Summary
The aerial epidermis of all land plants is covered with a hydrophobic cuticle that provides essential protection from desiccation, and so its evolution is believed to have been prerequisite for terrestrial colonization. A major structural component of apparently all plant cuticles is cutin, a polyester of hydroxy fatty acids. However, despite its ubiquity, the details of cutin polymeric structure and the mechanisms of its formation and remodeling are not well understood. We recently reported that cutin polymerization in tomato (Solanum lycopersicum) fruit occurs via transesterification of hydroxyacylglycerol precursors, catalyzed by the GDSL-motif lipase/hydrolase family protein (GDSL) Cutin Deficient 1 (CD1). Here we present additional biochemical characterization of CD1 and putative orthologs from Arabidopsis thaliana and the moss Physcomitrella patens, which represent a distinct clade of cutin synthases within the large GDSL super-family. We demonstrate that members of this ancient and conserved family of cutin synthase-like (CUS) proteins act as polyester synthases with negligible hydrolytic activity. Moreover, solution-state NMR analysis indicates that CD1 catalyzes the formation of primarily linear cutin oligomeric products in vitro. These results reveal a conserved mechanism of cutin polyester synthesis in land plants, and suggest that elaborations of the linear polymer, such as branching or cross-linking, may require additional, as yet unknown, factors.
Keywords: cuticle, cutin, cell wall, Solanum lycopersicum, Arabidopsis thaliana, Physcomitrella patens.
Introduction
The plant cuticle consists of two lipid constituents: a variety of primarily aliphatic, organic solvent-soluble compounds referred to as wax, and a structurally complex polymer of esterified oxygenated fatty acids that is known as cutin. Together, these components form a resilient barrier against environmental stresses, such as desiccation, pests and pathogens (Yeats and Rose, 2013). The monomeric composition of cutin varies between species, among organs within a species, and with ontogeny, but is typically dominated by ω-hydroxy C16 or C18 fatty acids, often with a mid-chain hydroxyl, ketone or epoxy group (Kolattukudy, 2001). However, beyond the level of monomeric composition, the structure of cutin polymers is poorly understood. Studies involving selective oxidation or mesylation of free hydroxyls in intact cutin, followed by analysis of the depolymerization products, indicate that in several species whose cutin is principally composed of dihydroxyhexadecanoic acid, nearly all of the primary hydroxyls and half of the secondary hydroxyls are involved in ester bonds (Deas and Holloway, 1976; Kolattukudy, 1976). Additionally, soluble oligomers derived from cutin by partial chemical or enzymatic depolymerization have ester linkages involving both primary and secondary hydroxyls (Ray et al., 1998; Tian et al., 2008; Graca and Lamosa, 2010). Together, these results suggest that the major topology of dihydroxyhexadecanoic acid-based cutin polymers consists of a mixture of linear and branched domains, wherein cutin monomers are coupled solely through the primary hydroxyl, or by both the primary and secondary hydroxyls, respectively.
A long-standing question in plant lipid biology has been the mechanism and location of cutin polymerization, particularly given its insoluble and extracellular nature (Pollard et al., 2008). Recently, we demonstrated that in tomato (Solanum lycopersicum) fruit, an emerging model for studies of cuticle biology, cutin synthesis is catalyzed by a GDSL-motif lipase/hydrolase family protein, encoded by Cutin Deficient 1 (CD1, Solyc11g006250). In wild type fruit, cutin is abundant and composed primarily of inter-esterified monomers of 10,16-dihydroxyhexadecanoic acid (DHHA). However, fruit of the cd1 tomato mutant show a severe reduction (>95%) in the amount of polymerized cutin, but accumulate the soluble glyceryl ester of the cutin monomer, 2-mono-(10,16-dihydroxyhexadecanoyl)glycerol (2-MHG). In vitro, CD1 catalyzes the formation of ester oligomers from 2-MHG, suggesting that cutin polymerization in planta proceeds via successive rounds of transesterification, leading to esterified DHHA groups and the release of free glycerol (Yeats et al., 2012).
Given that enzymes involved in earlier steps of the cutin biosynthetic pathway, including a cytochrome P450 fatty acid hydroxylase (Rensing et al., 2008) and a bifunctional glycerol-3-phosphate acyltransferase/phosphatase (Yang et al., 2012), are conserved among the genomes of land plants, or embryophytes, we hypothesized that CD1-related enzymes, which we now refer to as cutin synthase-like (CUS) proteins, also catalyze cutin polymerization. Supporting this notion, a cutin-related function of two close homologs of CD1 from Arabidopsis thaliana, At5g33370 and At3g04290 (LTL1), is suggested by the phenotype of plants expressing an artificial microRNA that silences both genes. In these plants, flower petals were fused and devoid of adaxial nanoridges (Shi et al., 2011), two phenotypes that have been previously associated with cutin deficiency (Li-Beisson et al., 2009). Alternatively, cutin polymerization may result from dehydrating conditions within the cuticle, driving CD1 or unrelated lipolytic enzymes to catalyze a synthetic, rather than hydrolytic reaction (Girard et al., 2012; Yeats et al., 2012;). This hypothesis would imply that a generic lipase is sufficient and that polymerization is instead driven by properties of the substrate and the environment within the cuticle.
In this report, we build on our original discovery of a tomato cutin synthase (CD1; here renamed SlCUS1) and describe a set of experiments to resolve the enzyme specificity, its primary mode of action and the nature of the linkages that it synthesizes during cutin polymerization. We also address the evolutionary origins of CUS through a phylogenetic survey of SlCUS1-related genes and a biochemical analysis of the corresponding activities, to test the idea that the CUS-catalyzed formation of linear cutin-like polymers evolved very early in the history of land plants.
Results
Kinetics and enzymatic properties of CD1
As with other acyltransferases of the α/β hydrolase superfamily, CD1 likely acts through a ping-pong bi-bi reaction mechanism (Jiang et al., 2011), although in vitro the substrate for each step is the same in the initial phase of the reaction. First, one molecule of 2-MHG reacts with CD1, forming an acyl-enzyme intermediate and releasing glycerol. In the second step, the acyl-enzyme intermediate reacts with a second molecule of 2-MHG to form a dimeric product (Figure 1). As the reaction proceeds, progressively larger oligomers act as substrates, with the enzyme receiving an additional acyl group derived from 2-MHG in each cycle. Presumably, in vivo, diverse cutin monomers are integrated into the polymer from their respective glyceryl esters via a similar mechanism.
