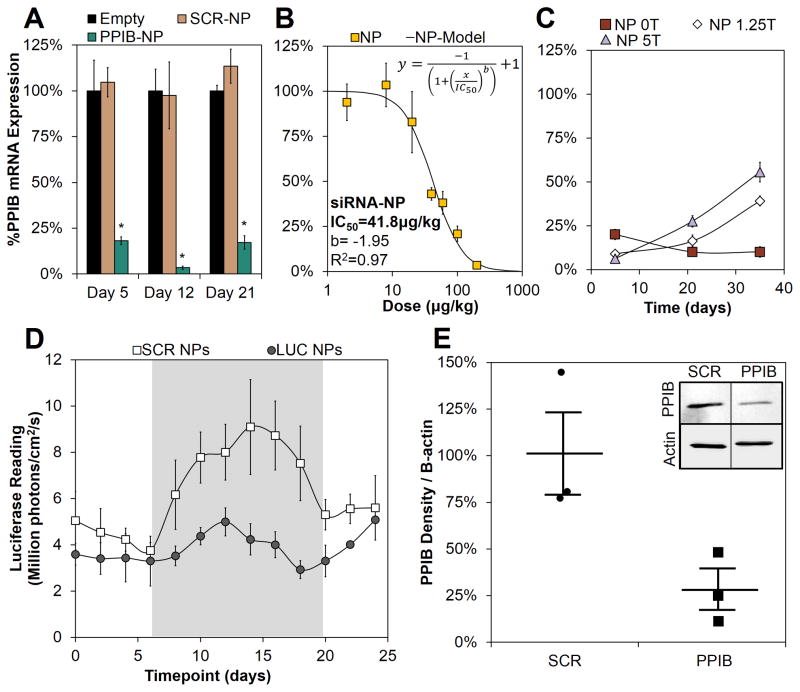
Dicer substrate siRNA (DsiRNA, see description with Supplemental Table S1) designed against the model gene PPIB was used to form si-NPs that were incorporated into PEUR scaffolds made with the “slow” release LTI formulation (0T in Fig 1O). The PPIB si-NPs efficiently reduced target gene expression within the subcutaneously implanted scaffolds, with 82%, 95%, 83% gene silencing achieved at days 5, 12, and 21, respectively (Fig 2A). Importantly, PEUR scaffolds loaded with si-NPs containing a scrambled sequence of DsiRNA showed no significant gene silencing at any time point relative to control scaffolds containing no si-NPs. This remarkable gene silencing was achieved with a relatively low dose of 200 μg DsiRNA/kg of mouse (300 pmol total dose). Next, the dose response behavior using this 0T LTI PEUR scaffold was thoroughly characterized at day 12. This study revealed a low IC50 of 41.8 μg/kg (mass siRNA/mass mouse; total dose of 62.7 pmol, Fig 2B) calculated from a 4-parameter model (Supplemental Equation S2). The potency and sustained action of siRNA with this system compares favorably with other recent regenerative applications of siRNA in vivo, for example, agarose depots loaded with 20 pmol siRNA achieved 76% knockdown of the target gene p53 at day 10, but this required 2 repeated applications and use of the commercial transfection reagent Lipofectamine 2000 to improve cell uptake.[12] Tissue regenerative siRNA delivery applications in vivo using synthetic biomaterials are limited, but a recent report using poly-D,L-lactic acid-p-dioxanone-polyethylene glycol block co-polymer (PLA-DX-PEG) pellets implanted into mouse dorsal muscle pouches served as a depot for the delivery of relatively high doses of 10 – 30 nmol siRNA per site and achieved ~75% gene silencing at day 1 that was sustained for 7 days, with ~50% silencing at day 7.[43] The current scaffold-based approach provided longer-term gene silencing with a 100-fold lower siRNA dose, while also providing a porous template that promotes tissue regeneration.
Figure 2.
