Abstract
TT-034 (PF-05095808) is a recombinant adeno-associated virus serotype 8 (AAV8) agent expressing three short hairpin RNA (shRNA) pro-drugs that target the hepatitis C virus (HCV) RNA genome. The cytosolic enzyme Dicer cleaves each shRNA into multiple, potentially active small interfering RNA (siRNA) drugs. Using next-generation sequencing (NGS) to identify and characterize active shRNAs maturation products, we observed that each TT-034–encoded shRNA could be processed into as many as 95 separate siRNA strands. Few of these appeared active as determined by Sanger 5′ RNA Ligase-Mediated Rapid Amplification of cDNA Ends (5-RACE) and through synthetic shRNA and siRNA analogue studies. Moreover, NGS scrutiny applied on 5-RACE products (RACE-seq) suggested that synthetic siRNAs could direct cleavage in not one, but up to five separate positions on targeted RNA, in a sequence-dependent manner. These data support an on-target mechanism of action for TT-034 without cytotoxicity and question the accepted precision of substrate processing by the key RNA interference (RNAi) enzymes Dicer and siRNA-induced silencing complex (siRISC).
Keywords: HCV, mechanism of action, RACE-seq, siRNA, shRNA
Introduction
Chronic hepatitis caused by hepatitis C virus (HCV), affects 170 million people worldwide, progresses to liver cirrhosis in about 1 in 4 cases and is associated with increased risk of hepatocellular carcinoma.1,2 The current standard of combination therapy-based care is not effective against all genotypes, exhibits variable cure rates and is associated with significant side effects. Moreover, even the latest management strategies require chronic dosing and have been already shown to be prone to viral escape.
It has been suggested that direct targeting of the HCV genome could provide a valuable alternative.3 High genomic variability is encountered in HCV due to error-prone replication. Because the HCV genome is propagated exclusively as RNA, several direct targeting approaches have been proposed,4,5 including RNA interference (RNAi).6 A high efficiency alternative to synthetic RNAi delivery systems,7 however, involves the use of recombinant viruses possessing capsid-specific tropism for different organs.8,9,10 In these systems, siRNA are delivered in the form of short hairpin RNA (shRNA) precursors expressed from virus-encoded DNA plasmids.
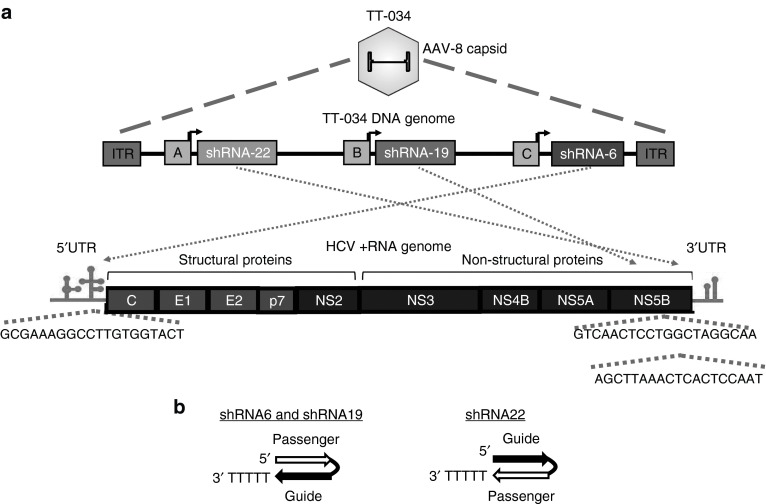
An example of such an approach is TT-034 (also known as PF-05095808), a recombinant adeno-associated virus (AAV) vector with hepatic tropism conferred by its serotype 8 capsid (AAV8).11,12 The virus has been modified to encode three shRNAs against HCV (Figure 1a), two of which were designed in a passenger strand-loop-guide strand orientation (Figure 1b). We previously reported dose-dependent activity of these shRNAs, delivered individually or together as TT-034, against a HCV genotype 1 replicon model in vitro.12 In those experiments, we did not observe activation of innate immune responses nor detect replicon inhibition following transduction with empty capsid vector confirming that the activity of TT-034 was specifically due to delivery of the shRNAs.
Figure 1.
Genome organization and target site location of TT-034, a recombinant AAV8 vector encoding three shRNA targeting the HCV genome. (a) The viral genome of AAV8 has been replaced with 3 expression cassettes separated by Pol III termination sequences and stuffer regions. The contents of the expression cassettes are from 5′ to 3′: modified U6-9 Pol III promoter (box A) driving shRNA22 expression targeted to the HCV NS5B region (viral RNA-dependent RNA polymerase), modified U6-1 Pol III promoter (box B) driving shRNA19 expression, also targeted to the NS5B region and modified U6-8 Pol III promoter (box C) driving shRNA6 targeted to the 5′ UTR of HCV. The expression cassettes are flanked by inverted tandem repeats derived from AAV4 at the 5′ end and AAV2 at the 3′ end of the genome. (b) Schematic representation of the expressed hairpin polarity. The guide strand is defined as the strand targeting the (+) RNA HCV genome.
Short hairpin RNAs are transcribed in the nucleus and are exported into the cytoplasm via Exportin 5 to be matured into RNAi-active siRNA.13 In agreement with reports on microRNAs (miRNAs),14 shRNA maturation involves multiple cleavages of the loop structure in the hairpin by the cytoplasmic RNAse III enzyme Dicer.15,16,17 Dicer is thought to cleave shRNA at 21–25 base pair intervals from the stem-end of the shRNA, resulting in loop removal and generation of double-strand siRNAs.18 One of the strands, known as the guide or active strand, is then loaded into one of four Argonaute (AGO) proteins forming the siRNA-induced silencing complex (siRISC; reviewed in ref. 13). Thermodynamic stability design of synthetic siRNA duplex ends can direct siRISC loading.19,20 Lack of such design flexibility, however, is valuable to TT-034 as HCV replication involves a negative polarity RNA intermediate. Thus, both strands of the three shRNAs encoded in TT-034 could be active against HCV.
After Ago is loaded, siRISC uses the guide strand to identify RNA harboring complementary sequences and, if AGO2 is involved, direct endonucleolytic cleavage of the target phosphodiester backbone specifically opposite nucleotides 10–11 from the 5′ end of the guide strand.21 The result is believed to be a single, novel 5′ end on the RNA target, typically confirmed by the Sanger sequencing-based method 5′ RNA Ligase-Mediated Rapid Amplification of cDNA Ends (5′ RACE). Importantly, data thereto are the only means of obtaining definitive proof of mechanism for candidate siRNA and shRNA therapeutics.
In this article, we explore the utility of next-generation sequencing (NGS) in scrutinizing shRNA maturation and siRNA cleavage, as a means of fully characterizing the potentially active drug products and specific mode of action of TT-034. Our investigations reveal that shRNA can yield an unexpectedly large diversity of putatively RNAi-active strands and an increased number of cleavage sites on targeted RNA. Moreover, we report that siRNA can direct target cleavage in more than one positions on the targeted RNA, in a sequence-dependent manner.
Results
TT-034–encoded shRNA are processed into an unexpectedly diverse panel of putative siRNA strand products
In order to characterize the diversity of siRNAs produced by the three anti-HCV shRNAs expressed from TT-034, we treated Con1b HCV replicon cells with TT-034. This resulted in replicon inhibition as measured by reporter activity (Table 1). RNA was therefore extracted and subjected to pair-end small RNA NGS (sRNA-seq). The sum of sRNA-seq reads aligning to the TT-034–encoded shRNAs represented an average of 0.063% of all sequenced RNA with 1712 (0.016%) molecules originating from shRNA6, 418 (0.004%) from shRNA19 and 3718 (0.042%) from shRNA22 (Supplementary Table S2). The relative frequency of these putative siRNA strands indicated uneven hairpin expression, corroborating previous data assessing production of guide strands against the (+) RNA strand of the HCV genome as determined by qPCR.12
Table 1. RNAi agents used to prepare samples for 5′ RACE and RACE-Seq.