Figure 1.
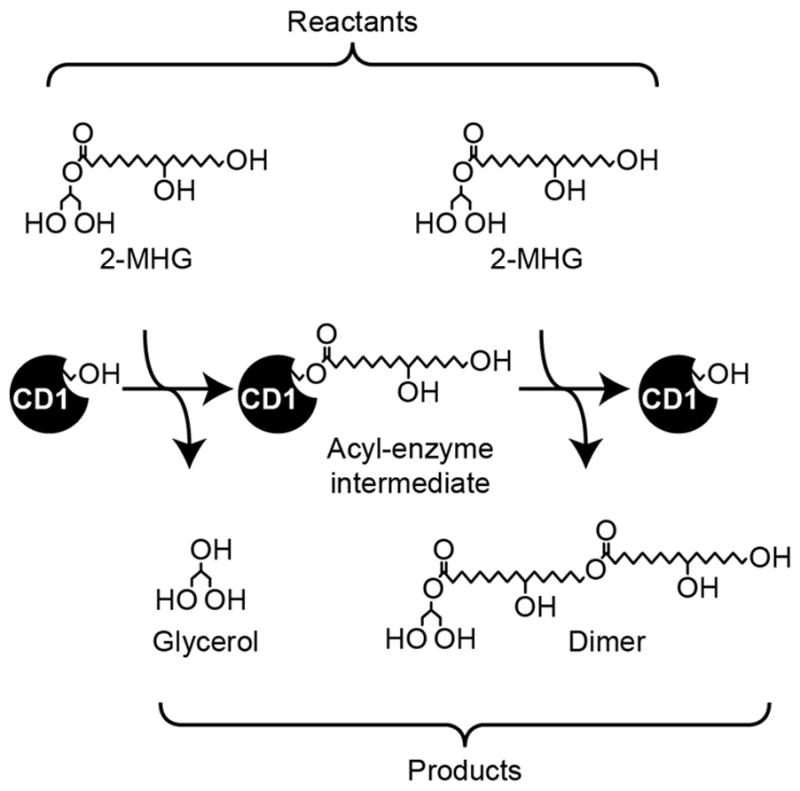
Reaction scheme for the formation of a 10,16-dihydroxyhexadecanoyl dimer from two molecules of 2-MHG in sequential steps. In subsequent reactions, the dimer serves as the substrate for the second step, yielding increasingly larger products. Minor amounts of acyl migrated product are also formed in vitro (see Figure 3 and Figure S1).
In order to determine the optimal conditions for the CD1 enzyme activity during the initial phase of oligomer formation, we assayed the formation of free glycerol, as this provided a means to quantify reaction progress while avoiding the complex technical challenges of quantifying a mixture of lipid products with diverse degrees of polymerization. As discussed below, nuclear magnetic resonance (NMR) spectroscopy and matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) analyses confirmed that there was negligible hydrolysis of 2-MHG, validating this approach to monitoring the progress of the reaction. Initial reaction rates were determined at a range of pH from 4.0 to 9.5. The enzyme exhibited maximum activity at acidic pH greater than 4.0, with optimal activity at a pH around 5.0 (Figure 2a), while notably, activity was minimal at pH ≥ 7.0. Together, these results are consistent with the known localization of CD1 in the acidic apoplast, rather than the neutral pH cytoplasm (Yeats et al., 2012; Girard et al., 2012). At pH 5.0, the enzyme exhibited substantial thermal stability, with decreased activity observed only at temperatures ≥50°C under our assay conditions (Figure 2b).
Figure 2.
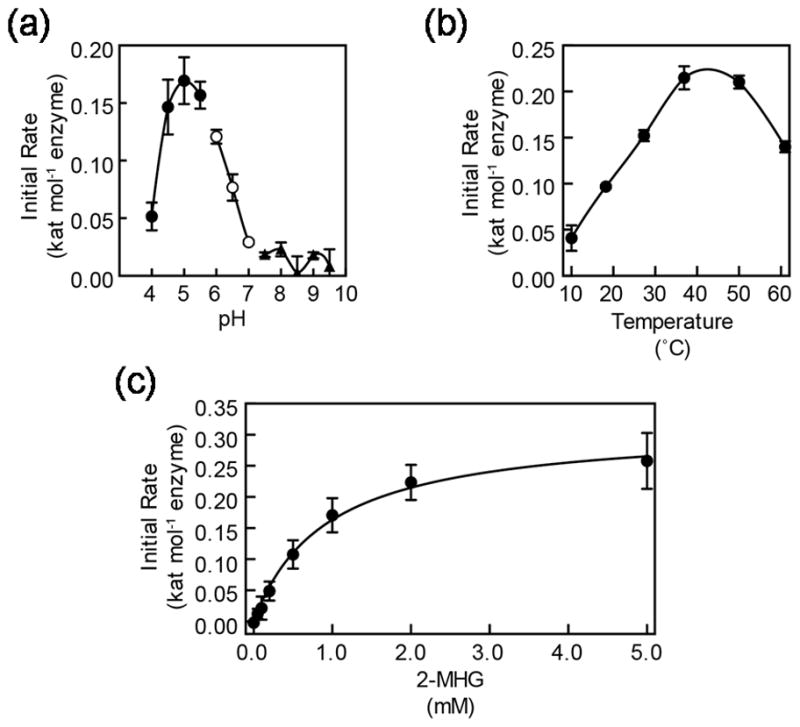
Kinetic characterization of CD1/SlCUS1. (a) The pH optimum of CD1/SlCUS1 assayed at 37°C for 2 h in buffers of respective pH values. Buffers used were sodium acetate, closed circles; sodium phosphate, open circles; Tris-HCl, closed triangles. (b) The temperature optimum of CD1/SlCUS1 assayed for 2 h at the respective temperatures in pH 5.0 buffer. (c) The effect of 2-MHG concentration on initial reaction rate. Data were fit to the Michaelis-Menten model as shown. Error bars represent standard deviation for triplicate reactions.