The si-NP-loaded PEUR scaffolds provide a potent and temporally-tunable gene silencing platform. A) PPIB mRNA was significantly silenced by siRNA-NP-PEUR at day 5, 12, and 21 (p<0.002 for all groups, n=4) in subcutaneous implants in mice at a siRNA dose of 200μg/kg. B) A dose response at day 12 demonstrated a low IC50 for siRNA-NP-PEUR of 41.8 μg/kg C) The temporal gene silencing profile was tuned through the use of trehalose to control release kinetics (day 5 p<.0005 0T vs 5T, day 35 p<.0005 0T vs 5T). D) A longitudinal study demonstrated significant luciferase reduction over the time course of wound healing, highlighted in gray, in a COL1A2 luciferase reporter mouse model (p<.01, n=5). E) Western blotting for PPIB at day 12 showed significant protein reduction in PPIB siRNA loaded scaffolds (n=3, p<.05).
Next, the correlation between si-NP release kinetics and the resulting time course of gene silencing was tested. For the LTI-based PEUR scaffolds, the fast releasing 5T formulation resulted in 94% PPIB silencing at day 5, compared to 80% for the slower release 0T scaffolds (p<0.005). However, the faster releasing 5T scaffolds had a more transient gene silencing effect and produced 45% PPIB silencing at day 35 compared to the slower releasing 0T scaffolds which produced 90% silencing at 35 days (p<0.0005). A similar analysis was performed for HDIT as reported in Supplemental Figure S3, and the results showed a similar correlation between si-NP release kinetics and the temporal gene silencing profile. To further validate our measurements, knockdown of PPIB protein from scaffold explants at day 12 was evaluated (200 μg DsiRNA/kg mouse, 0T/LTI). As shown in the western blot in the inset (Figure 2E) approximately 75% less PPIB protein was detected (p<0.05). The combination of low dose, sustained silencing effect, and tunability achieved here is unprecedented for in vivo delivery of siRNA from a regenerative tissue scaffold. The potency of our system may have been enhanced by direct, substrate-mediated transfection of si-NPs into cells migrating into the cell-inductive PEUR scaffold. For example, high local concentration of plasmid DNA, achieved through immobilization onto the surface of materials, has been shown to increase transfection efficiency 10–100 fold relative to plasmid polyplexes freely diffusing within the cell’s surroundings.[44–46] This uptake mechanism mimics the pathway hijacked by viruses that attach to extracellular matrix proteins to enhance their rate of cellular internalization.[47, 48]
Transgenic mice with a collagen α-2(I) chain (COL1A2) luciferase reporter were next utilized to assess the ability of the si-NP-PEUR platform to effectively silence a wound-related gene throughout the entire time course of healing. The COL1A2 reporter is upregulated between approximately days 7–14 in mouse incisional and laser irradiated wounds.[49, 50] A COL1A2 luciferase reporter mouse model was used to allow a longitudinal, quantitative, and protein-level readout of luciferase silencing using intravital bioluminescence imaging. The mice received subcutaneously-implanted PEUR scaffolds (OT, LTI) containing si-NPs loaded with DsiRNA against luciferase or a scrambled control sequence. In the control animals, the activity of the COL1A2 reporter was elevated between approximately days 8–20 post-wounding (highlighted in gray) (Figure 2D). However, incorporation of luciferase DsiRNA maintained the local luciferase activity at approximately baseline levels, and there was a significant reduction in luciferase activity in these scaffolds relative to scaffolds containing si-NPs loaded with scrambled siRNA (p<0.01, n=4). The efficient and sustained gene silencing achieved throughout the full time course of wound healing suggests that this platform can be utilized to abrogate the function of a therapeutic target gene throughout the healing process.
To exclude a nonspecific biological response to the si-NPs, histology of tissue explants and PCR against signal transducer and activator of transcription factor 1 (STAT-1, a readout for TLR activation[51]) and tumor necrosis factor α (TNFα) was performed. Histology revealed that there was no inflammation or toxicity associated with incorporation of the si-NPs into the PEUR scaffolds (Supplemental Figure S4), and PCR for STAT-1 and TNFα showed that the si-NPs did not increase these inflammatory markers relative to empty scaffolds (Supplemental Figure S5).