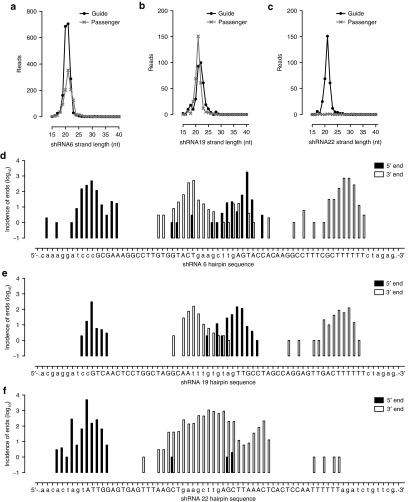
Differences in putative siRNA strand length were also observed (Figure 2a–c): the geometrical means of shRNA-aligning sequence lengths were similar between shRNA6 and shRNA19 (19.9 and 19.5 nt respectively), but significantly larger for shRNA22 (23.5 nt). Similarly, the range of putative siRNA strand lengths was comparable between shRNA6 and shRNA19 (15–32 nt and 15–30 nt respectively), with shRNA22 yielding sequences between 15–37 nt in length (Supplementary Table S2). This range was found to be on account of an unexpectedly large diversity of sequences matured from each hairpin: an average of 95, 67, and 89 different sequences corresponding to shRNA6, shRNA19, and shRNA22 were observed respectively (Supplementary Table S2). While some of these sequences were detected only once, the most predominant product was detected up to 677, 145, and 1,075 times for each of the shRNAs. Thus, while a single sequence represented up to 35% of all putative siRNA strands, the remainder were noteworthy for the their breadth of diversity: 5′ end variability ranged between 10 and 25 distinct positions, whereas sequences starting from the same 5′ end could terminate in up to 22 different 3′ ends, as observed for shRNA22 (Figure 2d–f and Supplementary Table S2). Moreover, while shRNA6 and shRNA19 were both processed to yield sequences from both hairpin strands (Supplementary Table S2), the shRNA22 hairpin was predominantly processed in favor of the guide strand (99.89% of shRNA22-aligning reads). Mechanistically, however, this shRNA product diversity meant that a 2.65%, 24.66%, and 6.19% fraction of the putative siRNA strands of shRNA6, shRNA19, and shRNA22, respectively, contained nucleotides from the TT-034 vector backbone or from the shRNA-loop and therefore bore mismatches in their seed sequence to their target in the HCV genome.
Figure 2.
Length and sequence diversity of siRNA strands processed out of the three HCV-targeting shRNA expressed from TT-034. Following in vitro transduction of Con1b HCV replicon cells with TT-034, small RNA NGS was performed on RNA extracts to determine the diversity of siRNA guide and passenger strands processed out of TT-034–encoded hairpins. (a–c) The distribution of guide (black circle) and passenger (grey X) strand lengths is indicated for each of the three shRNAs encoded in TT-034. (d–f) The cumulative incidence (y axis, logarithmic scale) of distinct 5′ ends (black bars) and 3′ ends (white bars) processed out of the three shRNAs relative to the TT-034–encoded hairpin sequence (x axis) is also shown as an indicator of sequence diversity.
Dicer is the key cytosolic enzyme responsible for both shRNA and miRNA maturation.17 To eliminate the possibility that the plethora of shRNA products encountered in our studies were the consequence of deficient DICER processing in this cell line, we queried our dataset for evidence of aberrant endogenous miRNA maturation. In stark contrast to our observations on shRNA processing and in accordance with previously published NGS data on mature miRNA diversity archived on miRbase22 and elsewhere,23 an average of 2.9 variants were observed among the 147 separate mature miRNA detected in these cells, with mature miRNA diversity essentially limited to the 3′ end (Supplementary Table S3).
TT-034–encoded shRNAs induce an increased number of specific cleavages on the HCV replicon RNA genome consistent with an RNAi mechanism of action
To understand the impact of this diversity of putative siRNA strands, we engaged in studies on the effect of the TT-034–encoded shRNA on the HCV replicon genome. A cursory examination of the sRNA-seq dataset identified a limited number of reads fully aligning to the HCV replicon, some of which could be a consequence of RNAi (Supplementary Table S4). This was an interesting result given the technology limitations in detecting sequences >80 nt, as direct action of TT-034 putative siRNA strands should yield HCV replicon genome fragments >400 nt.
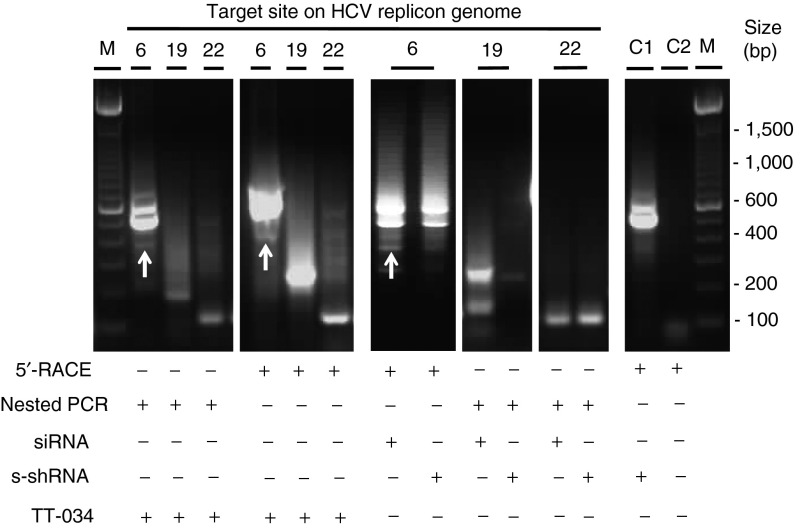
As the anticipated mode of action of TT-034 was RNAi, we developed 5′ RACE assays to examine if the three shRNA products could direct the generation of novel 5′ ends in the expected locations of the HCV replicon RNA genome. Electrophoretic analysis of the 5′ RACE products before and after nested PCR re-amplification was in agreement with this hypothesis, since all assays yielded bands of the expected size and no amplicons could be detected in the absence of TT-034 (Figure 3). Unexpectedly, reactions designed to detect shRNA6 activity also yielded high molecular weight bands. The size of these bands raised the possibility they might reflect 5′ RACE products from full-length HCV replicon RNA, given the target site of shRNA6 is proximal to the 5′ end of the HCV genome (Figure 1a). Sanger sequencing of these bands was in agreement with this proposition, indicating that the 5′ RACE RNA adapter had indeed ligated some 271 nucleotides upstream of the shRNA6 target site. On the other hand, Sanger sequencing of the 5′ RACE bands produced from target cleavage at the expected site of action yielded an interesting discrepancy; the first 44 nt at the 5′ end of the sequencing results were consistently ambiguous irrespective of the target site under examination (Supplementary Tables S5 and S6). Given the 5′ RACE adapter was also 44 nt long, we resolved this ambiguous region into superposed individual sequences composed of replicon fragments ligated to the 5′ RACE adapter. This analysis revealed that two different novel 5′ ends were generated on the HCV replicon +RNA genome in each site targeted by the three shRNA encoded in TT-034 (Table 2).