Determination of the Michaelis-Menten kinetics of the enzyme is complicated by the fact that in the initial phase of the reaction, both substrates are 2-MHG, reacting first to yield an acyl-enzyme intermediate and then the second molecule reacting to yield a dimeric product (Figure 1). Thus, the Km and kcat of each reaction step are not easily determined experimentally by saturating the rate of each individual step. Nevertheless, we conducted experiments to determine the initial rate of the reaction with varying concentrations of 2-MHG. Despite the anticipated complexity of the kinetics, the data fit a Michaelis-Menten saturation curve well, yielding an apparent Km of 925 ± 98 μM and apparent kcat of 0.314 ± 0.012 s−1 (Figure 2c).
CD1 catalyzes the synthesis of primarily linear cutin oligomers
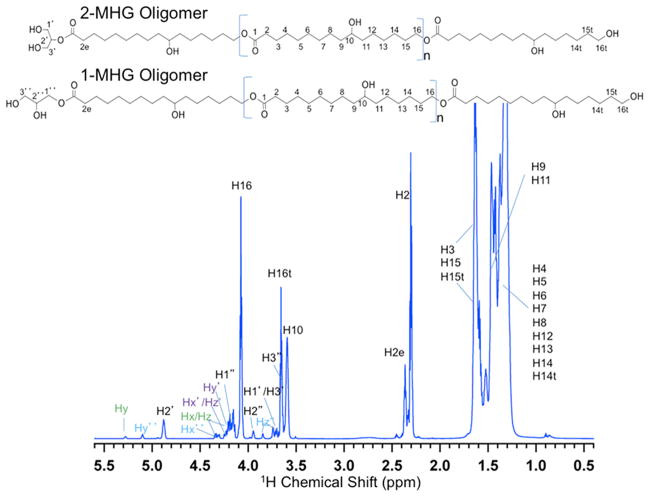
Previously, we demonstrated that CD1 catalyzes the formation of ester-linked oligomeric products in vitro using MALDI-TOF MS (Yeats et al., 2012). While that analysis provided evidence for the formation of ester linkages, it did not resolve the critical question of whether these linkages involved the midchain- or ω-hydroxyls of the cutin monomers, corresponding to branched or linear products, respectively. To determine the enzymatic specificity of CD1, we scaled up the in vitro reaction and extracted the mixture of products after 24 h at 37°C. Approximately 0.5 mg of material was recovered, dissolved in CDCl3 and subjected to 1D (1H) and 2D (1H-13C, 1H-1H) NMR spectroscopic characterization.
Tentative assignments for the 1H NMR spectrum (Figure 3) were made by comparison with semi-empirical predictions for a variety of CD1 oligomers with linear and/or branched architecture and systematically varying chain lengths (ACD NMR Predictor). Whereas the resonances from 1.2 to 1.7 ppm were attributed to chain methylene groups within each monomeric unit, the 1H signals at 2.3 and 2.4 ppm were assigned to H2 and H2e methylene groups that are downfield shifted by proximity to carboxyls of the repeating monomers and the terminal monomer linked to the glycerol group, respectively (Figure 3). The unique chemical shifts of these latter two protons permitted integration of the peak areas to estimate an average degree of polymerization of 4–5 for the CD1 products, in good agreement with our prior MALDI-TOF MS analysis (Yeats et al., 2012). Importantly, this provides independent confirmation that the majority of products of this enzyme are polymeric and not hydrolysis products. Analogously, the resonances at 4.1 and 3.6 ppm were assigned to H16 and H16t, downfield-shifted methylene protons (Figure 3) linked to the ester of interior monomers and hydroxyl groups of the terminal monomer, respectively (Tian et al., 2008; Wang et al., 2010). The resonance at 3.6 ppm is attributed to the H10 proton connected to a secondary alcohol of each monomeric unit (Tian et al., 2008).
Figure 3.
1H NMR spectrum of cutin oligomers with 1-MHG and 2-MHG terminal groups. Provisional chemical shift assignments were made by reference to semi-empirical spectral simulations (ACD, www.acdlabs.com) and confirmed by two-dimensional NMR experiments. The (CH2)n signal at 1.2 ppm is truncated to permit better visualization of the less abundant end groups. Backbone resonances for the minor glyceride products (x, y, z, x′, y′, z′, x″, y″, z″) are delineated in Figure S1 and Figure S2. This spectrum was well replicated by a second batch of CD1 reaction products.
The resonances at 4.9 and 3.7 ppm were assigned to methine H2′ and methylene H1′/H3′, respectively, within the glycerol end group of the 2-MHG oligomers (Figure 3). In addition, acyl-migrated oligomers terminated with 1-MHG were evidenced by 1H NMR signals at 4.1, 3.9 and 3.7 ppm (H1″, H2″, H3″; Figure 3). Comparison of the H2′ and H2″ integrated signal intensities suggests that 1-MHG oligomers represent ~30% of the isolated products, with no evidence for acyl migration found before treatment of freshly made substrate with the CD1 enzyme, but migration plausibly occurring after chain termination under the mildly acidic conditions of the reaction (Schuchardt et al., 1998). Very weak 1H resonances between 3.7 and 5.3 ppm were attributed to di- and triacylglycerols (Wang et al., 2010) that may form under these acidic conditions (Figure S1).
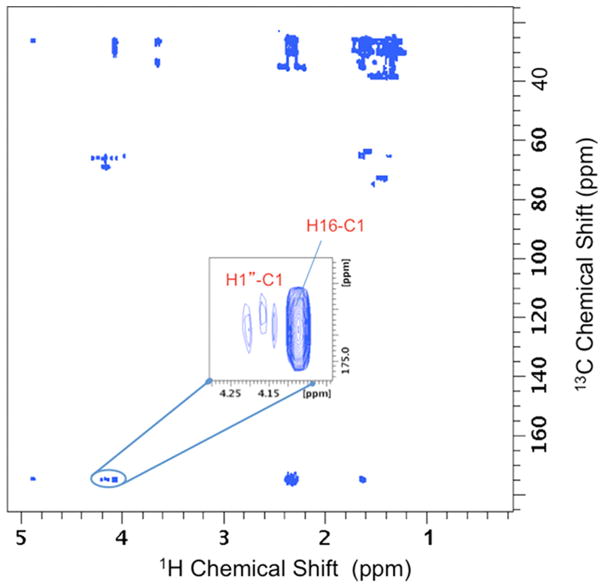
The peak assignments for the MHG oligomers were confirmed by 2D 1H-13C heteronuclear single quantum coherence (gHSQC; Palmer et al., 1991; Kay et al., 1992); Figure S1) and heteronuclear multiple bond correlation (gHMBC; Bax and Summers, 1986; Figure S2) NMR spectroscopy. These analyses showed direct and remote through-bond 1H-13C correlations, corresponding to the proposed structural features in the two oligomer types, and were confirmed independently in 2-MHG monomer samples. The 13C chemical shifts observed in the 2D correlation spectra were in excellent agreement with the ACD predictions. The expected 1H-1H connections within bonded networks were also found from 2D total correlation spectroscopy (TOCSY; Shaka et al., 1988). Independent evidence for inter-monomer linkages, indicative of a linear polymer chain, is illustrated by the HMBC contour plot in Figure 4, which shows a region of the spectrum in which a strong three-bond correlation is evident between H16 and C1. Notably, the spectrum displayed no HMBC crosspeaks between H10 and C1 nuclei, as would be expected for an ester linkage in a branched oligomer (Tian, 2005).