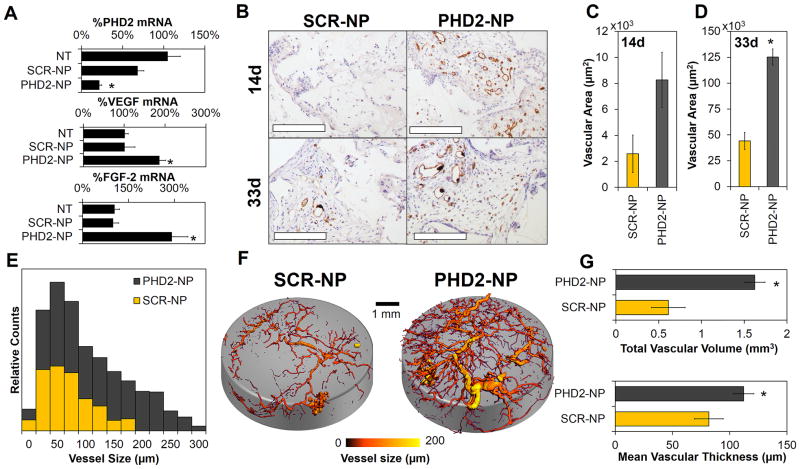
Prolyl hydroxylase 2 (PHD2) activity triggers degradation of the pro-angiogenic transcription factor hypoxia inducible factor 1α (HIF1α) during normoxia. When PHD2 is naturally inactivated (i.e., under hypoxic conditions) or silenced through RNAi, HIF1α mediates transcription of pro-angiogenic genes such as vascular endothelial growth factor (VEGF), fibroblast growth factor 2 (FGF-2), and others.[52] To demonstrate the therapeutic potential of our platform for promoting tissue regeneration, si-NPs were formulated with PHD2 DsiRNA (PHD2-NPs or scrambled siRNA (SCR-NP) and incorporated into PEURs scaffolds that were implanted subcutaneously. At 14d, PCR revealed an ~80% reduction in PHD2 levels (Figure 3A) which resulted in a ~200% increase in VEGF and ~300% increase in FGF-2 mRNA levels. For an evaluation of neovessel formation with the scaffolds, immunohistochemistry (IHC) for CD31 was done at 14d and 33d, and development of stable, functional vascular structures was imaged and quantified using micro-CT following systemic vascular perfusion with a contrast agent at 33d. CD31 IHC showed visually increased vessel density in PHD2-NP-containing scaffolds (Figure 3B) and a significant, 280% increase in vessel area at day 33 (Figure 3C–D). Scaffolds characterization with micro-CT provided quantitative histograms that demonstrated that PHD2-NPs increased both number and size of vessels within the scaffolds (Figure 3E, representative images in Figure 3F). Quantitative 3D image analysis[53] showed that PHD2-NPs increased the vascular volume by 300% and increased the mean vascular thickness by 137% (Figure 3G). These data convincingly demonstrate the regenerative potential of this platform, as formation of robust, mature vessels is one of the primary challenges in tissue regeneration. We anticipate that sustained RNAi-induced modulation of transcription factors, such as HIF1α, that control groups of related genes has the potential to produce better-orchestrated and more robust effects on tissue regeneration compared to delivery of a single growth factor (e.g., VEGF of FGF), which is the current standard. For example, VEGF has had limited therapeutic success because it produces immature vessels that suffer from instability and poor long-term function.[54]
Figure 3.
