Abstract
Purpose of the review
Epigenetic mechanisms have the ability to alter the phenotype without changing the genetic code. The science of epigenetics has grown considerably in recent years, and future epigenetically-based treatments or prevention strategies are likely. Epigenetic associations with asthma have received growing interest because genetic and environmental factors have been unable to independently explain the etiology of asthma.
Recent Findings
Recent findings suggest that both the environment and underlying genetic sequence variation influence DNA methylation, which in turn seems to modify the risk conferred by genetic variants for various asthma phenotypes. In particular DNA methylation may act as an archive of a variety of early developmental exposures which then can modify the risk related to genetic variants.
Summary
Current asthma treatments may control the symptoms of asthma but do not modify its natural history. Epigenetic mechanisms and novel explanatory models provide burgeoning approaches to significantly increase our understanding of the initiation and progression of asthma. This will lead to critical information to prevent or treat asthma not only in the current generation, but due to the epigenetic inheritance may also prevent asthma in future generations.
Keywords: Asthma, Epigenetics, DNA methylation, methylation quantitative trait loci, modifiable genetic variants
INTRODUCTION
In the 1940s, Conrad Waddington used the term epigenetics to describe how the genotype manifests itself as a phenotype (1). In 1958, David Nanney borrowed the term to describe inherited phenomena that could not be explained by conventional genetics (2). Recently, epigenetics has been defined concisely by Mark Ptashne in 2007 by three criteria: (I) a change in the activity of a gene that does not involve a mutation, (II) that is initiated by a signal, and (III) that is inherited (mitotically or meiotically) in the absence of the signal that initiated the change (3). Classically, four epigenetic mechanisms have been identified: (a) DNA methylation, (b) Histone modification, (c) chromatin remodeling, and (d) small (21- to 26-nt) and non-coding RNAs.
There is ample evidence that DNA methylation fulfills all three criteria required to be considered as an epigenetic mechanism (4–6). Histone modifications fulfill some of the criteria for being epigenetic mechanisms in that they can result from exogenous signals such as cigarette smoke and that they alter gene activity (7–9). However, meiotic inheritance has only been demonstrated in C. elegans, a transparent nematode (10). DNA methylation usually works hand in hand with histone modifications to activate or silence genes by influencing chromatin structure and its accessibility by transcription factors (11). So it is possible that DNA methylation constitutes a mechanism of inheritance for some histone modifications. Given the complex and ever-changing structure of chromatin, there is little information on chromatin remodeling regarding initiation, alteration of gene activity, and inheritance (12–14). MicroRNAs (miRNAs) also have been shown to be caused by exogenous factors and to alter gene activity by either inhibiting translation or degrading messenger RNAs (mRNA) (15, 16). For instance, in humans, miRNAs have been demonstrated to be differentially expressed in current and never smokers and to be related to particulate matter exposure (7, 17). Currently there is little evidence that miRNAs can be inherited (18). However, since miRNAs are part of the genetic code, it is possible that DNA methylation affects the activity of miRNAs and thus facilitates inheritance. Hence in the following we concentrate on the truly and well-establish epigenetic mechanism, DNA methylation.
DNA methylation and asthma phenotypes
Asthma is the most common chronic disease among children and it has a complex etiology including genetic and environmental factors. Human studies have investigated the role of DNA methylation more often than other epigenetic marks due to practical and biological reasons (19). Table 1 gives a summary of recent population-based studies investigating the association between DNA methylation and asthma. Sood et al investigated the role of DNA methylation of 12 genes selected due to their involvement in oxidative stress pathways in sputum of 695 older adults (20). They found that the PCDH20 gene coding protocadherin-20, a protein involved in cell adhesion and signal transduction, was statistically higher methylated in sputum cells from asthma patients. In a subsample of 36 of 637 children, Isidoro-Garcia and coworkers studied methylation of the D prostanoid receptor (PTGDR) gene. Prostaglandin D2, a metabolite of arachidonic acid, inhibits apoptosis, prolonging eosinophilic survival, and biases the development of naïve T lymphocytes to T helper 2 cells. Isidoro-Garcia et al showed that genetic variants of the PTGDR gene altered adjacent DNA methylation levels, which was related to hypomethylation of the promoter of the PTGDR gene among asthmatic participants (21). In gene expression analyses, the authors were able to demonstrate that hypomethylation caused by underlying sequence variants in patients was associated with increased PTGDR expression (21). A limitation of these two studies is that DNA methylation may not constitute a risk for asthma but may reflect a response due to the disease (reverse causation).