Figure 4.
2D 1H-13C HMBC NMR contour plot for MHG oligomers. The H16-C1 correlation indicates formation of a linear oligomeric product involving esterification of a primary alcohol. The weaker correlations in this region correspond to H1″-C1 connectivities in the 1-MHG oligomer. Additional carboxyl carbon correlations in this region are identified in Figure S2.
Identification of cutin synthase proteins (CUS) in evolutionarily distant land plants
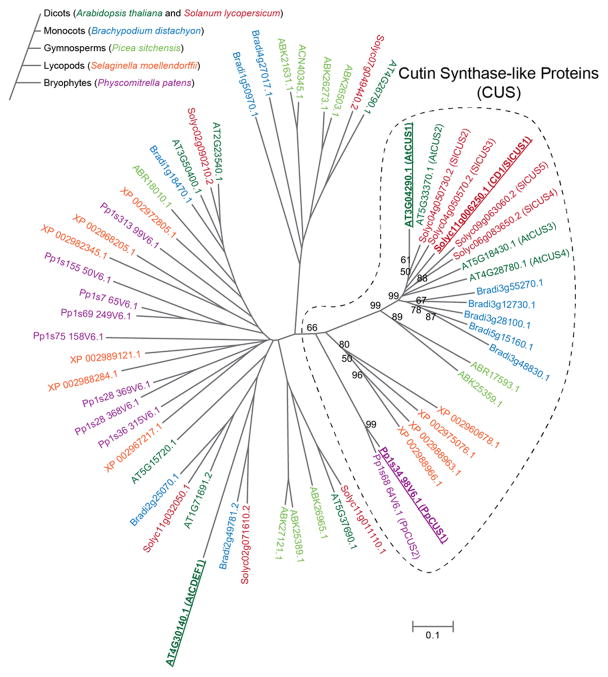
CD1 belongs to a very large super-family of plant proteins containing the GDSL domain (Akoh et al., 2004). In order to identify homologous proteins and putative orthologs in a taxonomically broad range of lineages that collectively represent both earlier and later diverging plant species, we constructed a multiple sequence alignment with a subset of amino acid sequences derived from complete genomes (Arabidopsis thaliana, S. lycopersicum, Brachypodium distachyon, Selaginella moellendorffii and Physcomitrella patens), as well as a unigene collection derived from deep sequencing of expressed sequence tags (Picea sitchensis). From this alignment, a distance tree was constructed using the neighbor-joining method (Figure 5).
Figure 5.
Unrooted phylogenetic tree of CD1-related sequences from representative embryophyte species. Genes were identified as described in the Materials and Methods section. The phylogeny of species considered is shown in the schematic tree to the left and genes are color coded by species as indicated. Bootstrap support >50% is indicated at each branch node within the putative cutin synthase-like clade (indicated by a dotted line). Genes corresponding to enzymes characterized in the present work are highlighted in bold and underlined.
Despite the diversity of the sequences considered, a well-supported and closely related clade of CD1-related putative CUS proteins was identified in the spermatophytes (Figure 5). In the three complete angiosperm genomes considered (A. thaliana, S. lycopersicum and B. distachyon), the putative CUS family consists of 4 or 5 closely related genes per genome (~70% or greater amino acid identity within each species). A clade with somewhat lower bootstrap support (66%) was also identified containing representatives from all the species, including the more distantly related and earlier diverging non-spermatophytes S. moellendorffii (spike moss) and P. patens (moss). This suggests that the proposed CUS-like family of GDSL-domain proteins may be anciently conserved across all land plants. Genes in this clade are therefore tentatively designated as cutin synthase-like (CUS) genes.
CUS proteins from evolutionarily distant plant species catalyze cutin polymerization
To test whether proteins from the proposed CUS clade possess the acyltransferase activity that would typify cutin synthases, we expressed and purified a putative ortholog of CD1 from each of A. thaliana and P. patens, and compared their activities to that of CD1/SlCUS1 (Figure 6). We also expressed and purified AtCDEF1, an A. thaliana GDSL protein that aligns outside this clade (33% sequence identity with CD1; Figure 5), which has previously been shown to have esterase activity and has been proposed to be an endogenous cutinase (Takahashi et al., 2010).
Figure 6.
