Abstract
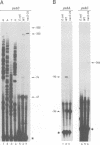
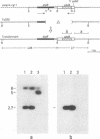
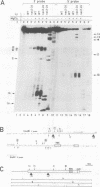
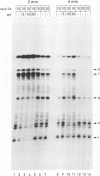
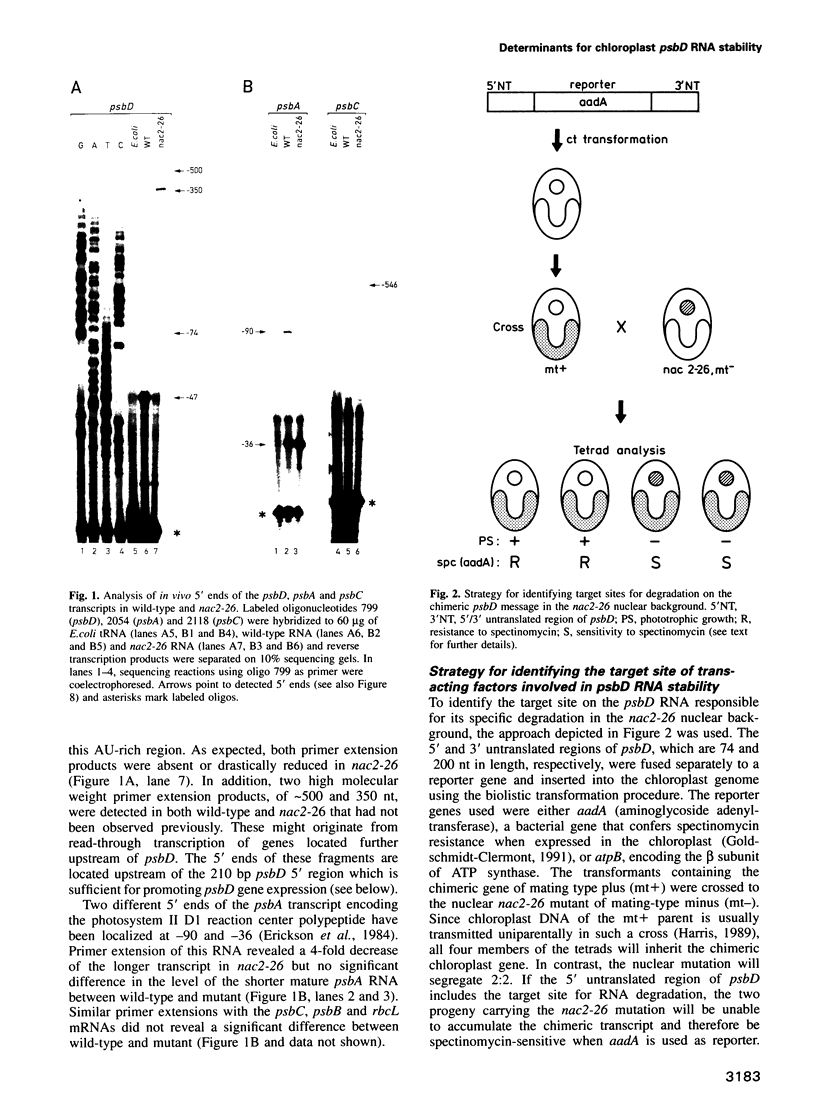
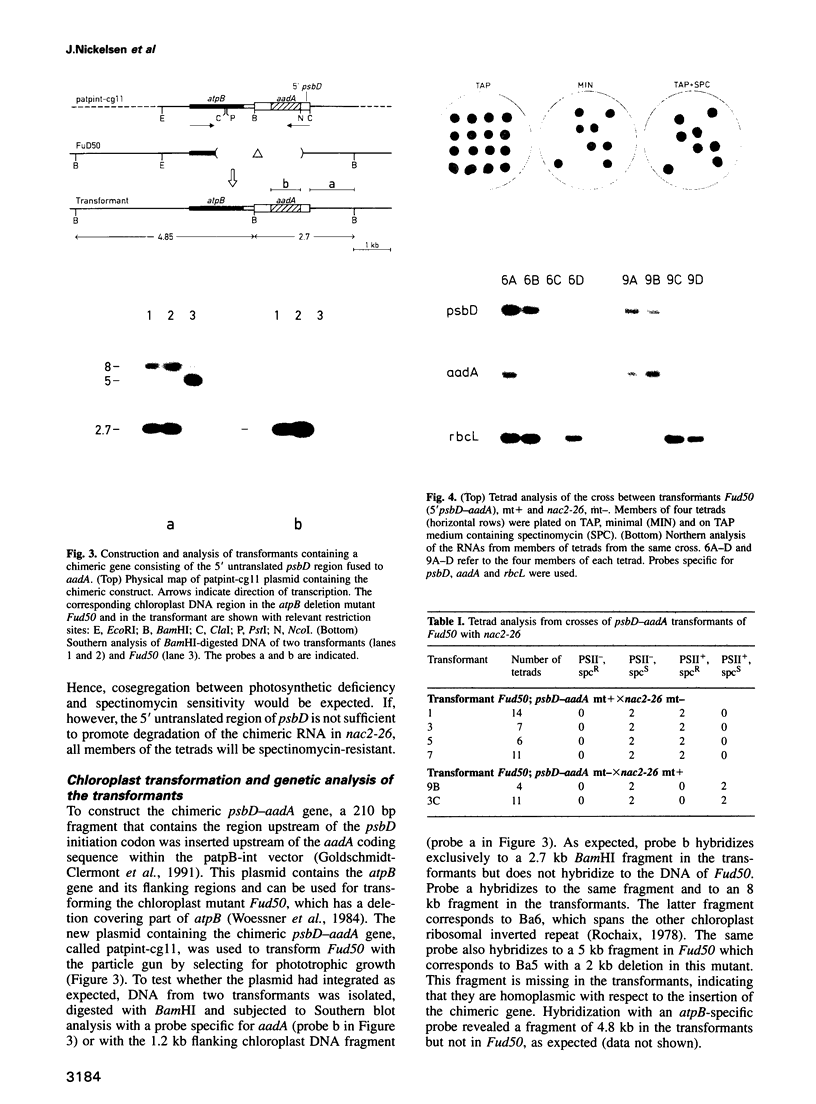
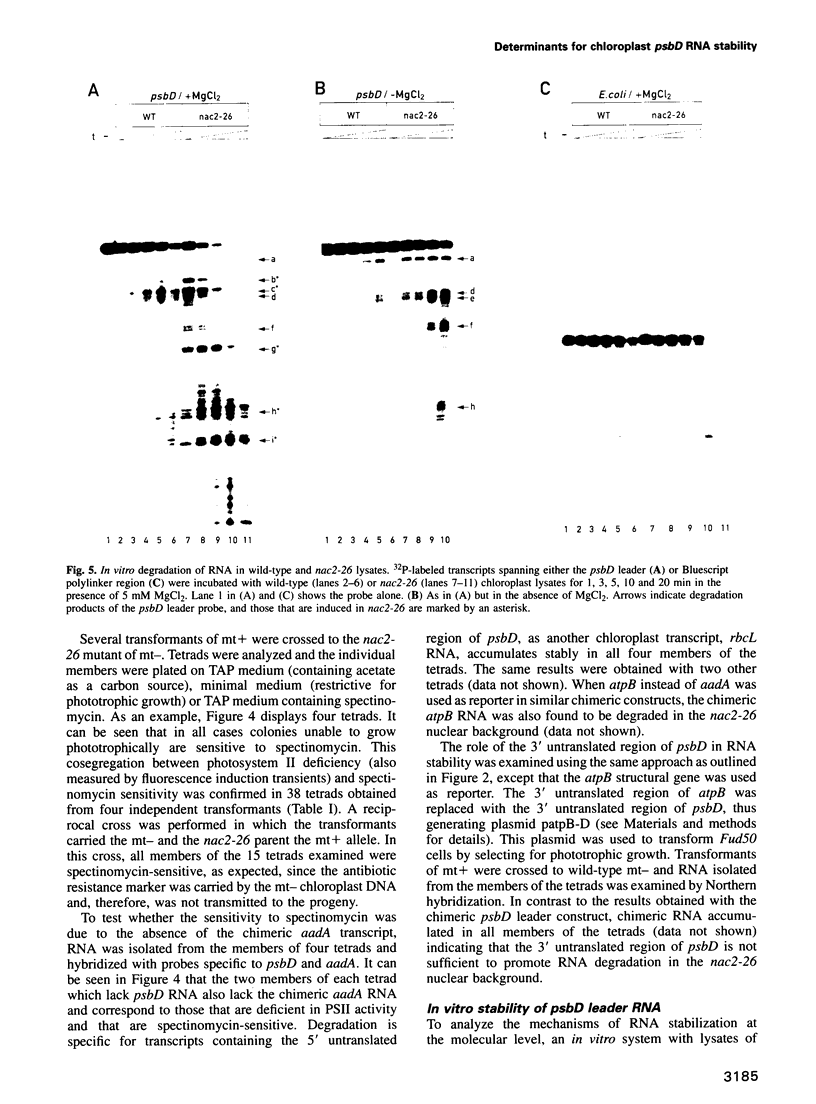
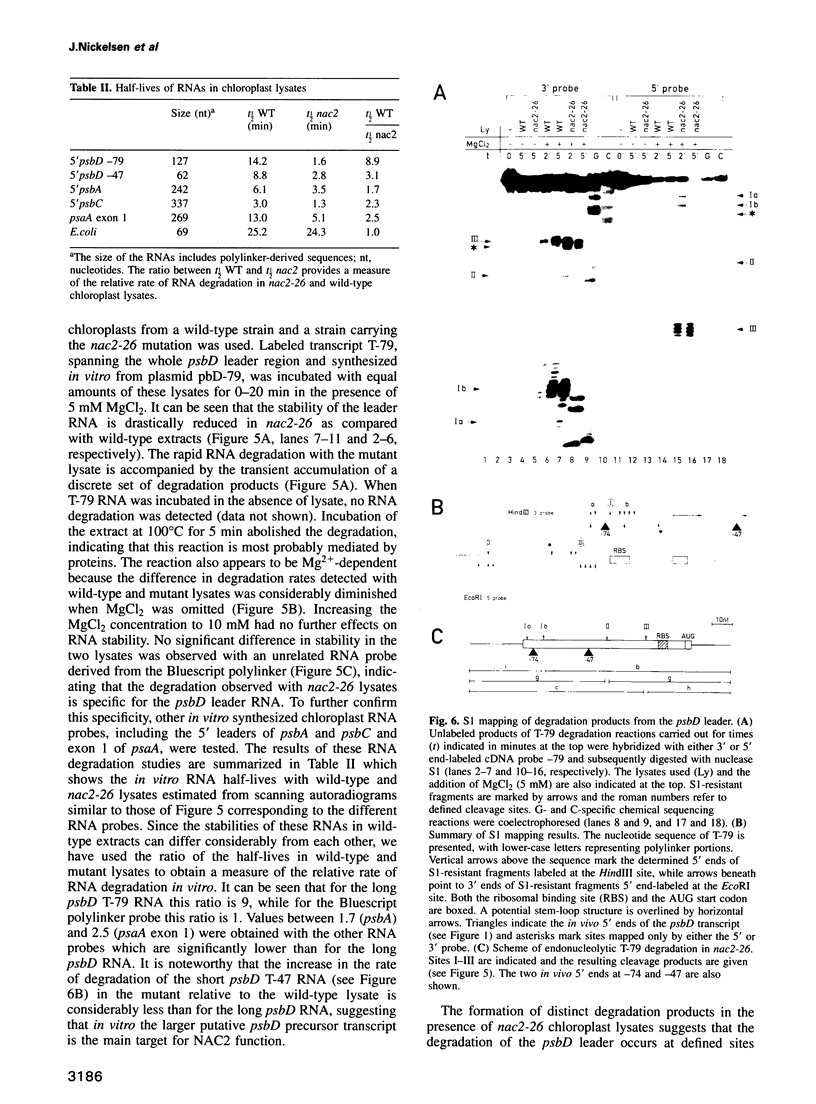
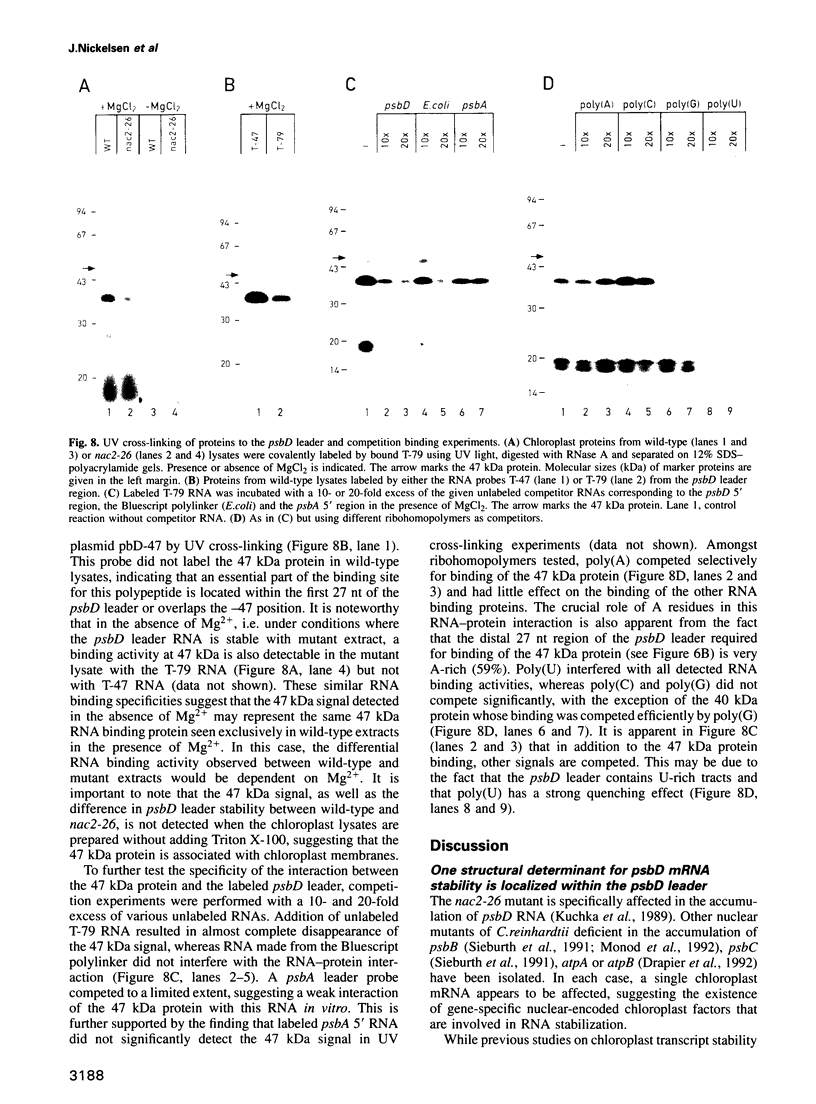
Stability of the chloroplast psbD mRNA encoding the D2 protein of the photosystem II reaction center is drastically decreased in the nuclear photosynthetic mutant nac2-26 of Chlamydomonas reinhardtii. Using biolistic transformation and genetic crosses we have introduced chimeric genes consisting of the psbD leader fused to a reporter gene into the chloroplast in both wild-type and mutant nuclear backgrounds. The chimeric message is destabilized in the latter, but not in the former case, indicating that the 74 nt psbD leader includes one of the target sites for psbD RNA degradation in the absence of wild-type NAC2 function. Increased instability of the psbD leader in mutant versus wild-type chloroplast lysates is also demonstrated in vitro and the primary cleavage sites have been mapped. The instability of the psbD RNA in the mutant correlates with the loss of binding of a 47 kDa protein to the psbD leader RNA, suggesting that this factor acts as message stabilizer in wild-type.