Table 1.
Recent studies investigating the role of DNA methylation on asthma-related phenotypes
| Reference | Study location & sample size | Gene symbol(s) | Exposure | Outcome | Summary of findings |
|---|---|---|---|---|---|
| Sood et al. 2012 | North America (n = 695) | PCDH20, SULF2, PAX5α | Smoking | Asthma |
|
| Isidoro-Garcia et al. 2011 | Spain (36 of 637 children) | PTGDR | Genetic variation in PTGDR promoter region | Asthma |
|
| Fu et al. 2012 | North America (n = 182) | ADRB2 | Indoor NO2 | Asthma severity |
|
| Salam et al. 2012 | North America (n = 940) | NOS2 | PM2.5, NOS2 promoter haplotyp es | FeNO |
|
| Brenton et al. 2011 | North America (n = 940) | NOS1, NOS2A, NOS3, ARG1, ARG2, | Asthma was considered as an effect modifier | FeNO |
|
| Morales et al. 2012 | Spain (discovery cohort: n = 122; replication: n = 236) | ALOX12 | Prenatal DDE, Genetic variation in ALOX12 | Wheezing |
|
FeNO: fractional concentration of exhaled nitric oxide which can be used as an indicator for airway inflammation. PM2.5: particulate matter with an aerodynamic diameter of 2.5 μm or less. PCDH20: protocadherin 20; SULF2: sulfatase 2; PAX5α: paired box 5; PTGDR: prostaglandin D2 receptor; ADRB2: adrenoceptor beta 2; NOS2: nitric oxide synthase 2; NOS1: nitric oxide synthase 1; NOS2A: nitric oxide synthase 2a; NOS3: nitric oxide synthase 3; ARG1: arginase-1, ARG2: arginase-2; ALOX12: arachidonate 12-lipoxygenase.
Among 182 children with asthma, high methylation levels of adrenergic-receptor beta-2 (ADRB2) gene, an important regulator of airway smooth muscle tone, have been associated with severe childhood asthma (22). Taking environmental exposures into account, an increased risk of severe asthma was associated with the joint effect of indoor NO2 exposure and high levels of ADRB2 methylation, which suggests that DNA methylation can act as an effect modifier for the association between NO2 levels and asthma severity (22). An environmental study focused on the effects of particulate matter (PM) conducted among 940 southern California school children. Salam et al investigated the fraction of exhaled Nitric Oxide (FeNO) produced by the bronchial epithelium and the NOS2 gene that codes the nitric oxide synthase (23). The results demonstrated two-way interactions between ‘PM2.5 exposure × NOS2 genetic variants’ and ‘PM2.5 exposure × NOS2 methylation’ and a three-way interaction between ‘PM2.5 exposure × NOS2 genetic variants × CpG methylation levels’ that jointly influenced FeNO levels (23). In another investigation of this cohort, Breton et al reported associations between differential DNA methylation of arginase-1 (ARG1) and ARG2 and significantly higher levels of FeNO in children with asthma (24). The authors suggest that differential methylation of ARG genes may play a role in modifying FeNO production in individuals whose inflammatory and oxidative stress pathways are already upregulated.
Morales et al addressed a burgeoning question (25), namely whether DDE, a metabolite of the pesticide DDT, is related to the development of asthma (26, 27). Their results suggest that prenatal DDE exposure and genetic variants were associated with DNA hypomethylation of ALOX12 gene. In turn, this hypomethylation was a risk for persistent wheezing up to 6 years of age (25).