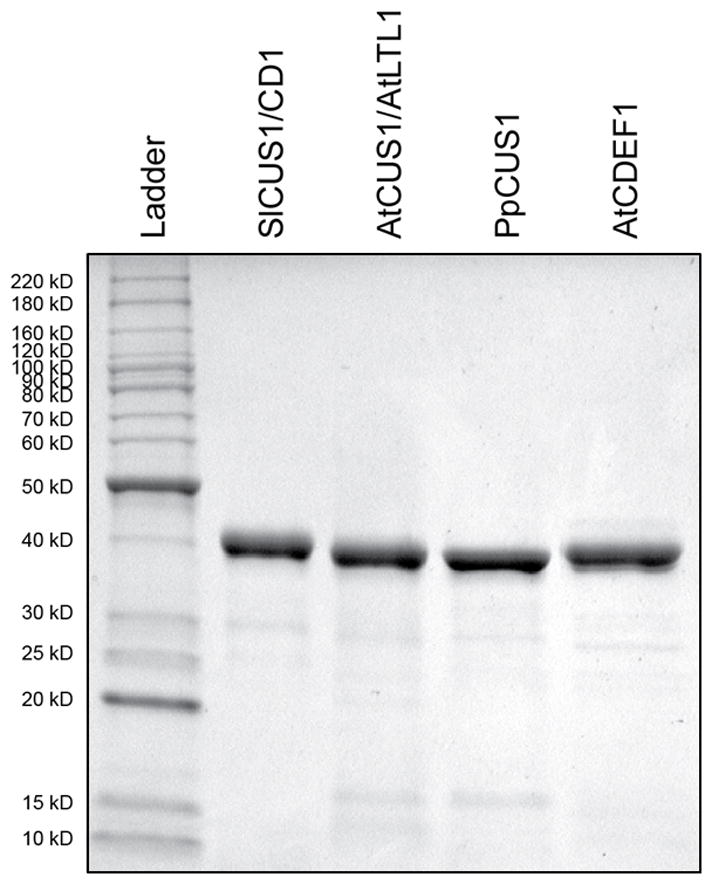
Coomassie Brilliant Blue stained SDS-PAGE gel of purified recombinant CUS enzymes and AtCDEF1. A ladder of molecular markers is shown with units in kilodaltons (kD).
After a reaction time of 24 h at 37 °C, the products were analyzed by MALDI-TOF MS. All of the putative CUS enzymes catalyzed the formation of cutin oligomers and lesser amounts of the hydrolysis product DHHA, while AtCDEF1 exclusively catalyzed hydrolysis of 2-MHG (Figure 7). Approximate quantification of the product profile by integration of mass spectra indicated that for the enzymes with acyltransferase activity, hydrolytic products represented a minor portion of the total products formed (Table 1). As the integrated ion abundance signal is proportional to the molar amount of each detected ion and not the mass amount, and thus the number of 2-MHG from which it is derived, this method likely overestimates the amount of hydrolysis that is taking place. More precise quantification of the in vitro products was not attempted as this would require synthesis of oligomeric standards of defined size. However, comparison of the product profile of CD1/SlCUS1, as determined by NMR and MALDI-TOF, lead us to conclude that MALDI-TOF is a valid means of confirming that the great majority of reaction products are oligomers and not DHHA for all of the CUS enzymes tested.
Figure 7.
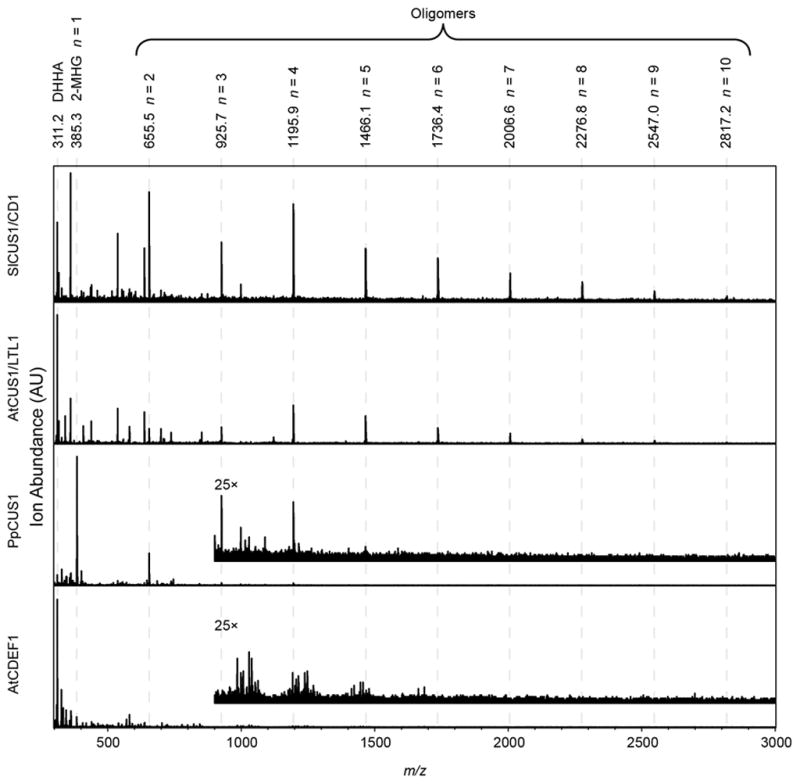
MALDI-TOF spectra of reaction products. Predicted masses of Na+ adducts of 2-MHG, its hydrolysis product DHHA, or oligomer products, are indicated at the top. Inset spectra, scaled 25× in the intensity axis, are shown for PpCUS1 and AtCDEF1.
Table 1.
Apparent Michaelis-Menten Kinetic Parameters and in vitro Product Distribution. Kinetic parameters were estimated by non-linear least-squares fitting of the data to the Michaelis-Menten equation and standard error of the parameter estimates is given.
| Enzyme | Apparent Kinetic Parameters | Products (Relative Ion Abundance) | |||
|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Polymerization | Hydrolysis | |
| SlCUS1/CD1 | 925 ± 98 | 0.314 ± 0.012 | 339 | 96% | 4% |
| AtCUS1/LTL1 | 903 ± 106 | 0.169 ± 0.007 | 187 | 78% | 22% |
| PpCUS1 | 543 ± 138 | 0.013 ± 0.001 | 24 | 87% | 13% |
| AtCDEF1s | <50* | 3.25 ± 0.17 | >65000 | 0% | 100% |
Precise determination of Km for this enzyme was limited by assay sensitivity.
To compare the efficiency of these enzymes, glycerol release assays were performed with varying concentrations of 2-MHG. As for CD1, the initial rates of each reaction fit a Michaelis-Menten saturation curve well and yielded apparent kcat and Km values (Table 1). PpCUS1 was notably less efficient than the other acyltransferases, although MALDI-TOF analysis of products indicated that acyltransferase activity greatly exceeded hydrolysis (Table 1). All CUS enzymes exhibited apparent Km values of < 1 mM (Table 1). The entirely hydrolytic AtCDEF1 was a much more efficient enzyme, with a much lower apparent Km and higher kcat than any of the CUS enzymes tested (Table 1).