Sustained silencing of PHD2 increases angiogenesis within PEUR tissue scaffolds. A) 80% silencing of PHD2 increased VEGF and FGF-2 expression by 200% and 290% respectively (*p<0.01). B) CD31 staining was significantly increased within PHD2 scaffolds at day 14 and day 33 (Scale = 200 μm, vessels appear read, nuclei are counterstained purple with hematoxylin, and white space represents residual PEUR scaffold). C–D) CD31 sections were quantified showing a significant increase in vessel area at day 33 (*p<0.01). E) Micro-CT of explanted PHD2-NP scaffolds showed a significant increase in both vessel number and vessel size for PHD2-NP scaffolds as shown in the histogram. F) Micro-CT images visually demonstrate the increased vasculature within the scaffolds. G) Quantitative analysis of 3D micro-CT vessel images revealed a significant increase in vascular volume and mean vascular thickness within PHD2-NP-loaded scaffolds.
This study validates that si-NP delivery from tissue inductive PEUR scaffolds provides a new, tunable platform technology for efficient, local gene silencing. The in vitro and in vivo data suggest that this platform is highly versatile for siRNA delivery in vitro or in vivo through tuning the quantity of trehalose added during PEUR scaffold fabrication and by alteration of the chemistry of the isocyanate. This provides the opportunity to tune this delivery system based on the desired expression profile of the therapeutically targeted gene or to optimally match rates of scaffold degradation, tissue growth, and siRNA delivery. PHD2 silencing studies demonstrated that this platform can promote angiogenesis in vivo. These proof-of-concept data validate that this platform provides a powerful research tool and also represents a technology with the potential to be utilized therapeutically for manipulation of genes whose silencing promotes tissue regeneration.
Experimental Section
si-NP synthesis and characterization
Dicer substrate siRNAs (DsiRNAs) were obtained from IDT and screened in vitro for activity before use in vivo. A diblock copolymer composed of 2-(dimethylamino)ethyl methacylate (DMAEMA), 2-propylacrylic acid (PAA), and butyl methacrylate (BMA) was synthesized using reversible addition-fragmentation chain transfer (RAFT) polymerization as described previously.[23, 30] NPs were fabricated by dissolving in ethanol, followed by slow addition of dH2O, which spontaneously triggered formation of micelles. Subsequently, siRNA was electrostatically loaded onto the surface of NPs. Dynamic light scattering (DLS, Zetasizer nano-ZS Malvern Instruments Ltd, Worcestershire, U.K.) was used to analyze size and zeta potential of the si-NPs.
si-NP-PEUR synthesis and characterization
The polymeric NPs (1mg) were mixed with siRNA (5 nmol, 0.08mg) in an RNAse free polypropylene tube and allowed to electrostatically condense for 30min. Trehalose was added to the si-NPs at varying concentrations from 0 to 5 wt% of PEUR and allowed to stabilize for 30 min. The solutions were frozen and then lyophilized. Lyophilized si-NP samples were suspended into a 900 Da polyester triol with a backbone comprised of 60 wt% ε-caprolactone, 30 wt% glycolide, and 10 wt% D,L-lactide. PEUR scaffolds were synthesized by reacting 67 μmol of the polyol component of PEUR with a slight excess of lysine triisocyanate (LTI, 193 μmol, 35mg) in the presence of 67 μmol water. The water reacts with the isocyanate to produce CO2 and serves as a blowing agent that creates the pores within the scaffold. The polyol and LTI were mixed using a Hauschild DAC 150 FVZ-K SpeedMixer (FlackTek, Inc., Landrum, SC). Alternatively, 42 μmol of polyol was reacted with a slight excess of hexamethylene diisocyanate trimer (HDIt, 111 μmol) in the presence of 63 μmol of water.
The resulting 100mg PEUR foams were sectioned into 6mm diameter × 1 mm thick discs and imaged with a fluorescent confocal microscope (Zeiss LSM 710 Meta Oberkochen, Germany) to analyze the distribution of fluorescently labeled si-NPs in the scaffold. Scaffold morphology was assessed with a scanning electron microscope (SEM Hitachi S4200, Tokyo, Japan) for structure and porosity. PEUR scaffolds were immersed in PBS, and releasate was collected and quantified using fluorescence for percent release. Released si-NPs were incubated on NIH3T3 mouse embryo fibroblasts at varying concentrations that were imaged with a fluorescent confocal microscope (Zeiss LSM 710 Meta) and measured for gene silencing by RT-PCR.