The interplay of genetic variants, environmental factors, and DNA methylation
The epidemiological investigations (Table 1) demonstrate that both environmental and genetic factors may influence DNA methylation levels and could act as effect modifiers for asthma-related phenotypes. Hence, environmental exposures and genetic factors are both essential elements that determine epigenetic state in asthma (28, 29). Multiple past and current exposures have been linked to levels of DNA methylation such as the Dutch famine (30, 31); low birth weight, and fetal alcohol syndrome (32, 33), maternal gestational stress in third trimester (34), gestational folate levels (35–38), early life socio-economic position (39), infections (40–43), and smoking (44–52). Similarly genetic variants have been shown to affect the susceptibility to DNA methylation, a process named allele-specific or genotype-dependent DNA methylation (53–59). Such genetic variants have recently been named methylation quantitative trait loci (methQTL) (60, 61).
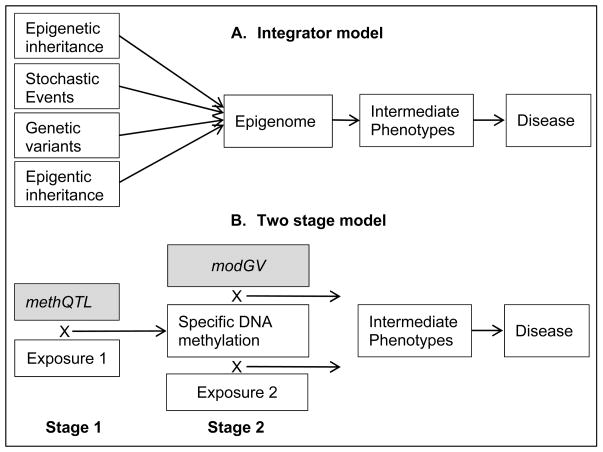
Hence, we do not only need to understand the mechanisms by which alterations in the epigenome alter phenotype but also to test different models of how genetic variants, environmental factors, and DNA methylation interplay in the etiology of asthma. A common idea is that the epigenome is an integrator of multiple signals in the pathway to diseases. Although different steps seem to be involved in structuring the DNA methylation profiles, the integrative role often remains a black box (Figure 1, Model A) (62, 63). Here, we propose a two-stage model (Figure 1, Model B), allowing that these stages develop in different life phases. In Stage 1, specific exposures and methQTLs interact within one gene and change the DNA methylation status of specific genetic elements (either promoter or intragenic). Once a methylation change close to a methQTL has been established, for instance at the promoter site, the gene may be differentially regulated. The response to additional exposures that interact with other genetic variants of the same gene depends on whether, e.g., the promoter is silenced or activated. To contrast these other genetic variants, whose response may be modified as a consequence of prior DNA methylation, from methQTLs, we call these modifiable genetic variants (modGV). The three-way interaction in the study by Salam et al in children in southern California showed that NOS2 genetic variants were modifiable (Table 1) (23). The study of asthma severity by Fu et al demonstrates the modifiable role of DNA methylation for the association of NO2 and asthma (22). Recently for eczema, Ziyab et al demonstrated that the haploinsufficiency of the filaggrin gene can be modified by DNA methylation within the intragenic region that worsens the insufficiency (64). Experimentally, in lymphoblastoid cell lines similar models have been identified by Berlivet et al for the asthma-associated locus 17q12-q21 (65).
Figure 1. Models of the interplay of exposures, genetic, and epigenetic elements in the disease etiology.
methQTL – methylation quantitative trait loci, genetic variants that change the susceptibility for DNA methylation
modGV – modifiable genetic variants, genetic variants that are modified by DNA methylation
Comparable to the studies described above that have focused on gene promoter methylation, these models also apply to intragenic methylation. DNA methylation is more frequent within gene-bodies (intragenic) than in promoters. Whereas hypermethylation of promoter sites has been associated with transcriptional silencing, intragenic methylation has been observed to be positively or bell-shaped correlated with gene expression (66). Recently, for the CD45 transcript, it has been demonstrated that intragenic DNA methylation is related to alternative pre-mRNA splicing (67). In particular, it has been suggested that a specific binding factor was involved in splicing regulation (67). We speculate that methQTLs and exposure may also affect intragenic DNA methylation (Stage 1) and then modify pre-mRNA splicing (Stage 2).