Discussion
Despite the ubiquity of polymeric cutin among land plants and the clear association between its structure and resistance to terrestrial environmental stresses, the nature of its biosynthesis has remained largely enigmatic. Tomato fruit provide an excellent experimental system in which to study cutin biochemistry, structure and function and we previously demonstrated that tomato fruit cutin polymerization occurs via CD1 (SlCUS1) catalyzed transesterification of 2-MHG (Yeats et al., 2012). Here, we show that homologous enzymes with the same activity are present across the taxonomic spectrum of extant land plants, indicating that a conserved cutin biosynthesis pathway evolved prior to the divergence of bryophytes and tracheophytes.
The biochemical characterization of CD1 presented here revealed some of the enzymatic features of cutin synthases. Consistent with its previously demonstrated extracellular localization, CD1 has a pH optimum of 5.0 (Figure 2a), which is in accordance with apoplastic pH values that have been reported for various plant tissues (Felle, 2001). Notably, CD1 activity is greatly reduced above pH 7 (Figure 2A) and we propose that this pH specificity may provide a regulatory mechanism that prevents premature formation of insoluble polymers in the neutral endoplasmic reticulum, the likely site of 2-MHG biosynthesis (Beisson et al., 2012). Additional characterization of the pH microenvironment of the outer epidermal cell wall and cuticle may inform our understanding of the spatial and temporal regulation of cutin synthesis.
Remarkable thermal stability of CD1 was also noted, with reduced activity during a 2 hour period occurring only at ~50°C (Figure 2b). This stability could be particularly important in the case of cutin synthases that must maintain activity at the outer surfaces of plants, which are often subjected to high temperatures in direct sunlight. The initial rate-limiting Michaelis-Menten parameters of CD1 and its putative orthologs were determined, and although they cannot be assigned to specific steps of the enzymatic reaction (Figure 1), they provide a means of comparing the efficiency of the respective enzymes (Table 1). The turnover numbers of all the enzymes were relatively low, and apparent Km values are relatively high, although we note that this reflects the in vitro formation and elongation of relatively small oligomers. In planta, chain elongation within polymeric cutin may proceed at a greater rate, possibly aided by molecular crowding or the uniquely hydrophobic environment of the cuticle. On the other hand, the observed high abundance of CD1 in the tomato fruit exocarp (Girard et al., 2012) suggests that high enzymatic efficiency may not be required for effective cutin polymerization. The observed efficiencies (kcat/Km) of CD1/SlCUS1 and LTL1/AtCUS1 were similar, perhaps reflecting the function of both enzymes as 2-MHG ‘specialists’, in that both tomato fruit cutin and A. thaliana floral cutin are rich in DHHA (Li-Beisson et al., 2009). The PpCUS1 enzyme was markedly less efficient using 2-MHG as a substrate, indicating that its native substrate may be the monoacylglyceryl ester of a different cutin monomer. Indeed, we recently reported that in P. patens, polymerized DHHA is detected in delipidated residues at lower levels than other putative cutin monomers (Buda et al., 2013). Nevertheless, our results clearly indicate that PpCUS1 can function as a 2-MHG acyltransferase, and has negligible hydrolytic activity. Given the basal position of PpCUS1 within a clade of GDSLs that spans the evolutionary history of extant land plants, we propose a family of cutin synthase-like (CUS) proteins that are likely responsible for the formation of protective cutin polymers. While we have demonstrated this at the level of in vitro synthesis of cutin-like oligomers, true orthology of these genes will require additional genetic tests of their function. Indeed, we note that these enzymes may be involved in the biosynthesis of other apoplastic polyesters, such as suberin.
Finally, the observation that CD1 catalyzes the formation of an exclusively linear product from 2-MHG in vitro indicates that there may be additional factors required for the formation of branched or cross-linked cutin. Whether these modifications are catalyzed by GDSL cutin synthases or other extracellular enzymes can now be addressed, providing further insight into the evolution of cutin structures.
Experimental Procedures
Protein Expression
The coding sequences of AtCUS1 and AtCDEF1 were amplified by PCR from A. thaliana var. Columbia flower cDNA and cloned into the vector pEAQ-HT (Sainsbury et al., 2009) using restriction enzyme sites introduced in the PCR primers. The coding sequence of PpCUS1 from the moss P. patens was similarly cloned into pEAQ-HT following PCR amplification from cDNA prepared from P. patens gametophore tissue.
Proteins were transiently expressed in Nicotiana benthamiana leaves and purified as previously described (Yeats et al., 2012), except that size exclusion chromatography was performed with 100 mM ammonium acetate buffer (pH 7.0) and sample concentration was performed with a Vivaspin 6 ultrafiltration spin column (10,000 MWCO, Sartorius Stedim Biotech, www.sartorius.com) that was previously washed extensively with distilled water to remove glycerol from the filter.
Enzyme kinetic assays
For determination of the pH optimum, 25 μL assays containing 250 ng of CD1/SlCUS1, 120 mM NaCl, 40 mM buffer and 800 μM 2-MHG, synthesized as described previously (Yeats et al., 2012), were incubated for 2 hours at 37°C. The buffers used were: sodium acetate (pH 4.0– 5.5), sodium phosphate (pH 6.0– 7.0), Tris-HCl (pH 7.5– 9.5). The reaction was terminated by boiling for 10 minutes. 10 μL from each assay were transferred to a microtiter plate and analyzed for glycerol content by addition of 90 μL of Free Glycerol Reagent (Sigma-Aldrich, www.sigmaaldrich.com), a coupled enzyme-based assay in which formation of quinoneimine dye is proportional to the free glycerol concentration. Absorbance at 560 nm was measured and compared to that of a standard curve. For determination of the temperature optimum, 20 μL assays containing 250 ng of CD1/SLCUS1, 1 mM 2-MHG, 150 mM NaCl, and 50 mM of sodium acetate (pH 5.0) were prepared and incubated at the indicated temperatures for 2 hours. The reactions were terminated and glycerol was quantified as above.