Figure 3.
Evidence of on-target mode of action for TT-034 by 5′ RACE. Electrophoretic analysis of 5′ RACE products and nested PCR re-amplification of 5′ RACE products generated on the HCV replicon RNA genome on account of the action of TT-034, transfected synthetic siRNA analogues or transfected synthetic shRNA analogues (s-shRNA) of shRNA6 (target 6), shRNA19 (target 19), and shRNA22 (target 22). The control lanes correspond to 5′ RACE amplification using primers specific for target site 6 in the presence of synthetic shRNA19 (lane C1), or in the absence of TT-034 using primers for the target site of shRNA19 (lane C2). White arrows identify 5′ RACE products generated by shRNA6 versus full-length HCV replicon RNA (large molecular weight bands).
Table 2. Resolution of the Sanger sequenced 5′ RACE amplicons generated by TT-034, s-shRNA analogues of the TT-034–encoded shRNAs or synthetic siRNA representing the most abundant shRNA products yielded from TT-034 expression in Con1b HCV replicon cells.
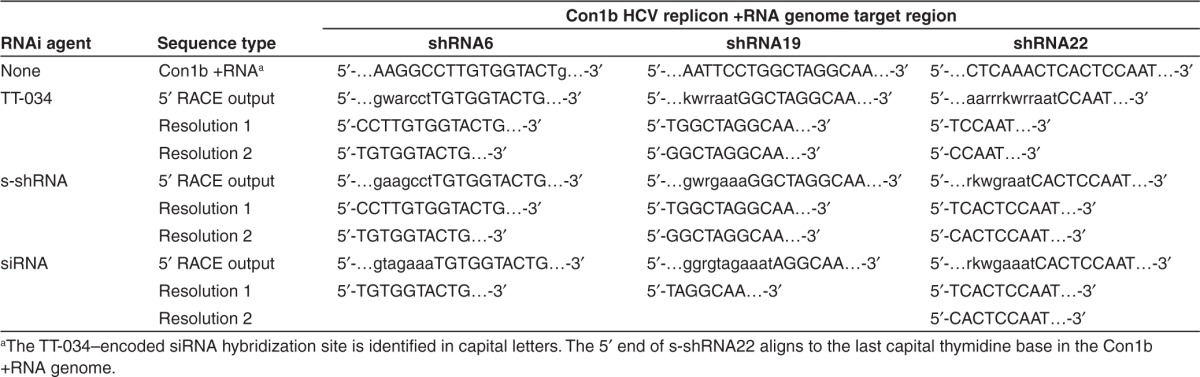
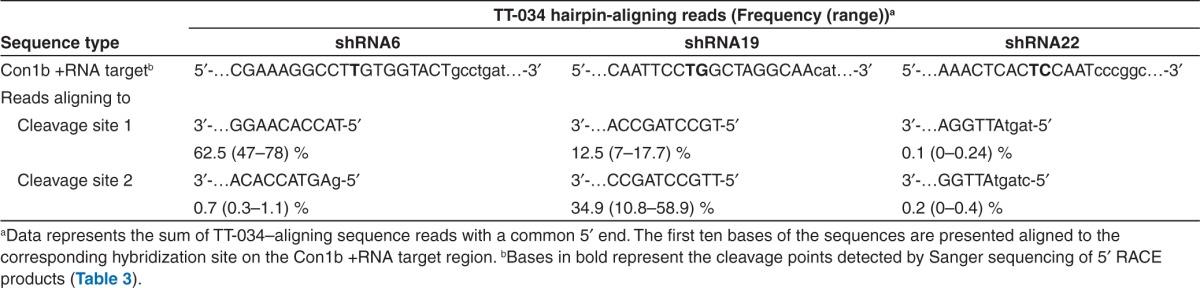
siRNA-directed RNAi has been reported to direct cleavage of complementary RNA targets opposite positions 10–11 from the 5′ end of the siRNA guide strand loaded in RISC.21 To assess if the observed sequencing results could be a consequence of an RNAi mode of action, we re-examined the sRNA-seq data set for TT-034 shRNA products that could account for the Sanger 5′ RACE results. Such putative RNAi mediators were successfully identified for all the novel 5′ ends on the HCV replicon RNA genome (Table 3). Interestingly, the relative frequency of these sequences varied substantially both within and between biological replicates. Moreover, despite the presence of as many as six continuous mismatches in the 5′ ends of these putative guide strands (e.g., shRNA22; Table 3), activity consistent with an RNAi mode of action appeared to be retained.
Table 3. Frequency of sequences processed out of TT-034–encoded shRNAs detected by pair-end small RNA NGS that could account for novel 5′ ends generated on the Con1b HCV replicon +RNA genome by an RNAi mechanism of action.
Synthetic analogues of the TT-034–encoded shRNA exhibit comparable activity consistent with an RNAi mechanism of action
To clarify further whether the TT-034–encoded shRNA were indeed acting via an RNAi mechanism of action, we evaluated the inhibitory potential and cleavage activity of three synthetic shRNA (s-shRNA) mimics of the hairpins encoded in TT-034 (Table 1). All three s-shRNAs were found to be active against the HCV replicon (Table 1) and therefore RNA was extracted and subjected to Sanger 5′ RACE analysis. Interestingly, s-shRNA6 and s-shRNA19 directed restriction of the HCV replicon RNA genome at the precisely the same positions as the TT-034–encoded shRNAs (Figure 3; Table 2), suggesting a common mechanism of action. However, shRNA22 resulted in two novel cleavage sites 4 bases upstream of those detected after treatment with TT-034 (Table 2). This apparent inconsistency was attributed to the different orientation design of the three hairpins (Figure 1b): Maturation of shRNA22 was shown to yield putative guide strands with up to 6 nucleotides upstream of the shRNA22 hairpin encoded in TT-034 (Figure 2f): these upstream sequences were absent from s-shRNA22 (Table 1), eliminating putative guide strand production that could incorporate these nucleotides. In contrast, the shRNA6 and shRNA19 guide strand populations incorporated loop sequences, which were retained in the s-shRNA analogues. Notably, the novel cleavage sites directed by s-shRNA22 were 10 and 9 nt from the 5′ end of the hairpin, suggesting that at least one of these cleavage sites was generated by a process consistent with an RNAi mode of action. We interpreted the second 5′ end as the result of 5′→3′ degradation of the primary cleavage product. These data, therefore, further supported the precept that the cleavages induced by TT-034 on the HCV replicon RNA genome might indeed be on account of an RNAi mechanism, despite the imprecision of shRNA processing observed.