Subcutaneous Implant of si-NP-PEUR
The animal studies were conducted with adherence to the guidelines for the care and use of laboratory animals of the National Institutes of Health (NIH). All experiments with animals were approved by Vanderbilt University’s Institutional Animal Care and Use Committee (IACUC). Cy5-labled DsiRNA was complexed into NPs and loaded into PEUR scaffolds in the same quantities outlined above. Scaffolds were sectioned into approximately 6 mm × 1mm discs and sterilized by ethylene oxide treatment. 8–10 week balb/c mice were purchased from Charles River Laboratories. The animals were fed a standard chow diet ad libitium and had free access to water. The mice were anesthetized with 1.5–2% isoflurane and maintained at 37°C. The mice abdomen was shaved and sterilized. A 1 cm incision was made in the ventral side of the skin in the abdomen of the mice. A pocket was made with sterilized haemostatic forceps on each side of the midline and 6 mm scaffolds were implanted subcutaneously. The incision was sutured, and the mice were allowed to recover at 37°C. Analgesic agent (ketoprofen, 5 mg/kg) was injected as needed.
In vivo release kinetics
Release of si-NPs from the scaffold was quantified by measuring the loss of Cy5 fluorescence over time in regions of interest (ROIs) defined by the PEUR implant using an IVIS 200® imaging system (Caliper Life Sciences, Hopkinton, Massachusetts). Mice were anesthetized with 1.5–2% isoflurane and maintained at 37°C and measured with constant image settings every 2–3 days.
In vivo gene silencing
siRNA against cyclophilin B (PPIB) was formulated into si-NPs, incorporated into PEUR scaffolds, and implanted subcutaneously for 5, 12, 21, or 35 days using the procedure described above. At defined endpoints, the mice were anesthetized heavily with isoflurane and sacrificed by cervical dislocation. The scaffolds were collected postmortem and bisected in half for preparation for both histology and PCR. RNA was extracted with TRIZOL (Invitrogen, Carlsbad, CA) and purified with RNEasy spin column (Qiagen, Venlo, Netherlands). The expression of PPIB was evaluated by RT-PCR using the ΔΔCt method normalizing to GAPDH. Histological sections were used to evaluate the host response to the implants through H&E staining. RT-PCR for inflammatory markers was also performed to evaluate immune response and activation of toll like receptor signaling. A western blot was used to confirm protein level silencing using primary antibodies anti-PPIB (Sigma) and anti-B-actin (Santa Cruz Biotechnology). The full method is described in the supplemental information.
In vivo gene silencing during the time course of wound healing
For a longitudinal, protein level readout, firefly luciferase siRNA was formulated into si-NPs, incorporated into PEUR scaffolds, and implanted into transgenic balb/c mice with a COL1A2 luciferase reporter using the same procedure described above. The bioluminescence at the scaffold site was evaluated every 2 days for 24 days using an IVIS® 100 bioluminescence imaging system.
In vivo silencing of PHD2
PHD2 or scrambled siRNA was formulated into si-NPs that were incorporated into PEUR scaffolds and implanted subcutaneously into balb/c mice. Mice were sacrificed at day 14 and scaffolds were evaluated for gene expression by real-time RT-PCR. At day 33, scaffold vascularization was assessed with microCT using established methods[53, 55–57] (full method can be found in the supplemental information). H&E staining and CD31 immunohistochemistry were done on scaffolds explanted at days 14 and 33.