For determination of apparent Michaelis-Menten kinetic parameters, 100 μL assays were prepared with the indicated concentration of 2-MHG, 150 mM NaCl, and 50 mM of sodium acetate (pH 5.0) at 37 °C. The reactions were initiated by the addition of an appropriate dilution of each enzyme to ensure a linear initial rate after 2 hours (80 ng for CD1/SlCUS1 and AtCUS1, 320 ng for PpCUS1, 4 ng for AtCDEF1). Reactions were terminated as above and the glycerol assay was modified to increase its sensitivity without exceeding the buffering capacity of the reagent: 80 μL of each reaction was withdrawn, 20 μL of 1 M sodium phosphate (pH 7.0) and 100 μL of Free Glycerol Reagent was added and absorbance at 560 nm was measured using a plate reader. Non-linear least squares fitting of the data to the Michaelis-Menten equation was performed with SigmaPlot 9 software (Systat Software). For all assays, background was determined for assays conducted with boiled enzyme and subtracted. As the background signal was strictly linear with respect to 2-MHG concentration, it was attributed to non-enzymatic hydrolysis of 2-MHG, likely during termination of the reaction by boiling. In all experiments, background hydrolysis accounted for <3% of the initial substrate concentration.
NMR analysis
For large scale oligomer production, 2.5 mL reactions containing 25 μg of protein, 1 mM 2-MHG (from a 100 mM stock in dimethyl sulfoxide [DMSO]) and 50 mM ammonium acetate, pH 5 were incubated for 24 h at 37°C with rotary shaking (250 rpm). Products were extracted twice with 2.5 mL of 2:1 (v/v) chloroform/methanol and the pooled products were back extracted twice with distilled water. The organic phase was dried over anhydrous sodium sulfate and evaporated under a gentle stream of nitrogen, yielding ~0.5 mg of product.
Solution-state 1H NMR spectra of ~0.5 mg of an MHG oligomer mixture in 50 μL of CDCl3 were acquired at 298 K on an AVANCE I spectrometer (Bruker, www.bruker.com) operating at a 1H frequency of 800.129995 MHz and equipped with a 1.7-mm cryogenic triple resonance microprobe. Analogous reference spectra of the 2-MHG monomer were obtained in CD3OD. For both samples, typical acquisition conditions included a 10.7 μs 90° pulse width, 32K complex data points defining a spectral width of 9.6 kHz (~12 ppm), 128 scans, and a 1.0-s relaxation delay added to a 1.7-s acquisition time, inserted between successive acquisitions to ensure quantitative reliability of the resulting spectra. Topspin 2.1 software (Bruker, www.bruker.com) software was used to condition and Fourier transform the NMR data, for which the resonances were referenced to tetramethylsilane as an internal chemical shift standard and integrated to determine the relative numbers of each structural moiety and the average degree of polymerization. Error limits for these quantitative analyses were estimated from duplicate reaction mixtures, which yielded visually indistinguishable 1H spectra.
Provisional structural assignments of the spectra were made by reference to semi-empirical predictions (ACD NMR Predictor, v9, Advanced Chemistry Development, www.acdlabs.com) with confidence limits of < 0.3 ppm (1H) and < 3 ppm (13C). These assignments were confirmed with two-dimensional NMR experiments that identified pairwise (1H-1H and 1H-13C) interactions between directly or remotely bonded nuclei, both within and between monomer units. Heteronuclear single quantum coherence (gHSQC) experiments (Palmer et al., 1991; Kay et al., 1992) were carried out to detect directly bonded 1H-13C pairs, using a 12 μs 90° 13C hard pulse, polarization transfer time corresponding to a coupling constant 1JCH = 145 Hz, relaxation delays of 2 s between 96 successive transients, with 9.6 and 18.1 kHz spectral widths in the proton and carbon (aliphatic region) dimensions defined by 1K data points and 512 time increments, respectively. 1H-13C heteronuclear multiple bond correlation (gHMBC) spectra (Bax and Summers, 1986) were acquired with a polarization transfer time corresponding to 3JCH of 10 Hz, relaxation delays of 2 s between 128 transients, with 8.8 and 42 kHz spectral widths in the proton and carbon dimensions defined by 8K data points and 256 time increments in the indirect dimension. Total correlation spectroscopy (TOCSY) (Shaka et al., 1988) was used to confirm through-bond 1H coupling networks within the glycerol backbone and along the acyl chains of the oligomers.
Bioinformatic analysis
Sequences were retrieved by BLASTP search of the NCBI (P. sitchensis and S. moellendorffii; ncbi.nlm.nih.gov), Cosmoss (P. patens; www.cosmoss.org), TAIR (A. thaliana; www.arabidopsis.org), SGN (S. lycopersicum; www.solgenomics.net) and Brachypodium.org (B. distachyon; www.brachypodium.org) databases, using the amino acid sequence of SlCUS1/CD1 as a query. For each species, the top 10 hits (Bit score) were retained after discarding very close duplicates (>95% similarity) from the draft genome of S. moellendorffii and EST-derived sequences of P. sitchensis. The putative endogenous cutinase, AtCDEF1 (Takahashi et al., 2010), was also included in this analysis. Amino acid sequences were aligned using Clustal W (Thompson et al., 1994), and after discarding the predicted secretory signal peptide region of each sequence, a neighbor–joining tree was constructed using MEGA4 (Tamura et al., 2007), with the default settings of the MEGA4 package.
MALDI-TOF mass spectrometry analysis
For analysis of reaction products, 50 μL reactions were prepared containing 0.5 μg of protein, 1 mM 2-MHG (from a 100 mM stock in DMSO) and 50 mM ammonium acetate, pH 5. Reactions were incubated at 37°C for 24 hours and then frozen until further analysis. The thawed samples were diluted with three volumes of methanol, then spotted with DHB matrix and analyzed by MALDI-TOF MS (matrix assisted laser desorption ionization time of flight mass spectrometry) as previously described (Yeats et al., 2012). Background spectra were also collected from reactions containing no 2-MHG substrate and subtracted using mMass 3 software (Strohalm et al., 2010). Semi-quantitative analysis of reaction products was based on integration of the isotope envelope of each compound.