DNA methylation is likely to have contributed to discrepancies found among genome-wide association studies. Both methQTL and modGV are part of the set of genetic variants (e.g., SNP, haplotypes), that are the focus of genome-wide association studies. The detection of associations between such genetic variants and phenotypes may therefore depend on other modifiers of DNA methylation levels such as environmental exposure. For instance, a SNP may facilitate DNA methylation in an exposed study group but not in the unexposed group. Since Stage 1 changes may to some extent penetrate through Stage 2, a methQTL in the exposed group may be associated with increased risk of the disease. However, in another study group, a different genetic sequence (non-risk genotype) in the same methQTL may not be favoring DNA methylation, and thus not establishing a risk for a disease. Also modGV with the same genetic code may be masked by DNA methylation in one study group but unmasked in another study. Such settings lead to disagreements between genetic studies and reduce the chance to replicate candidate genes (68). Hence, a methQTL cannot be assessed without knowing the exposure and modGV cannot be assessed without taking the methylation of other SNPs/haplotypes into account that may influence gene regulation or splicing.
The role of different life phases
As exemplified by the study by Morales et al (25), it is important to consider the timing of exposure, of measurement of DNA methylation, and of phenotypic outcome assessment with reference to the life course. In the Morales et al study, firstly change in methylation related to prenatal DDE in DNA obtained at 4 years of age was assessed (replication study: cord blood), and secondly, the altered DNA methylation was linked to wheezing (25). This approach avoids the problem of reverse causation that can result if DNA methylation may either result as a response to the disease or may be considered as a risk factor. The concept of the “developmental programming” has been well accepted (69–71) and there is increasing awareness of its importance in asthma (72). Environmental pollutants may influence crucial cellular functions during critical periods of fetal development and permanently alter the structure or function of specific organ systems.
Some studies suggest that intrauterine and early life exposures to a farming environment are associated with decreased risk of allergic disorders, including asthma (73, 74). This protective effect is believed to be associated with epigenetic mechanisms that are induced during early developmental stages. Recently, Slaats et al demonstrated that profiles of promoter DNA methylation of CD14 gene measured in placentas were different among mothers living on a farm compared with mothers not living on a farm (75). However, this finding need to be replicated in a larger sample and the biological pathway underlying the protective effect needs further elucidation.
Another example of prenatal exposure is maternal smoking. Using the Norwegian Mother and Child Cohort Study (cord blood), Joubert et al. reported that DNA methylation in cord blood derived DNA of genes including the cytochrome P450 aryl-hydrocarbon-hydroxylase CYP1A1 gene and the aryl hydrocarbon receptor repressor gene (AHRR) are differentially methylated after gestational exposure to cigarette smoking (76). The CYP1A1 gene codes an enzyme that catalyzes the conversion of chemical species into reactive intermediates such as quinones; AHRR competes with AHR for binding at xenobiotic response elements and is related to active smoking. In addition, Karmaus et al. have demonstrated that these genes were also differentially methylated in individuals exposed to in utero cigarette smoke in blood DNA samples at age 18 years in the Isle of Wight Birth cohort (77). Given that early life DNA methylation leads to a cell memory (78, 79), children may be programmed to metabolize xenobiotics differently, which can increase their disease risk due to smoke exposure later in life. Hence, DNA methylation builds gene-activation memories during key periods of development (e.g., in utero and adolescence) producing aberrant activation patterns later in life which may elevate disease risk.
Conclusions
To date, only a few studies have reported associations between epigenetic marks and the diverse asthma-related phenotypes. Although most studies have focused on different candidate genes, they have shown similar models of interactions between genetic variants (methQTLs and modGV), environmental exposures, and DNA methylation. There is a need to improve our knowledge about the black box of epigenetics with regard to exposures and diseases. The biological mechanisms that lead to specific changes in gene regulation in response to specific exposures are not known. We need to determine whether epigenetics should be considered as a major integrator of multiple signals, or, alternatively, whether DNA methylation acts differently at various developmental stages conditional on genetic variants and exposures, such as in the proposed two-stage model. In addition, since there is a lack of critical knowledge on which genes are programmed or re-programmed at what time during gestation and in which developmental phase, birth cohort studies need to trace DNA methylation over time, and ideally over generations. This will provide critical information about which phases in the course of life are most suitable to prevent deviant DNA methylation (preventive epigenomics) or intervene to normalize DNA methylation to prevent disease (pharmaco-epigenomics) (80). Given that patterns of DNA methylation can be inherited through meiosis, future research will provide a unique chance not only to prevent and treat asthma in the current generation, but also prevent it in subsequent generations.