Supplementary Material
Acknowledgments
The authors thank Garrett Rettig (IDT) for advice on designing dsiRNAs. Fluorescence, bioluminescence, and microCT imaging (supported by NIH S10 RR027631) were performed through the Vanderbilt University Institute for Imaging Science (VUIIS). Confocal Imaging was performed in part through the use of the VUMC Cell Imaging Shared Resource, (supported by NIH grants CA68485, DK20593, DK58404, HD15052, DK59637 and Ey008126). DLS and TEM were conducted at the Vanderbilt Institute of Nanoscale Sciences and Engineering (VINSE). qRT-PCR was conducted at the Vanderbilt University Molecular Cell Biology Resource Core. Histology sections were prepared through the use of Translation Pathology Shared Resource (TPSR). This work was supported by a training grant from the Vanderbilt Diabetes Research Training and Training Center (NIH DK20593), a Vanderbilt Discovery Grant, NIH R21EB012750, and NIH 1R01AR056138-01A2.
Footnotes
Supporting Information is available online from Wiley Online Library.
References
- 1.White PJ. Clin Exp Pharmacol P. 2008;35:1371–1376. doi: 10.1111/j.1440-1681.2008.04992.x. [DOI] [PubMed] [Google Scholar]
- 2.Dominska M, Dykxhoorn DM. J Cell Sci. 123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 3.Rettig GR, Behlke MA. Mol Ther. 2012;20:483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabernero J, Shapiro GI, Lorusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, Gounder MM, Falzone R, Harrop J, Seila White AC, Toudjarska I, Bumcrot D, Meyers RE, Hinkle G, Svrzikapa N, Hutabarat RM, Clausen VA, Cehelsky J, Nochur SV, Gamba-Vitalo C, Vaishnaw AK, Sah DW, Gollob JA, Burris HA., 3rd Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser PK, Symons RC, Shah SM, Quinlan EJ, Tabandeh H, Do DV, Reisen G, Lockridge JA, Short B, Guerciolini R, Nguyen QD. Am J Ophthalmol. 2010;150:33–39. doi: 10.1016/j.ajo.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Zamora MR, Budev M, Rolfe M, Gottlieb J, Humar A, Devincenzo J, Vaishnaw A, Cehelsky J, Albert G, Nochur S, Gollob JA, Glanville AR. Am J Resp Crit Care. 2011;183:531–538. doi: 10.1164/rccm.201003-0422OC. [DOI] [PubMed] [Google Scholar]
- 9.Leachman SA, Hickerson RP, Schwartz ME, Bullough EE, Hutcherson SL, Boucher KM, Hansen CD, Eliason MJ, Srivatsa GS, Kornbrust DJ, Smith FJ, McLean WI, Milstone LM, Kaspar RL. Mol Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dykxhoorn DM, Palliser D, Lieberman J. Gene Ther. 2006;13:541–542. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen PD, Tutela JP, Thanik VD, Knobel D, Allen RJ, Chang CC, Levine JP, Warren SM, Saadeh PB. Wound Repair Regen. 2010;18:553–559. doi: 10.1111/j.1524-475X.2010.00638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JW, Tutela JP, Zoumalan RA, Thanik VD, Nguyen PD, Varjabedian L, Warren SM, Saadeh PB. Arch Otolaryngol. 2010;136:714–719. doi: 10.1001/archoto.2010.107. [DOI] [PubMed] [Google Scholar]
- 14.Wetterau M, George F, Weinstein A, Nguyen PD, Tutela JP, Knobel D, Cohen O, Warren SM, Saadeh PB. Wound Repair Regen. 2011;19:481–486. doi: 10.1111/j.1524-475X.2011.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs MD, Jeon O, Alsberg E. J Am Chem Soc. 2009;131:9204–9206. doi: 10.1021/ja9037615. [DOI] [PubMed] [Google Scholar]