Supplementary Material
Acknowledgments
We thank Dr. George Lomonossoff (John Innes Centre) and Plant Bioscience Limited for the pEAQ vector, Dr. Hsin Wang for assistance with the NMR experiments, and Dr. Sheng Zhang and Robert Sherwood (Institute of Biotechnology, Cornell University) for assistance with MALD-TOF MS. This work was supported by grants from the US National Science Foundation (Plant Genome Program; DBI-0606595), the United States–Israel Binational Agricultural Research and Development Fund (IS-4234-09); the US Department of Agriculture Cooperative State Research, Education and Extension Service (2011-04197); and the Danish Council for Strategic Research (10-093465). The 800 MHz NMR spectrometer is operated by the New York Structural Biology Center, a STAR center supported by the New York State Office of Science, Technology, and Academic Research. Additional infrastructural support was provided at The City College of New York by grants from the National Center for Research Resources (2G12RR03060) and the National Institute on Minority Health and Health Disparities (8G12MD007603) of the National Institutes of Health.
Abbreviations
- 2-MHG
2-mono-(10,16-dihydroxyhexadecanoyl)glycerol
- CD1
Cutin Deficient 1
- CUS
cutin synthase-like
- DHHA
10,16-dihydroxyhexadecanoic acid
- GDSL
GDSL-motif lipase/hydrolase
- MALDI-TOF MS
matrix assisted laser desorption ionization time-of-flight mass spectrometry
- NMR
nuclear magnetic resonance
Footnotes
Conflict Of Interest
The authors declare no conflict of interest
Contributor Information
Trevor H. Yeats, Email: tyeats@berkeley.edu.
Wenlin Huang, Email: whuang2@ccny.cuny.edu.
Subhasish Chatterjee, Email: subhas1012@gmail.com.
Hélène M-F. Viart, Email: hmvi@kemi.dtu.dk.
Mads H. Clausen, Email: mhc@kemi.dtu.dk.
Ruth E. Stark, Email: stark@sci.ccny.cuny.edu.
Jocelyn K.C. Rose, Email: jr286@cornell.edu.
References
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. GDSL family of serine esterases/lipases. Prog Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Bax A, Summers MF. H-1 and C-13 assignments from sensitivity-enhanced detection of heteronuclear multiple-bond connectivity by 2D multiple quantum NMR. J Am Chem, Soc. 1986;108:2093–2094. [Google Scholar]
- Buda GJ, Barnes WJ, Fich EA, Park S, Yeats TH, Zhao L, Domozych DS, Rose JKC. An ABCG transporter is required for cuticular wax deposition and desiccation tolerance in the moss Physcomitrella patens. Plant Cell. 2013 doi: 10.1105/tpc.113.117648. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr Opin Plant Biol. 2012;15:329–337. doi: 10.1016/j.pbi.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Deas AHB, Holloway PJ. The intermolecular structure of some plant cutins. In: Tevini M, Lichtenthaler HK, editors. Lipids and Lipid Polymers in Higher Plants. Berlin: Springer-Verlag; 1976. pp. 293–299. [Google Scholar]
- Felle HH. pH: signal and messenger in plant cells. Plant Biol. 2001;3:577–591. [Google Scholar]
- Girard AL, Mounet F, Lemaire-Chamley M, et al. Tomato GDSL1 is Required for Cutin Deposition in the Fruit Cuticle. Plant Cell. 2012;24:3119–3134. doi: 10.1105/tpc.112.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graca J, Lamosa P. Linear and branched poly(omega-hydroxyacid) esters in plant cutins. J Agric Food Chem. 2010;58:9666–9674. doi: 10.1021/jf1015297. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Morley KL, Schrag JD, Kazlauskas RJ. Different active-site loop orientation in serine hydrolases versus acyltransferases. Chembiochem. 2011;12:768–776. doi: 10.1002/cbic.201000693. [DOI] [PubMed] [Google Scholar]
- Kay L, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- Kolattukudy P. Lipid polymers and associated phenols, their chemistry, biosynthesis, and role in pathogenesis. In: Loewus FA, Runeckles VC, editors. Recent Advances in Phytochemistry. New York: Plenum; 1976. pp. 185–246. [Google Scholar]
- Kolattukudy PE. Polyesters in higher plants. Adv Biochem Eng Biotechnol. 2001;71:1–49. doi: 10.1007/3-540-40021-4_1. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA. 2009;106:22008–22013. doi: 10.1073/pnas.0909090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AG, Cavanagh J, Wright PE, Rance M. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J Magn Reson (1969) 1991;93:151–170. [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Ray AK, Chen ZJ, Stark RE. Chemical depolymerization studies of the molecular architecture of lime fruit cuticle. Phytochemistry. 1998;49:65–70. [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J. 2009;7:682–693. doi: 10.1111/j.1467-7652.2009.00434.x. [DOI] [PubMed] [Google Scholar]
- Schuchardt U, Sercheli R, Vargas RM. Transesterification of vegetable oils: a review. J Brazil Chem Soc. 1998;9:199–210. [Google Scholar]
- Shaka AJ, Lee CJ, Pines A. Iterative schemes for bilinear operators - application to spin decoupling. J Magn Reson. 1988;77:274–293. [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 2011;7:e1001388. doi: 10.1371/journal.pgen.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohalm M, Kavan D, Novak P, Volny M, Havlicek V. mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem. 2010;82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Shimada T, Kondo M, Tamai A, Mori M, Nishimura M, Hara-Nishimura I. Ectopic expression of an esterase, which is a candidate for the unidentified plant cutinase, causes cuticular defects in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:123–131. doi: 10.1093/pcp/pcp173. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S. PhD thesis. City University of New York; 2005. Molecular structures of natural polymers: Cutin, suberin, and melanin. [Google Scholar]
- Tian S, Fang X, Wang W, Yu B, Cheng X, Qiu F, Mort AJ, Stark RE. Isolation and identification of oligomers from partial degradation of lime fruit cutin. J Agric Food Chem. 2008;56:10318–10325. doi: 10.1021/jf801028g. [DOI] [PubMed] [Google Scholar]
- Wang W, Tian S, Stark RE. Isolation and identification of triglycerides and ester oligomers from partial degradation of potato suberin. J Agric Food Chem. 2010;58:1040–1045. doi: 10.1021/jf902854y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: Substrate specificity, sn-2 preference, and evolution. Plant Physiol. 2012;160:638–652. doi: 10.1104/pp.112.201996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Martin LB, Viart HM, et al. The identification of cutin synthase: formation of the plant polyester cutin. Nat Chem Biol. 2012;8:609–611. doi: 10.1038/nchembio.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JK. The formation and function of plant cuticles. Plant Physiol. 2013;163:5–20. doi: 10.1104/pp.113.222737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.