Not all of the putative siRNA guide strands processed out of TT-034 can induce cleavage of the HCV replicon RNA
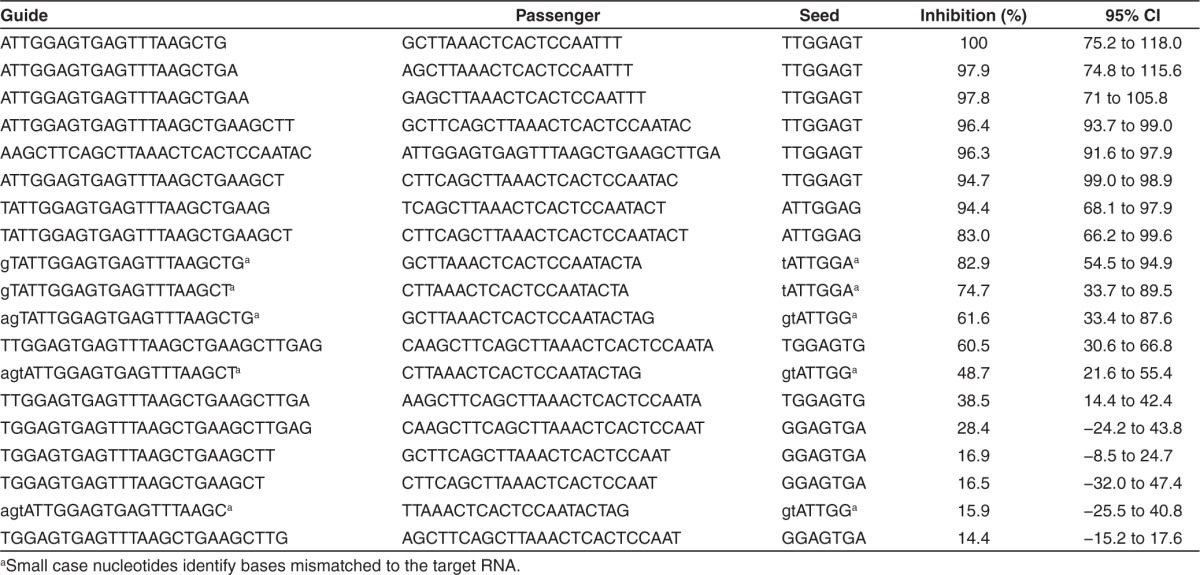
To understand which of the putative siRNA guide strands processed out of TT-034 harbored RNAi activity, we carried out focused in vitro studies on synthetic siRNA analogues of shRNA22 maturation products. To select candidates for these investigations, we ranked the putative guide strands produced out of shRNA22 (Figure 2c) by incidence and selected the top 20 sequences (Table 4). Of these, 11 (60%) exhibited >50% inhibitory activity in the HCV replicon reporter assay when transfected at 30 nmol/l concentrations, representing 4 out of 6 (66%) of the different 5′ ends tested. Interestingly, variability at the 5′ end through addition of off-target nucleotides resulted in progressive activity reduction (17–39.5%). On the other hand, removal of nucleotides from the 5′ end had a more substantial effect (51.3–88.2% activity reduction). In contrast, 3′ end variability did not appear to drive or impact robust inhibitory potential. Thus, the activity of the seven, most potent sequences with a common 5′ end but different 3′ ends varied by less than 6%.
Table 4. Activity of siRNAs (30 nmol/l; n of 5–8) directed against the shRNA22 target site.
To validate that the observed replicon inhibition was on account of an RNAi mechanism of action, 5′ RACE was performed following transfection with siRNA analogues of the most prominent maturation products of shRNA6, shRNA19 and shRNA22 which exhibited comparable potency (Table 1). These studies identified only one predominant 5′ RACE product for siRNA6 and siRNA19 consistent with an RNAi-induced mechanism of action;17 curiously, limited sequence ambiguity was also observed in the 5′ RACE product of siRNA19 (Supplementary Table S5). In contrast, siRNA22 resulted in two juxtaposed 5′ RACE products identical to those obtained with the s-shRNA22 hairpin (Table 2). These results reinforced our confidence that TT-034 activity was on account of RNAi. The true nature of the secondary 5′ RACE product encountered with siRNA22 and s-shRNA22, though suspected to be a degradation product, remained unresolved.
RACE-seq reveals siRNA-directed cleavage events can occur beyond position 10–11
Perplexed over this apparent cleavage imprecision demonstrated by possibly two of the synthetic siRNAs tested by Sanger 5′ RACE, we sought to study the outcome of their molecular action in more detail. We therefore designed new sets of gene-specific reverse transcription primers (Supplementary Table S1) that would yield short 5′ RACE cDNA products, suitably sized for Illumina pair-end NGS (<80 bp), or RACE-seq. Thus, we carried out independent studies on each of the three synthetic siRNA analogues of the TT-034–encoded shRNAs by transfecting the siRNAs separately into HCV replicon cells and subjecting total RNA extracts to RACE-seq.
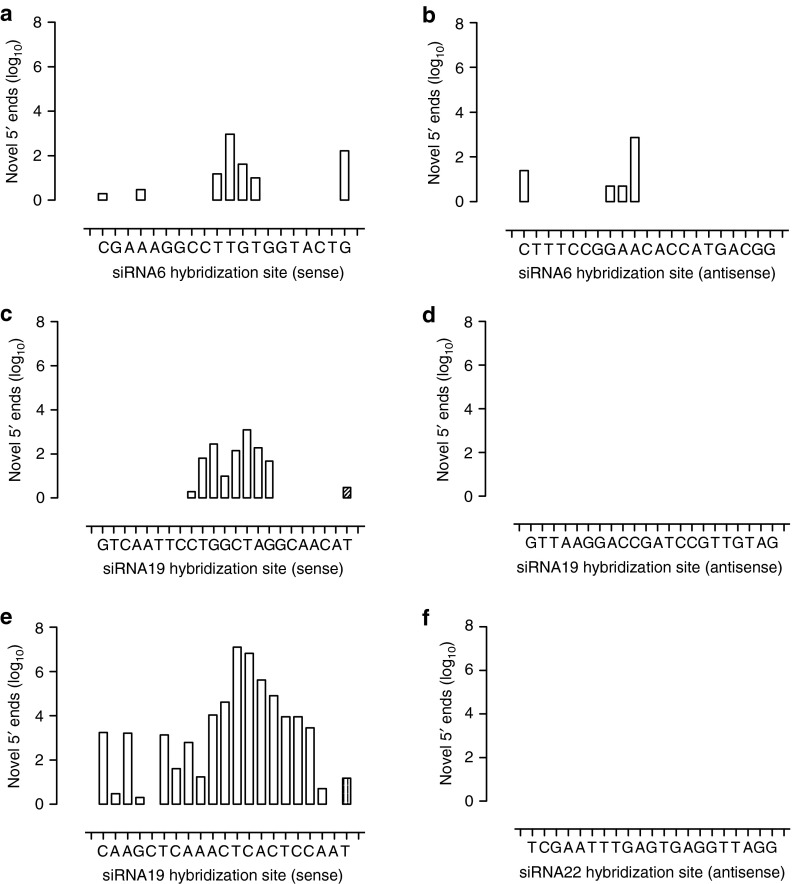
Taking into account the possibility of 5′→3′ degradation of RNAi products, we expected the higher sensitivity of RACE-seq to yield a profile of progressive novel 5′ ends starting from position 10–11 and heading downstream on the HCV genome target (i.e., −1, −2, etc. towards the 5′ end of the siRNA guide strand; Supplementary Figure S1). Accordingly, analysis of the novel 5′ ends generated from siRNA6 (Figure 4a,b), siRNA19 (Figure 4c,d) and siRNA22 (Figure 4e,f) yielded evidence of cleavage occurring opposite position 10–11 with variable detection of shorter products, consistent with an RNAi mechanism of action. To our surprise, however, all siRNAs yielded evidence of cleavage occurring beyond position 10, i.e., at +1, +2, etc. away from the accepted site of siRNA-directed, AGO2-induced cleavage. Particularly, in the case of siRNA19 (Figure 4c), the profile obtained hinted at the possibility that at least two main cleavage sites might be favored by RISC at positions 10–11 and 13–14 for this siRNA. Apart from these, the majority of other novel 5′ ends encountered in the siRNA target region occurred at frequencies below 5%. Detection of these low frequency novel 5′ ends appeared sensitive to both 5′ RACE primer optimization (Supplementary Figure S2a) and sequencing depth (Supplementary Figure S2b), however their relative incidence remained largely unaffected. In stark contrast, neither of the RACE-Seq reactions performed on the no-transfection control and the mock transfection control yielded any reads within the targeted amplification regions. Furthermore, for each siRNA under investigation, no novel 5′ end reads could be detected in the other two target regions, confirming that novel 5′ end generation was specific to the target site in question and on account of siRNA-directed activity, rather than off-target effects or RNA degradation. Similarly, no novel 5′ ends were detected in the antisense RNA replicon genome hybridization sites for siRNA19 (Figure 4d) and siRNA22 (Figure 4f), in accordance with previous data indicating activity of TT-034 on the antisense HCV replicon genome intermediate only at the hybridization site of siRNA6.11
Figure 4.