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Stern D. B. Control of mRNA stability in chloroplasts by 3' inverted repeats: effects of stem and loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990 Oct 25;18(20):6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Blowers A. D., Klein U., Ellmore G. S., Bogorad L. Functional in vivo analyses of the 3' flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol Gen Genet. 1993 Apr;238(3):339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Danon A., Mayfield S. P. Light regulated translational activators: identification of chloroplast gene specific mRNA binding proteins. EMBO J. 1991 Dec;10(13):3993–4001. doi: 10.1002/j.1460-2075.1991.tb04974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Dieckmann C. L., Mittelmeier T. M. Nuclearly-encoded CBP1 interacts with the 5' end of mitochondrial cytochrome b pre-mRNA. Curr Genet. 1987;12(6):391–397. doi: 10.1007/BF00434815. [DOI] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Wollman F. A. Evidence for Nuclear Control of the Expression of the atpA and atpB Chloroplast Genes in Chlamydomonas. Plant Cell. 1992 Mar;4(3):283–295. doi: 10.1105/tpc.4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Erickson J. M., Rahire M., Malnoë P., Girard-Bascou J., Pierre Y., Bennoun P., Rochaix J. D. Lack of the D2 protein in a Chlamydomonas reinhardtii psbD mutant affects photosystem II stability and D1 expression. EMBO J. 1986 Aug;5(8):1745–1754. doi: 10.1002/j.1460-2075.1986.tb04422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J. M., Rahire M., Rochaix J. D. Chlamydomonas reinhardii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 1984 Dec 1;3(12):2753–2762. doi: 10.1002/j.1460-2075.1984.tb02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M., Choquet Y., Girard-Bascou J., Michel F., Schirmer-Rahire M., Rochaix J. D. A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell. 1991 Apr 5;65(1):135–143. doi: 10.1016/0092-8674(91)90415-u. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M., Girard-Bascou J., Choquet Y., Rochaix J. D. Trans-splicing mutants of Chlamydomonas reinhardtii. Mol Gen Genet. 1990 Sep;223(3):417–425. doi: 10.1007/BF00264448. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991 Aug 11;19(15):4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski K., Roch J. M., Prentki P., Krisch H. M. The stability of bacteriophage T4 gene 32 mRNA: a 5' leader sequence that can stabilize mRNA transcripts. Cell. 1985 Dec;43(2 Pt 1):461–469. doi: 10.1016/0092-8674(85)90176-x. [DOI] [PubMed] [Google Scholar]
- Gruissem W. Chloroplast gene expression: how plants turn their plastids on. Cell. 1989 Jan 27;56(2):161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- Kuchka M. R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J. D. Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell. 1989 Sep 8;58(5):869–876. doi: 10.1016/0092-8674(89)90939-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li Y. Q., Sugiura M. Nucleic acid-binding specificities of tobacco chloroplast ribonucleoproteins. Nucleic Acids Res. 1991 Jun 11;19(11):2893–2896. doi: 10.1093/nar/19.11.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Q., Sugiura M. Three distinct ribonucleoproteins from tobacco chloroplasts: each contains a unique amino terminal acidic domain and two ribonucleoprotein consensus motifs. EMBO J. 1990 Oct;9(10):3059–3066. doi: 10.1002/j.1460-2075.1990.tb07502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis U., Körte A., Rödel G. Association of cytochrome b translational activator proteins with the mitochondrial membrane: implications for cytochrome b expression in yeast. Mol Gen Genet. 1991 Nov;230(1-2):177–185. doi: 10.1007/BF00290666. [DOI] [PubMed] [Google Scholar]
- Mieszczak M., Klahre U., Levy J. H., Goodall G. J., Filipowicz W. Multiple plant RNA binding proteins identified by PCR: expression of cDNAs encoding RNA binding proteins targeted to chloroplasts in Nicotiana plumbaginifolia. Mol Gen Genet. 1992 Sep;234(3):390–400. doi: 10.1007/BF00538698. [DOI] [PubMed] [Google Scholar]
- Monod C., Goldschmidt-Clermont M., Rochaix J. D. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol Gen Genet. 1992 Feb;231(3):449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Link G. Interaction of a 3' RNA region of the mustard trnK gene with chloroplast proteins. Nucleic Acids Res. 1989 Dec 11;17(23):9637–9648. doi: 10.1093/nar/17.23.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J., Link G. RNA-protein interactions at transcript 3' ends and evidence for trnK-psbA cotranscription in mustard chloroplasts. Mol Gen Genet. 1991 Aug;228(1-2):89–96. doi: 10.1007/BF00282452. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Link G. The 54 kDa RNA-binding protein from mustard chloroplasts mediates endonucleolytic transcript 3' end formation in vitro. Plant J. 1993 Apr;3(4):537–544. doi: 10.1046/j.1365-313x.1993.03040537.x. [DOI] [PubMed] [Google Scholar]