Key Points.
Of the potential mechanisms, only DNA methylation fulfills all three epigenetic criteria: (I) A change in the activity of a gene that does not involve a mutation. (II) It is initiated by a signal or exposure. (III) It is inherited (mitotically or meiotically) in the absence of the signal that initiated the change.
Various studies demonstrate for multiple genes that different asthma phenotypes are associated with DNA methylation; however, a clear time order of DNA methylation and asthma is not always established.
To distinguish whether DNA methylation precedes asthma and results from early exposures, birth cohort studies are needed to trace DNA methylation over time, and ideally over generations.
The effect of environmental exposure seems to be conditional on genetic variants (methylation quantitative trait loci); and the risk related to genetic variants is modified by adjacent DNA methylation (modifiable genetic variants).
Acknowledgments
Funding source:
Research reported in this review was supported by the National Institute of Allergy and Infectious Diseases under Award Number R01 AI091905-01 (PI: Wilfried Karmaus).
The Authors work was funded by a supported by the National Institute of Allergy and Infectious Diseases under award number R01 AI091905.
Footnotes
Conflicts of Interest:
The Authors have no conflicts of interest to declare in connection with this work.
References
- 1.Ledford H. Language: Disputed definitions. Nature. 2008;455(7216):1023–8. doi: 10.1038/4551023a. Epub 2008/10/25. [DOI] [PubMed] [Google Scholar]
- 2.Nanney DL. Epigenetic Control Systems. Proc Natl Acad Sci U S A. 1958;44(7):712–7. doi: 10.1073/pnas.44.7.712. Epub 1958/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R6. doi: 10.1016/j.cub.2007.02.030. This paper provides a concise definition of epigenetics. [DOI] [PubMed] [Google Scholar]
- 4.Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13(3):153–62. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- 5.Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011;3(3):267–77. doi: 10.2217/epi.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 7.Lovinsky-Desir S, Miller RL. Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr Allergy Asthma Rep. 2012;12(3):211–20. doi: 10.1007/s11882-012-0257-4. Epub 2012/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal histone methylation is responsible for increased vascular endothelial growth factor 165a secretion from airway smooth muscle cells in asthma. J Immunol. 2012;189(2):819–31. doi: 10.4049/jimmunol.1103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Royce SG, Karagiannis TC. Histone deacetylases and their role in asthma. J Asthma. 2012;49(2):121–8. doi: 10.3109/02770903.2011.648298. [DOI] [PubMed] [Google Scholar]
- 10.Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479(7373):365–71. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet. 2006;15(Spec No 1):R95–101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- 12.Travers AA, Vaillant C, Arneodo A, Muskhelishvili G. DNA structure, nucleosome placement and chromatin remodelling: a perspective. Biochem Soc Trans. 2012;40(2):335–40. doi: 10.1042/BST20110757. [DOI] [PubMed] [Google Scholar]
- 13.Berr A, Menard R, Heitz T, Shen WH. Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cellular microbiology. 2012;14(6):829–39. doi: 10.1111/j.1462-5822.2012.01785.x. [DOI] [PubMed] [Google Scholar]
- 14.Grigoryev SA, Woodcock CL. Chromatin organization - the 30 nm fiber. Experimental cell research. 2012;318(12):1448–55. doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Angulo M, Lecuona E, Sznajder JI. Role of MicroRNAs in Lung Disease. Arch Bronconeumol. 2012;48(9):325–30. doi: 10.1016/j.arbres.2012.04.011. Epub 2012/05/23. Rol de los microARN en las enfermedades pulmonares. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su WY, Xiong H, Fang JY. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem Biophys Res Commun. 2010;396(2):177–81. doi: 10.1016/j.bbrc.2010.04.147. [DOI] [PubMed] [Google Scholar]
- 17.Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med. 2011;183(10):1295–301. doi: 10.1164/rccm.201010-1579PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012 doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, et al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J. 2010;24(9):3135–44. doi: 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]