- 16.Vinas-Castells R, Holladay C, di Luca A, Diaz VM, Pandit A. Bioconjugate Chem. 2009;20:2262–2269. doi: 10.1021/bc900241w. [DOI] [PubMed] [Google Scholar]
- 17.Mountziaris PM, Tzouanas SN, Sing DC, Kramer PR, Kasper FK, Mikos AG. Acta Biomater. 2012;8:3552–3560. doi: 10.1016/j.actbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YM, Park MR, Song SC. Biomaterials. 2013;34:4493–4500. doi: 10.1016/j.biomaterials.2013.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Han HD, Mora EM, Roh JW, Nishimura M, Lee SJ, Stone RL, Bar-Eli M, Lopez-Berestein G, Sood AK. Cancer Biol Ther. 2011;11:839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen K, Dang PN, Alsberg E. Acta Biomater. 2013;9:4487–4495. doi: 10.1016/j.actbio.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rujitanaroj PO, Wang YC, Wang J, Chew SY. Biomaterials. 2011;32:5915–5923. doi: 10.1016/j.biomaterials.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 22.Chen ML, Gao S, Dong MD, Song J, Yang CX, Howard KA, Kjems J, Besenbacher F. ACS Nano. 2012;6:4835–4844. doi: 10.1021/nn300106t. [DOI] [PubMed] [Google Scholar]
- 23.Nelson CE, Gupta MK, Adolph EJ, Shannon JM, Guelcher SA, Duvall CL. Biomaterials. 2012;33:1154–1161. doi: 10.1016/j.biomaterials.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hafeman AE, Zienkiewicz KJ, Zachman AL, Sung HJ, Nanney LB, Davidson JM, Guelcher SA. Biomaterials. 2011;32:419–429. doi: 10.1016/j.biomaterials.2010.08.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Davidson JM, Guelcher SA. Biomaterials. 2009;30:3486–3494. doi: 10.1016/j.biomaterials.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Yoshii T, Hafeman AE, Nyman JS, Wenke JC, Guelcher SA. Biomaterials. 2009;30:6768–6779. doi: 10.1016/j.biomaterials.2009.08.038. [DOI] [PubMed] [Google Scholar]
- 27.Guan J, Stankus JJ, Wagner WR. J Control Release. 2007;120:70–78. doi: 10.1016/j.jconrel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adolph EJ, Hafeman A, Davidson J, Nanney L, Guelcher S. J Biomed Mater Res A. 2012;100A:450–461. doi: 10.1002/jbm.a.33266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson DM, Baraniak PR, Ma Z, Guan J, Mason NS, Wagner WR. Pharm Res. 2011;28:1282–1293. doi: 10.1007/s11095-011-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Convertine A, Benoit D, Duvall C, Hoffman A, Stayton P. J Control Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HM, Yu SS, Miteva M, Nelson CE, Werfel T, Giorgio TD, Duvall CL. Adv Funct Mater. 2013;23:3040–3052. doi: 10.1002/adfm.201202215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta KM, Duvall CL. ACS Nano. 2013 doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moad G, Chiefari J, Chong YK, Ercole F, Krstina J, Jeffery J, Le TPT, Mayadunne RTA, Meijs GF, Moad CL, Rizzardo E, Thang SH. Macromolecules. 1998;31:5559–5562. [Google Scholar]
- 34.Boyer C, Bulmus V, Davis TP, Ladmiral V, Liu J, Perrier S. Chem Rev. 2009;109:5402–5436. doi: 10.1021/cr9001403. [DOI] [PubMed] [Google Scholar]
- 35.Duvall CL, Convertine AJ, Benoit DS, Hoffman AS, Stayton PS. Mol Pharm. 2010;7:468–476. doi: 10.1021/mp9002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer C, Teo J, Phillips P, Erlich RB, Sagnella S, Sharbeen G, Dwarte T, Duong HT, Goldstein D, Davis TP, Kavallaris M, McCarroll J. Mol Pharm. 2013;10:2435–2444. doi: 10.1021/mp400049e. [DOI] [PubMed] [Google Scholar]