Detection of single siRNA-mediated cleavage points in the sense and antisense strands of the HCV replicon RNA genome by RACE-Seq. Following in vitro transfection of HCV replicon cells with individual synthetic siRNAs, RACE-Seq was performed using gene-specific reverse transcription primers hybridizing immediately downstream of the target sites of (a,b) siRNA6, (c,d) siRNA19, or (e,f) siRNA22 on either (a,c,e) the sense or (b,d,f) the antisense RNA genomes of the HCV replicon. The incidence of novel 5′ ends (y axis) on the HCV replicon genome within each siRNA hybridization site in a given orientation (x axis) is described. The orientation of potential 5′→3′ RNA degradation is indicated using a right or left facing triangle.
Discussion
Though considerable resources have been invested in understanding the biochemistry of RNAi,15,16,17,18,24 our efforts to develop a novel shRNA therapeutic necessitated full characterization of pro-drug (TT-034) maturation into an active drug form (siRNA guide strands). To date, NGS is the only methodology available to discriminate and relatively quantify single base pair differences in closely related sequences within a pure, but complex matrix of closely related nucleic acid materials. For this reason, we resolved to this approach to characterize TT-034. Our results indicate that the core processes of shRNA maturation and siRNA mode of action might be considerably more elaborate than previously thought.
Dicer cleavage of shRNA precursors has been reported to occur at nucleotide positions 21 to 25 from the hairpin ends.24 Such results are typically obtained using northern blotting leading to the commonly held assumption that the variety of putative siRNAs processed out of an shRNA was equivalent to the handful of bands observed under electrophoresis. Moreover, as 5′ RACE experiments suggested a single cleavage site on target RNAs, it was commonly assumed that the guide strands of these few siRNAs shared the same 5′ end but varied in their 3′ end. Instead, by implementing NGS, we were able to show that an shRNA can be processed into up to nearly 100 different putative siRNA strands. The clue to this apparent contention between the two techniques is not the length of the strands processed from shRNAs, but rather the dramatic variability in the sequences represented within each of these lengths (Supplementary Table S2). Thus, in their vast majority, the strands processed out of each shRNA might vary substantially in terms of 5′ and 3′ ends, but yield a strand population with sum strand lengths of ~21 nt length (Figure 2).
At first glance, these results could be dismissed as artefacts. After all, while double stranded RNA is partially protected from nuclease activity in cells,25 sample preparation for NGS provides ample opportunity for degradation.26 Yet comparative analysis of the reads aligning to either TT-034 or miRNA precursors within the same sequencing run suggest otherwise. Thus, the distributions of the 5′ ends of putative siRNA strands was both wide and asymmetrical, as opposed to the consistent Gaussian distribution of 3′ ends in strands with common 5′ ends (Supplementary Table S2). This irregularity suggested degradation might indeed be the source of variability at the 3′ end of putative siRNA strands, but not at the 5′ end. In stark contrast, microRNA-aligning sequences demonstrated distinct 5′ ends but variable 3′ ends in agreement with previous reports,23 indicating lack of degradation amongst miRNAs. Together, these data pointed towards both lack of sample degradation, and an intact Dicer mechanism in our model.
Recently, a novel, Dicer-independent pathway of miRNA maturation has been elegantly demonstrated around miR-451 biogenesis. Interestingly, the maturation of miR-451 is characterized by strong guide strand processing bias,27,28,29,30 a feature also of shRNA22 maturation (Figure 2c,f). This could indicate that shRNA22 might also be processed in a Dicer-independent manner, that the passive strand of shRNA22 is unusually unstable, or that the processing products are beyond the sensitivity of NGS. Although studies have been undertaken to assess if a miR-451 scaffold, which is crucial for Dicer-independent processing,31 can be used for shRNA design purposes30 there is only partial evidence that such constructs are matured in a Dicer-independent manner.31 More importantly, however, the decisive feature that drives miR-451 maturation independently of Dicer is the requirement for a short, 17 nt long hairpin stem, which is absent in the TT-034 shRNAs.31 The same, short stem feature was also corroborated recently in shRNA maturation engineering studies, which implicated AGO2 in Dicer-independent <19 bp stem shRNA (or AgoshRNA) maturation.32 Elsewhere, the re-examination of a dataset from a single in vitro Dicer knockdown study has raised the prospect that more miRNAs as well as other small noncoding RNAs might undergo Dicer-independent maturation.33 Thus, despite the limited structural similarity of shRNA22 to the primary transcript of miR-451 or published AgoshRNAs, a Dicer-independent pathway might be involved in the processing of this hairpin. The increased insight of strand processing afforded by sRNA-seq compared to northern blotting studies might serve to further clarify shRNA tailoring for strand selection and processing mechanism access control. Nevertheless, given that irrespective of maturation pathway all TT-034–encoded hairpins are processed into tens of different potential guide strands, we sought to understand how this might affect the RNAi potential of the ensuing siRNAs.
To address this conundrum, we resorted to classical 5′ RACE experiments. We reasoned that such an increased number of putative siRNAs would result in an accordingly higher number of cleavage points on the targeted RNA. Contrary to our expectations, electrophoretic analysis suggested that only a single siRNA-directed restriction event was mediated per shRNA, yet Sanger sequencing consistently resolved this band into at least two novel 5′ ends per target site. These results were reproduced both between biological replicates and with s-shRNA analogues, even where the s-shRNA design forced cleavage upstream from its' vector-encoded counterpart. Importantly, putative guide strands that could be responsible for the generation of these novel 5′ ends on the HCV replicon were identified amongst the sRNA-seq data for all hairpins. Hence, these results raised the possibility that our observations were not the outcome of degradation of the target RNA in a 5′→3′ direction, but the consequence of more than one putative siRNA guide strand being active through an RNAi mechanism.
Perplexingly, these findings were reproduced even with high purity synthetic siRNA analogues. Closer inspection of the capillary sequencing trace indicated that a third 5′ RACE product might indeed be present, but at levels below those acceptable in Sanger population sequencing analysis. We therefore concluded that a more sensitive method was required. Thus, we engaged in developing RACE-seq, reasoning that at sufficient depth the technique would be able to resolve all novel 5′ ends generated on an RNA target after exposure to RNAi mediators. To maximize fidelity, we opted for pair-end sequencing, and took care to generate cDNA compatible with the read length limitations of this approach (i.e., <80 bp).34 Thus, the need for mechanic/enzymatic shearing of long cDNA that could give rise to spurious 5′ ends or read elimination on account of sequencing errors due to cDNA length was avoided. Our control reactions confirmed the specificity of our approach, and the validity of the method was further demonstrated in the replication of previous findings on preferential strand activity (Figure 4d,f) observed in reporter construct and qPCR studies.12 In stark contrast, an excess of 108 reads corresponding to a primary novel 5′ end at the canonical restriction site directed by siRNA22 were detected (Supplementary Figure S2). In addition, substantial reads corresponding to additional novel 5′ ends juxtaposed to the primary cleavage site in the 5′→3′ orientation were detected for all siRNAs tested. The frequency of these secondary and tertiary reads was incrementally lower per nucleotide step (−1, −2, etc) in a Gaussian fashion, alluding to 5′→3′ RNA exonuclease or endonuclease activity (degradation). Intriguingly, novel 5′ ends in the HCV replicon genome were also detected at position +1 upstream of the expected cleavage point for siRNA6 (Figure 4a) and siRNA22 (Figure 4e) at apparently negligible frequencies (0.2 and 1.3%). Yet the most unexpected finding was obtained with siRNA19 (Figure 4c). In this case a cumulative 25% frequency of novel 5′ ends at positions +1 to +5 were encountered, with position +3 representing >14% of reads- an amount notable both at logarithmic and linear data representation scales. These results indicated that siRNA19 might direct on-target RNAi activity up to 5 bases beyond position 10–11 from the 5′ end of the guide strand, with position 13–14 being a secondary cleavage point.