- 20.Sood A, Petersen H, Blanchette CM, Meek P, Picchi MA, Belinsky SA, et al. Methylated Genes in Sputum among Older Smokers with Asthma. Chest. 2012 doi: 10.1378/chest.11-2519. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isidoro-Garcia M, Sanz C, Garcia-Solaesa V, Pascual M, Pescador DB, Lorente F, et al. PTGDR gene in asthma: a functional, genetic, and epigenetic study. Allergy. 2011;66(12):1553–62. doi: 10.1111/j.1398-9995.2011.02685.x. [DOI] [PubMed] [Google Scholar]
- 22.Fu A, Leaderer BP, Gent JF, Leaderer D, Zhu Y. An environmental epigenetic study of ADRB2 5′-UTR methylation and childhood asthma severity. Clinical and experimental allergy: journal of the British Society for Allergy and Clinical Immunology. 2012 doi: 10.1111/j.1365-2222.2012.04055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, et al. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2012;129(1):232–9. e1–7. doi: 10.1016/j.jaci.2011.09.037. This is the first paper that investigates the inteaction between genetic variants, exposures, and DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. American journal of respiratory and critical care medicine. 2011;184(2):191–7. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Morales E, Bustamante M, Vilahur N, Escaramis G, Montfort M, de Cid R, et al. DNA hypomethylation at ALOX12 is associated with persistent wheezing in childhood. American journal of respiratory and critical care medicine. 2012;185(9):937–43. doi: 10.1164/rccm.201105-0870OC. This is paper provides an excellent example how to analyze DNA methylation, addressing two steps: (1) from exposure to DNA methylation and (2) from DNA methylation to the phenotype. [DOI] [PubMed] [Google Scholar]
- 26.Karmaus W. Infections and atopic disorders in childhood and organochlorine exposure. Arch Environ Health. 2001;56(6):485–92. doi: 10.1080/00039890109602896. [DOI] [PubMed] [Google Scholar]
- 27.Sunyer J, Torrent M, Munoz-Ortiz L, Ribas-Fito N, Carrizo D, Grimalt J, et al. Prenatal dichlorodiphenyldichloroethylene (DDE) and asthma in children. Environ Health Perspect. 2005;113(12):1787–90. doi: 10.1289/ehp.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppelman GH, Nawijn MC. Recent advances in the epigenetics and genomics of asthma. Current opinion in allergy and clinical immunology. 2011;11(5):414–9. doi: 10.1097/ACI.0b013e32834a9573. [DOI] [PubMed] [Google Scholar]
- 29.Blumenthal MN. Genetic, epigenetic, and environmental factors in asthma and allergy. Annals of allergy, asthma & immunology: official publication of the American College of Allergy, Asthma, & Immunology. 2012;108(2):69–73. doi: 10.1016/j.anai.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105(44):17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18(21):4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobi EW, Heijmans BT, Kremer D, Putter H, Delemarre-van de Waal HA, Finken MJ, et al. DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics. 2011;6(2):171–6. doi: 10.4161/epi.6.2.13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biol Reprod. 2009;81(4):607–17. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- 34.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 35.Boeke CE, Baccarelli A, Kleinman KP, Burris HH, Litonjua AA, Rifas-Shiman SL, et al. Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood: Prospective results from a folate-replete population. Epigenetics. 2012;7(3):253–60. doi: 10.4161/epi.7.3.19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominguez-Salas P, Cox SE, Prentice AM, Hennig BJ, Moore SE. Maternal nutritional status, C(1) metabolism and offspring DNA methylation: a review of current evidence in human subjects. Proc Nutr Soc. 2012;71(1):154–65. doi: 10.1017/S0029665111003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoyo C, Murtha AP, Schildkraut JM, Jirtle RL, Demark-Wahnefried W, Forman MR, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6(7):928–36. doi: 10.4161/epi.6.7.16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowdhury S, Cleves MA, MacLeod SL, James SJ, Zhao W, Hobbs CA. Maternal DNA hypomethylation and congenital heart defects. Birth Defects Res A Clin Mol Teratol. 2011;91(2):69–76. doi: 10.1002/bdra.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borghol N, Suderman M, McArdle W, Racine A, Hallett M, Pembrey M, et al. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41(1):62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12(3 Pt 1):989–95. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 41.Yanagawa N, Osakabe M, Hayashi M, Tamura G, Motoyama T. Detection of HPV-DNA, p53 alterations, and methylation in penile squamous cell carcinoma in Japanese men. Pathology international. 2008;58(8):477–82. doi: 10.1111/j.1440-1827.2008.02259.x. [DOI] [PubMed] [Google Scholar]