- 37.Convertine AJ, Diab C, Prieve M, Paschal A, Hoffman AS, Johnson PH, Stayton PS. Biomacromolecules. 2010;11:2904–2911. doi: 10.1021/bm100652w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaushik JK, Bhat R. Journal of Biological Chemistry. 2003;278:26458–26465. doi: 10.1074/jbc.M300815200. [DOI] [PubMed] [Google Scholar]
- 39.Nugent M, McLaren A, Vernon B, McLemore R. Clin Orthop Relat R. 2010;468:2101–2106. doi: 10.1007/s11999-010-1264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hafeman AE, Li B, Yoshii T, Zienkiewicz K, Davidson JM, Guelcher SA. Pharm Res. 2008;25:2387–2399. doi: 10.1007/s11095-008-9618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papadopoulou V, Kosmidis K, Vlachou M, Macheras P. Int J Pharm. 2006;309:44–50. doi: 10.1016/j.ijpharm.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 42.Kim YM, Park MR, Song SC. ACS Nano. 2012;6:5757–5766. doi: 10.1021/nn300842a. [DOI] [PubMed] [Google Scholar]
- 43.Manaka T, Suzuki A, Takayama K, Imai Y, Nakamura H, Takaoka K. Biomaterials. 2011;32:9642–9648. doi: 10.1016/j.biomaterials.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 44.Shea LD, Smiley E, Bonadio J, Mooney DJ. Nat Biotechnol. 1999;17:551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 45.Luo D, Saltzman WM. Nat Biotechnol. 2000;18:893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- 46.Segura T, Shea LD. Bioconjugate Chem. 2002;13:621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 47.Hanenberg H, Xiao XL, Dilloo D, Hashino K, Kato I, Williams DA. Nat Med. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- 48.Lei P, Bajaj B, Andreadis ST. J Virol. 2002;76:8722–8728. doi: 10.1128/JVI.76.17.8722-8728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bou-Gharios G, Garrett LA, Rossert J, Niederreither K, Eberspaecher H, Smith C, Black C, Crombrugghe B. J Cell Biol. 1996;134:1333–1344. doi: 10.1083/jcb.134.5.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu N, Jansen ED, Davidson JM. J Invest Dermatol. 2003;121:926–932. doi: 10.1046/j.1523-1747.2003.12497.x. [DOI] [PubMed] [Google Scholar]
- 51.Larange A, Antonios D, Pallardy M, Kerdine-Romer S. J Leukocyte Biol. 2009;85:673–683. doi: 10.1189/jlb.0808504. [DOI] [PubMed] [Google Scholar]
- 52.Shen X, Wan C, Ramaswamy G, Mavalli M, Wang Y, Duvall CL, Deng LF, Guldberg RE, Eberhart A, Clemens TL, Gilbert SR. J Orthop Res. 2009;27:1298–1205. doi: 10.1002/jor.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duvall CL, Taylor WR, Weiss D, Guldberg RE. Am J Physiol-Heart C. 2004;287:H302–H310. doi: 10.1152/ajpheart.00928.2003. [DOI] [PubMed] [Google Scholar]
- 54.van Weel V, van Tongeren RB, van Hinsbergh VW, van Bockel JH, Quax PH. Ann Vasc Surg. 2008;22:582–597. doi: 10.1016/j.avsg.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 55.Guldberg RE, Duvall CL, Peister A, Oest ME, Lin AS, Palmer AW, Levenston ME. Biomaterials. 2008;29:3757–3761. doi: 10.1016/j.biomaterials.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duvall CL, Weiss D, Robinson ST, Alameddine FMF, Guldberg RE, Taylor WR. Arterioscl Throm Vas. 2008;28:290–295. doi: 10.1161/ATVBAHA.107.158485. [DOI] [PubMed] [Google Scholar]
- 57.Phelps EA, Landazuri N, Thule PM, Taylor WR, Garcia AJ. P Natl Acad Sci USA. 2010;107:3323–3328. doi: 10.1073/pnas.0905447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.