One possible explanation for this result could be that the guide strand of siRNA19 carries two mismatches to the HCV replicon genome at its 5′ end. However, this contrasts the +3 position of the secondary cleavage point and thus might not account for our observations, even after cytosolic 5′ end processing of siRNAs.35 Alternatively, the presence of 5′ mismatches to the target sequence might alter the “molecular ruler” properties of siRNA in AGO2 to define a single cut site. After all, the sense strand of siRNA6, which is active against the antisense RNA strand of the HCV replicon genome (Figure 4b and ref. 12), does not contain any 5′ end mismatches and does not display any inexplicable cleavages. On the other hand, the active strand of siRNA22 also lacks 5′ mismatches to its' target yet still was found to result in target cleavage beyond the expected position (Figure 4e). Thus, our results indicate that AGO2 slicer activity might not be as precise as previously proposed,21 and that this precision might be driven, at least in part, by a siRNA sequence component.
siRNA6 and especially siRNA22 also exhibited the capacity to generate novel 5′ ends well beyond position 10–11, proximal to the siRNA guide strand 3′ end. Importantly, while the detection and extent of these additional products appeared to be relative to RACE-seq depth (Supplementary Figure S2c,d), they were found at levels 102–103 lower than the primary product. One possible explanation for these cleavages could involve RNase H, a double stranded nucleic acid endonuclease typically exploited in antisense approaches.36 However, such a mechanism is unlikely: RNAse H loading would lead to the destruction of both siRNA strands. Moreover, no cleavages were detected in the antisense RNA intermediate of the HCV replicon genome at the hybridization sites of siRNA19 and siRNA22. Another possibility would involve AGO1 loading of the siRNAs. This would direct sequestration and eventual degradation of the replicon genome in p-bodies, in a microRNA-like mechanism currently hypothesized to be a low turnover process.37 The scarcity of these events in our data is in support of this tenet. RACE-seq might therefore be a useful tool in experimental validation of computationally predicted miRNA recognition elements. Such an approach would be complementary to Cross-Linking Immuno-Precipitation NGS (CLIP-seq),38 which provides a more physiologically relevant solution to overexpression reporter constructs,39 but gives only a global overview of potential mRNA::miRNA interactions.
One of the main risks of RNAi, especially in a therapeutics context, is its strong potential for off-target activity. Does the unexpected plethora of putative siRNA guide strands produced from shRNA precursors present such a risk? Our data indicate that strand prevalence may not necessarily drive potency. Thus, putative shRNA22 guide strands with the intended 5′ end were the most commonly observed ones according to sRNA-seq (Figure 2). Analogues of these were also shown capable of inducing potent AGO2-mediated RNAi according to both reporter and 5′ RACE assays (Tables 1, 2, and 4). Still, they were not the primary drivers of shRNA22 activity according to 5′ RACE (Table 2). Collectively, these results implicate preferential guide strand loading onto RISC as the crucial step in enabling potency. Dicer is known to play a fundamental role in siRNA selection prior to activation of the RISC complex.40,41 As Dicer is also involved in shRNA maturation, it is conceivable that only a fraction of the putative guide strands it produces exhibit the appropriate characteristics for RISC loading. These properties are still unclear although they appear sequence-dependent,42 as our focused studies on shRNA22 maturation products indicate (Table 4). Further investigations on the fundamental mechanisms of putative shRNA strand loading onto AGO 1 and 2 are likely to require techniques capable of identifying sequences associated with the RISC complex, such as co-immunoprecipitation.43,44 With respect to the clinical potential of TT-034, bioinformatic analysis of the putative guide strands generated by the three shRNAs indicates that these do not align to either the human or mouse transcriptome suggesting that off-target effects by these sequences, even if RISC-loaded, should be limited.
Accordingly, TT-034 has been repeatedly found to be nontoxic in vitro, in rodents and in nonhuman primates even at high doses ensuring comprehensive hepatocyte transduction.11 Moreover, our replicon efficacy studies have demonstrated strong antiviral activity,12 which, as presented herein, is in accordance with a siRNA mechanism of action. This efficacy is sustained despite the existence of single mismatches in the replicon genome in two of the three TT-034 target sites (Supplementary Table S7), or up to four 5′ end mismatches between the most active shRNA22 guide strands and the replicon genome. Mismatch tolerance was also observed with both the s-shRNA and siRNA analogues (Tables 1 and 4) within an AGO2-mediated RNAi context (Table 2). Our data therefore suggest that TT-034 might have a wide spectrum of activity not limited to HCV genomes carrying at least one fully homologous target sequence.
The capacity to tolerate mismatches is important for an antiviral RNAi-based agent, particularly against a pathogen exquisitely equipped for resistance development through mutational escape. This is a well-described problem in HCV therapy on account of it's error-prone RNA-dependent RNA polymerase.45 Herein, perhaps, might lie the biggest advantage of TT-034, in its capacity to generate a diverse panel of putative guide strands. Thus, mutations arising within the shRNA-targeted region might be tolerated by TT-034, allowing it to continue to suppress the virus effectively. Our in vitro observations so far suggest this might be the case,12 especially if one takes into account the mismatches in the replicon system and the inherent positive selection bias towards viral escape for cell propagation under G418 antibiotic selection. Nevertheless, our view is that the genomic adaptation of HCV required for maintenance in culture restricts extrapolation to the patient milieu. Similarly, we would caution direct comparisons to observations with other viral targets of entirely different nature such as the host genome-integrating HIV retrovirus.
Interestingly, RNAi susceptibility studies on HCV using reporter constructs have indicated that the (−) strand RNA intermediate of the HCV genome may not be accessible to RNAi pathways.46 Our RACE-Seq results, however, suggest otherwise for TT-034 at least for shRNA6, introducing additional therapeutic advantages by extending the number of loci targeted to four, and involving both versions of the HCV genome. On the other hand, the discrepancy between these two studies raises important questions on the extent to which explicit 5′-RACE or RACE-seq data confirming an RNAi mode of action might be necessary in drawing robust conclusions out of RNAi experiments.
In conclusion, in this work, we have applied NGS to scrutinize the biological processing and mechanism of action of the novel RNAi agent TT-034. To our knowledge, this is the first study engaging into such a detailed survey of these core mechanisms of therapeutic RNAi mediators. Our studies have revealed unexpected complexity in both shRNA maturation and siRNA activity supporting further studies on the basic processes governing Dicer-dependent RNAi agent processing and siRISC bioactivity. However, our results have also demonstrated an on-target mechanism of action for TT-034 against both the (+) and (−) strands of the HCV RNA genome. Coupled to its established safety in rodents and nonhuman primates11 and its apparent capacity to suppress HCV from resistance development,12 these findings justify progression of TT-034 to the clinic to investigate its potential benefit for the treatment of chronically infected HCV patients.