- Petersen C. Control of functional mRNA stability in bacteria: multiple mechanisms of nucleolytic and non-nucleolytic inactivation. Mol Microbiol. 1992 Feb;6(3):277–282. doi: 10.1111/j.1365-2958.1992.tb01469.x. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D. Post-transcriptional steps in the expression of chloroplast genes. Annu Rev Cell Biol. 1992;8:1–28. doi: 10.1146/annurev.cb.08.110192.000245. [DOI] [PubMed] [Google Scholar]
- Rochaix J. D. Restriction endonuclease map of the chloroplast DNA of Chlamydomonas reinhardii. J Mol Biol. 1978 Dec 25;126(4):597–617. doi: 10.1016/0022-2836(78)90011-6. [DOI] [PubMed] [Google Scholar]
- Salvador M. L., Klein U., Bogorad L. 5' sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast gene transcripts. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1556–1560. doi: 10.1073/pnas.90.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. Chloroplast mRNA 3' end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991 Jun;10(6):1493–1502. doi: 10.1002/j.1460-2075.1991.tb07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L. E., Berry-Lowe S., Schmidt G. W. Chloroplast RNA Stability in Chlamydomonas: Rapid Degradation of psbB and psbC Transcripts in Two Nuclear Mutants. Plant Cell. 1991 Feb;3(2):175–189. doi: 10.1105/tpc.3.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Jones H., Gruissem W. Function of plastid mRNA 3' inverted repeats. RNA stabilization and gene-specific protein binding. J Biol Chem. 1989 Nov 5;264(31):18742–18750. [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]