- 42.Bonvicini F, Manaresi E, Di Furio F, De Falco L, Gallinella G. Parvovirus b19 DNA CpG dinucleotide methylation and epigenetic regulation of viral expression. PLoS One. 2012;7(3):e33316. doi: 10.1371/journal.pone.0033316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70(4):1430–40. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 44.Ally MS, Al-Ghnaniem R, Pufulete M. The relationship between gene-specific DNA methylation in leukocytes and normal colorectal mucosa in subjects with and without colorectal tumors. Cancer Epidemiol Biomarkers Prev. 2009;18(3):922–8. doi: 10.1158/1055-9965.EPI-08-0703. [DOI] [PubMed] [Google Scholar]
- 45.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88(4):450–7. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ehrlich S, Walton E, Roffman JL, Weiss D, Puls I, Doehler N, et al. Smoking, But Not Malnutrition, Influences Promoter-Specific DNA Methylation of the Proopiomelanocortin Gene in Patients With and Without Anorexia Nervosa. Canadian journal of psychiatry Revue canadienne de psychiatrie. 2012;57(3):168–76. doi: 10.1177/070674371205700306. [DOI] [PubMed] [Google Scholar]
- 48.Flom JD, Ferris JS, Liao Y, Tehranifar P, Richards CB, Cho YH, et al. Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2518–23. doi: 10.1158/1055-9965.EPI-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suter M, Ma J, Harris AS, Patterson L, Brown KA, Shope C, et al. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6(11) doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philibert RA, Beach SR, Gunter TD, Brody GH, Madan A, Gerrard M. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):619–28. doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17(9):2306–10. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, Kornhuber J, et al. Global DNA methylation is influenced by smoking behaviour. European neuropsychopharmacology: The Journal of the European College of Neuropsychopharmacology. 2008;18(4):295–8. doi: 10.1016/j.euroneuro.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011;7(8):e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergman Y, Cedar H. Epigenetic control of recombination in the immune system. Semin Immunol. 2010;22(6):323–9. doi: 10.1016/j.smim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bosviel R, Garcia S, Lavediaux G, Michard E, Dravers M, Kwiatkowski F, et al. BRCA1 promoter methylation in peripheral blood DNA was identified in sporadic breast cancer and controls. Cancer Epidemiol. 2012 doi: 10.1016/j.canep.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Naghibalhossaini F, Zamani M, Mokarram P, Khalili I, Rasti M, Mostafavi-Pour Z. Epigenetic and genetic analysis of WNT signaling pathway in sporadic colorectal cancer patients from Iran. Molecular biology reports. 2012;39(5):6171–8. doi: 10.1007/s11033-011-1434-6. [DOI] [PubMed] [Google Scholar]
- 57.Tycko B. Allele-specific DNA methylation: beyond imprinting. Hum Mol Genet. 2010;19(R2):R210–20. doi: 10.1093/hmg/ddq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bock C, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet. 2006;2(3):e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20(7):883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, et al. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86(3):411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–41. doi: 10.1038/nrg3000. This paper provides an excellent overview of epigenetic association studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Relton CL, Davey Smith G. Is epidemiology ready for epigenetics? Int J Epidemiol. 2012;41(1):5–9. doi: 10.1093/ije/dys006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–8. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 64*.Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. Journal of the European Academy of Dermatology and Venereology. 2012 doi: 10.1111/jdv.12000. This is the first epidemiologic paper that demonstrates the genetic variants (haploinsufficiency) can be modified by DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, Al Tuwaijri A, et al. Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines. Hum Genet. 2012;131(7):1161–71. doi: 10.1007/s00439-012-1142-x. Experimentally, this paper examines different models of the interaction of genetic and epigenetic variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. On the presence and role of human gene-body DNA methylation. Oncotarget. 2012;3(4):462–74. doi: 10.18632/oncotarget.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Genuneit J, Cantelmo JL, Weinmayr G, Wong GW, Cooper PJ, Riikjarv MA, et al. A multi-centre study of candidate genes for wheeze and allergy: the International Study of Asthma and Allergies in Childhood Phase 2. Clin Exp Allergy. 2009;39(12):1875–88. doi: 10.1111/j.1365-2222.2009.03364.x. [DOI] [PubMed] [Google Scholar]
- 69.Chavatte-Palmer P, Tarrade A, Levy R. Developmental origins of health and disease in adults: Role of maternal environment. Gynecologie, obstetrique & fertilite. 2012;40(9):517–9. doi: 10.1016/j.gyobfe.2012.07.010. Origines developpementales de la sante et des maladies de l’adulte: role de l’environnement maternel. [DOI] [PubMed] [Google Scholar]
- 70.Gluckman PD, Hanson MA, Bateson P, Beedle AS, Law CM, Bhutta ZA, et al. Towards a new developmental synthesis: adaptive developmental plasticity and human disease. Lancet. 2009;373(9675):1654–7. doi: 10.1016/S0140-6736(09)60234-8. [DOI] [PubMed] [Google Scholar]
- 71.Hanson M, Gluckman P. Developmental origins of noncommunicable disease: population and public health implications. Am J Clin Nutr. 2011;94(6 Suppl):1754S–8S. doi: 10.3945/ajcn.110.001206. [DOI] [PubMed] [Google Scholar]
- 72.Krauss-Etschmann S, Bush A, Bellusci S, Brusselle GG, Erik KDS, Dehmel S, et al. Of flies, mice and men: a systematic approach to understanding the early life origins of chronic lung disease. Thorax. 2012 doi: 10.1136/thoraxjnl-2012-201902. [DOI] [PubMed] [Google Scholar]
- 73.Lampi J, Canoy D, Jarvis D, Hartikainen AL, Keski-Nisula L, Jarvelin MR, et al. Farming environment and prevalence of atopy at age 31: prospective birth cohort study in Finland. Clin Exp Allergy. 2011;41(7):987–93. doi: 10.1111/j.1365-2222.2011.03777.x. [DOI] [PubMed] [Google Scholar]
- 74.Ege MJ, Herzum I, Buchele G, Krauss-Etschmann S, Lauener RP, Roponen M, et al. Prenatal exposure to a farm environment modifies atopic sensitization at birth. J Allergy Clin Immunol. 2008;122(2):407–12. 12 e1–4. doi: 10.1016/j.jaci.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 75.Slaats GG, Reinius LE, Alm J, Kere J, Scheynius A, Joerink M. DNA methylation levels within the CD14 promoter region are lower in placentas of mothers living on a farm. Allergy. 2012 doi: 10.1111/j.1398-9995.2012.02831.x. [DOI] [PubMed] [Google Scholar]
- 76**.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 450K Epigenome-Wide Scan Identifies Differential DNA Methylation in Newborns Related to Maternal Smoking During Pregnancy. Environ Health Perspect. 2012 doi: 10.1289/ehp.1205412. This is the first paper that analyzes the effects of maternal smoking during pregnancy on DNA methylation of specific genes using a epigenome-wide approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karmaus WZ, HJWH, Tong X, Kurukulaaratchy R, Arshad SH. Maternal smoking during pregnancy is associated with DNA methylation in female offspring at age 18 –results of an epigenome-wide scan. Environmental Health Perspectives. 2012 submitted. [Google Scholar]
- 78.Kabesch M, Adcock IM. Epigenetics in asthma and COPD. Biochimie. 2012 doi: 10.1016/j.biochi.2012.07.017. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 79.Tammen SA, Friso S, Choi SW. Epigenetics: The link between nature and nurture. Molecular aspects of medicine. 2012 doi: 10.1016/j.mam.2012.07.018. Epub 2012/08/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ingelman-Sundberg M, Gomez A. The past, present and future of pharmacoepigenomics. Pharmacogenomics. 2010;11(5):625–7. doi: 10.2217/pgs.10.59. [DOI] [PubMed] [Google Scholar]