Materials and methods
Cells. Con1b replicon cells (licensed from RebLikon GmbH, Schriesheim, Germany) refer to a Huh-7-derived cell line supporting the replication of subgenomic replicon RNA from the Con1 strain of HCV genotype 1b. The replicon encodes the neomycin phosphotransferase gene for selection as well as the firefly luciferase reporter gene to monitor its expression in cells. Con1b replicon cells were grown in Dulbecco's modified Eagle's medium (Gibco, Paisley, UK) supplemented with 10% fetal calf serum, 1 mmol/l sodium pyruvate, 1× nonessential amino acids, 1× penicillin/streptomycin and 500 µg/ml G418.
TT-034 and synthetic RNAs. TT-034 was produced by triple transfection of HEK293 cells using an AAV helper virus system (Stratagene, Stockport, UK). Viruses were purified using cesium chloride gradient centrifugation as previously described47 and the titre was determined by quantitative real time PCR [Forward primer: 5′-AGCTCCACCCTCACTGGTTTTT-3′, reverse primer 5′-CAAGCATGATCAGAACGGTTGA-3′ and probe: 5′-FAM- TTGTTGAACCGGCCAAGCTGCTG-TAMRA-3′ (Applied Biosystems)]. The titer was expressed in vector genomes/millilitre (vg/ml) and the dose used in term of vector genomes / Huh7 cells (vg/cells). s-shRNA and siRNA (Table 1) were manufactured by Integrated DNA Technologies and purified with PAGE and ion-exchange HPLC. Synthetic RNA purity (>99%) was confirmed internally by mass spectrometry as previously described.36
TT-034, s-shRNA and siRNA activity in Con1b replicon cells. Con1b replicon cells were transduced with TT-034 by incubating the appropriate amount of vector with the cells in a 2 ml volume. After 4 hours, an additional 8 ml of complete medium were added and the cells were incubated further for 68 hours until harvested. s-shRNA and siRNA were reverse-transfected into cells using Dharmafect3 (Dharmacon, Epsom, UK) as summarized in Table 1. To ensure consistent nucleic acid concentration during transfection, up to 1 µmol/l of nonspecific oligonucleotide [5′-GACCACTTGCCACCCATC-3′] was added. Upon transduction or transfection, the Con1b replicon cells were incubated for 48 hours then harvested using 0.25% Trypsin-EDTA (Gibco). An aliquot was lysed to measure the luciferase activity and cytotoxicity. Luciferase activity was measured using the BriteLite assay system (Perkin Elmer, Cambridge, UK) as previously described.12 Cytotoxicity measurements were conducted using CellTiter-Glo according to manufacturer's instruction (Promega, Southampton, UK).
NGS of shRNA and miRNA maturation and data analysis. Con1b replicon cells were transduced with TT-034 at a dose of 30,000 vg/cell. After 3 days incubation, the cells were harvested and RNA was extracted using Trizol. Small RNA libraries were constructed (DNAVision, Charleroi, Belgium) using the Small RNA Sample Prep kit (Illumina, San Diego, CA) according to the manufacturer's instructions. Briefly, small RNA was gel purified from 6 µg of total RNA, 5′ and 3′ adaptors were ligated, followed by reverse transcription and pair-end library enrichment by amplification (Illumina). The cDNA library was purified and validated for size, quality and concentration using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Each small RNA cDNA library was denatured, hybridized to an 8-lane flow cell, followed by cluster generation using isothermal amplification with a DGE-Small RNA Cluster Generation Kit v1.0 (Illumina). The prepared flowcell was sequenced for 75 cycles using a Genome Analyzer IIx (Illumina), according to the manufacturer's instructions.
In order to obtain interpretable data, only sequences with full-length adapters at both termini were included in downstream analyses. Low quality 3′ tails, 3′ polyA tail (a common artefact of the Illumina sequencing technology)48 and adapter sequences were trimmed from the reads. Moreover, we filtered out sequencing reads of less than 15 nucleotides in order to eliminate siRISC-restricted siRNA passenger strand fragments. Given that both the forward and reverse reads covered the entirety of the mature species sequenced, reads with any mismatches between the two were discarded to yield a high-confidence data set. To identify shRNA-derived sequences, assembled species were aligned to the TT-034 reference sequence using Bowtie v0.12.5.49 To investigate microRNA (miRNA) diversity, sequencing data was similarly aligned to the 1527 miRNA hairpin sequences in mirBase v. 18.22 Only species with perfect match to TT-034 or pre-miRNA hairpins were included in the downstream analysis. The results presented are representative of the analysis of two independent samples.
Sanger 5′ RACE and NGS-adapted 5′ RACE (RACE-seq) analysis. Con1b replicon cells were transduced with TT-034 or reverse-transfected with synthetic siRNAs or shRNAs at the concentrations indicated in Table 1. After 48 hours and following luminometric confirmation of antiviral activity and absence of cytotoxicity, RNA was extracted using the Qiagen RNA miniprep kit (Qiagen, Crawley, UK). For Sanger sequencing, 5′ RACE analysis was performed on 2 µg of total RNA using the GeneRacer kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions with the exception that the RNA were directly ligated to the kit 5′ adapter. cDNA generation and RACE amplification were performed using the kit adapter and site-specific primers listed in Supplementary Table S1. Following amplification, each RACE mixture was loaded and resolved on a 1.6% agarose gel. Fragments of the approximate expected size were excised from the gel, extracted using the Qiaquick Extraction kit (Qiagen) and directly submitted for Sanger sequencing with 20% resolution per base (Lark Technologies, Takeley, UK). Sequence analysis was performed using FinchTV v1.4.0 (PerkinElmer).
For RACE-seq, a truncated GeneRacer 5′ RACE adapter (5′-GGACACUGACAUGGACUGAAGGAGUAGAAA-3′) was used in a 5′ RACE reaction with a specific set of primers suitable for high fidelity pair-end sequencing (Supplementary Table S1). Those primers were designed to produce amplicons of less than 80 bp with no overlap with the siRNA hybridization site, thereby avoiding accidental siRNA sequencing. Primers were designed for the (+) RNA and (−) RNA orientation of the HCV genome, as both might be targeted by TT-034–encoded shRNA. Total RNA 5′-end ligation and cDNA generation was performed using the GeneRacer kit according to the manufacturer's instructions. A touchdown qPCR protocol was used to improve amplicon specificity. Briefly, GeneRacer kit PCR reagents were supplemented with primer to the truncated 5′-RNA adapter and gene-specific NGS PCR primers at 900 nmol/l concentrations each: 95 °C denaturation (15 seconds) was followed by annealing-extension at 70–60 °C over 20 cycles (0.5 °C steps, 15 seconds), followed by a further 20 cycles of amplicon re-amplification at 60 °C (15 seconds) and a 72 °C final extension step for 1 minute. PCR products were submitted to DNAVision (Charleroi, Belgium) for library construction and pair-end NGS as described above. Resulting data were quality processed as indicated above, the adapter sequences were removed and sequences with >10 bp length and no mismatches to the Con1b genome, as determined by Bowtie, were selected. Due to the high incidence of complementary sequences in the 5′ end of the HCV genome, the alignments were forced according to the orientation of the HCV genome targeted by the 5′ RACE primer under investigation ((+) RNA or (−) RNA). Results were expressed as the incidence of novel 5′ ends generated on the target sequence within the siRNA hybridization site in a logarithmic scale on account of Gaussian distribution profiles at logarithmic versus linear scales (e.g., Figure 4 versus Supplementary Figure S2).
SUPPLEMENTARY MATERIAL Figure S1. Predicted profile of RACE-Seq products generated by siRNA experiments. Figure S2. Impact of 5′ RACE reaction optimization and NGS depth on detection sensitivity of novel 5′ ends by RACE-Seq. Table S1. Primer sequences for 5' RACE and RACE-Seq on the (A) shRNA6, (B) shRNA19, and (C) shRNA22 target sites. Table S2. Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing. Table S3. Diversity of mature miRNA sequences in TT-034–transduced, Con1b replicon Huh7 cells as determined by Illumina pair-end small RNA sequencing. Table S4. Number and sequence of Con1b HCV replicon aligning reads obtained by paired-end small RNA NGS following transduction of Con1b HCV replicon cells with TT-034. Table S5. 5'-RACE outputs of TT-034 target regions following treatment of Con1b HCV replicon cells with TT-034, synthetic shRNA analogues (s-shRNA) or siRNA analogues of TT-034–encoded shRNA6, shRNA19, and shRNA22 as compared to the HCV replicon target sequence (cleavage site identified by “*”). Table S6. Full 5'-RACE outputs of TT-034 target regions following treatment of Con1b HCV replicon cells with TT-034 (ambiguous 44 nt region in red). Table S7. TT-034 shRNA homology to Con1b strain (Genbank AJ238799).
Acknowledgments
We thank Bernard Souberbielle, Boris Savic, and Edward Murray for constructive comments in the 5′-RACE and shRNA product activity studies; Patrice Godard for technical support in RACE-seq assay design; David Collins and Andrew Dalby for lending their statistical insight and expertise on the best possible approach to data curation and presentation for the sRNA-seq and RACE-seq studies. This work was supported by Pfizer Limited. Funding for open access charge: Pfizer Limited. The authors declare no conflict of interest.
Supplementary material
Predicted profile of RACE-Seq products generated by siRNA experiments.
Impact of 5′ RACE reaction optimization and NGS depth on detection sensitivity of novel 5′ ends by RACE-Seq.
Primer sequences for 5' RACE and RACE-Seq on the (A) shRNA6, (B) shRNA19, and (C) shRNA22 target sites.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
References
- Fassio E. Hepatitis C and hepatocellular carcinoma. Ann Hepatol. 2010;9:119–122. [PubMed] [Google Scholar]
- Di Bisceglie AM, Goodman ZD, Ishak KG, Hoofnagle JH, Melpolder JJ, Alter HJ. Long-term clinical and histopathological follow-up of chronic posttransfusion hepatitis. Hepatology. 1991;14:969–974. doi: 10.1016/0270-9139(91)90113-a. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- Hanecak R, Brown-Driver V, Fox MC, Azad RF, Furusako S, Nozaki C, et al. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J Virol. 1996;70:5203–5212. doi: 10.1128/jvi.70.8.5203-5212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarczak D, Korf M, Beger C, Manns MP, Krüger M. Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons. FEBS J. 2005;272:5910–5922. doi: 10.1111/j.1742-4658.2005.04986.x. [DOI] [PubMed] [Google Scholar]
- Seo MY, Abrignani S, Houghton M, Han JH. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J Virol. 2003;77:810–812. doi: 10.1128/JVI.77.1.810-812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan SM, Nasheri N, Luebbert C, Pezacki JP. The efficacy of siRNAs against hepatitis C virus is strongly influenced by structure and target site accessibility. Chem Biol. 2010;17:515–527. doi: 10.1016/j.chembiol.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Kim SI, Shin D, Lee H, Ahn BY, Yoon Y, Kim M. Targeted delivery of siRNA against hepatitis C virus by apolipoprotein A-I-bound cationic liposomes. J Hepatol. 2009;50:479–488. doi: 10.1016/j.jhep.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Suhy DA, Kao SC, Mao T, Whiteley L, Denise H, Souberbielle B, et al. Safe, long-term hepatic expression of anti-HCV shRNA in a nonhuman primate model. Mol Ther. 2012;20:1737–1749. doi: 10.1038/mt.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender H, Brady K, Burden F, Delpuech-Adams O, Denise H, Palmer A, et al. In vitro characterization of the activity of PF-05095808, a novel biological agent for hepatitis C virus therapy. Antimicrob Agents Chemother. 2012;56:1364–1375. doi: 10.1128/AAC.05357-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschos SA.2013Jones L, McKnight AJ.eds). Biotherapeutics: Recent Developments Using Chemical and Molecular Biology. Royal Society of Chemistry Publishing; London, UK; 176–223. [Google Scholar]
- Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Fuchs U, Damm-Welk C, Borkhardt A. Silencing of disease-related genes by small interfering RNAs. Curr Mol Med. 2004;4:507–517. doi: 10.2174/1566524043360492. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39 Database issue:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerter JA, Krishnan V, Lionberger TA, Walter NG. siRNA-like double-stranded RNAs are specifically protected against degradation in human cell extract. PLoS ONE. 2011;6:e20359. doi: 10.1371/journal.pone.0020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Lai EC. Functional parameters of Dicer-independent microRNA biogenesis. RNA. 2012;18:945–957. doi: 10.1261/rna.032938.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Schopman NC, Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res. 2013;41:3723–3733. doi: 10.1093/nar/gkt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenberger D, Çakir MV, Hoffmann S, Stadler PF. Dicer-processed small RNAs: rules and exceptions. J Exp Zool B Mol Dev Evol. 2013;320:35–46. doi: 10.1002/jez.b.22481. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Wei CL. Multiplex parallel pair-end-ditag sequencing approaches in system biology. Wiley Interdiscip Rev Syst Biol Med. 2010;2:224–234. doi: 10.1002/wsbm.40. [DOI] [PubMed] [Google Scholar]
- Snead NM, Rossi JJ. Biogenesis and function of endogenous and exogenous siRNAs. Wiley Interdiscip Rev RNA. 2010;1:117–131. doi: 10.1002/wrna.14. [DOI] [PubMed] [Google Scholar]
- Moschos SA, Frick M, Taylor B, Turnpenny P, Graves H, Spink KG, et al. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock GJ, Moschos S, Spivack SD, Hurteau GJ. The 3 prime paradigm of the miR-200 family and other microRNAs. Epigenetics. 2011;6:268–272. doi: 10.4161/epi.6.3.14635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Amarzguioui M, Kim DH, Alluin J, Heale B, Song MS, et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 2011;39:1510–1525. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowski L, Elazar M, Chu K, Glenn JS, Kay MA. The anti-genomic (negative) strand of Hepatitis C Virus is not targetable by shRNA. Nucleic Acids Res. 2013;41:3688–3698. doi: 10.1093/nar/gkt068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog RW, Fields PA, Arruda VR, Brubaker JO, Armstrong E, McClintock D, et al. Influence of vector dose on factor IX-specific T and B cell responses in muscle-directed gene therapy. Hum Gene Ther. 2002;13:1281–1291. doi: 10.1089/104303402760128513. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Goodhead I, Bähler J. Defining transcribed regions using RNA-seq. Nat Protoc. 2010;5:255–266. doi: 10.1038/nprot.2009.229. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted profile of RACE-Seq products generated by siRNA experiments.
Impact of 5′ RACE reaction optimization and NGS depth on detection sensitivity of novel 5′ ends by RACE-Seq.
Primer sequences for 5' RACE and RACE-Seq on the (A) shRNA6, (B) shRNA19, and (C) shRNA22 target sites.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.
Diversity of ssRNA strands processed out of TT-034–encoded shRNA hairpins expressed in TT-034–treated Con1b HCV replicon cells as determined by Illumina pair-end small RNA sequencing.