Abstract
During Tn10 transposition, the transposon is fully excised from the donor site by double strand cleavages at the two ends of the element prior to integration at a new target site. Results presented here demonstrate that an interaction between the two transposon ends is required for double strand cleavage at either end. Furthermore, despite this essential interaction of ends, subsequent cleavages at the two ends can occur at observably distinct times prior to occurrence of strand transfer at either end. Moreover, the time between cleavages at the two ends is exaggerated by the presence of an appropriate mutation at one end of the element. Biological rationales for this constellation of mechanistic features are suggested. Additional results demonstrate that mutations at the three terminal basepairs of Tn10 confer defects subsequent to interaction of ends, in confirmation of inferences from genetic analysis. More specifically, mutations in bp 1-3 confer strong defects during conversion of the full excision intermediate to a complete strand transfer product; mutations in bp 1 and 2 also confer more subtle defects subsequent to interaction of ends but prior to full excision. Such defects might reflect roles for these basepairs in the chemical steps of transposition per se, the positioning of terminal residues for those chemical steps, and/or the coupling of cleavage(s) to subsequent conformational changes.
Full text
PDF

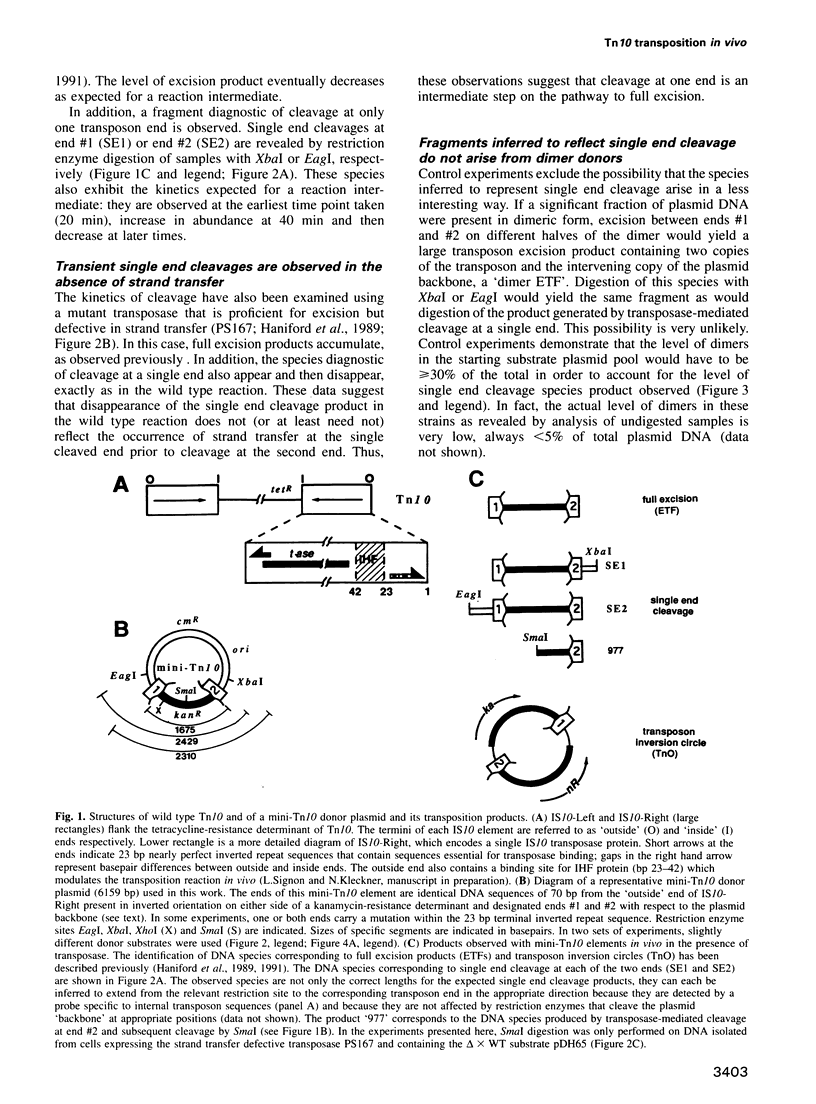
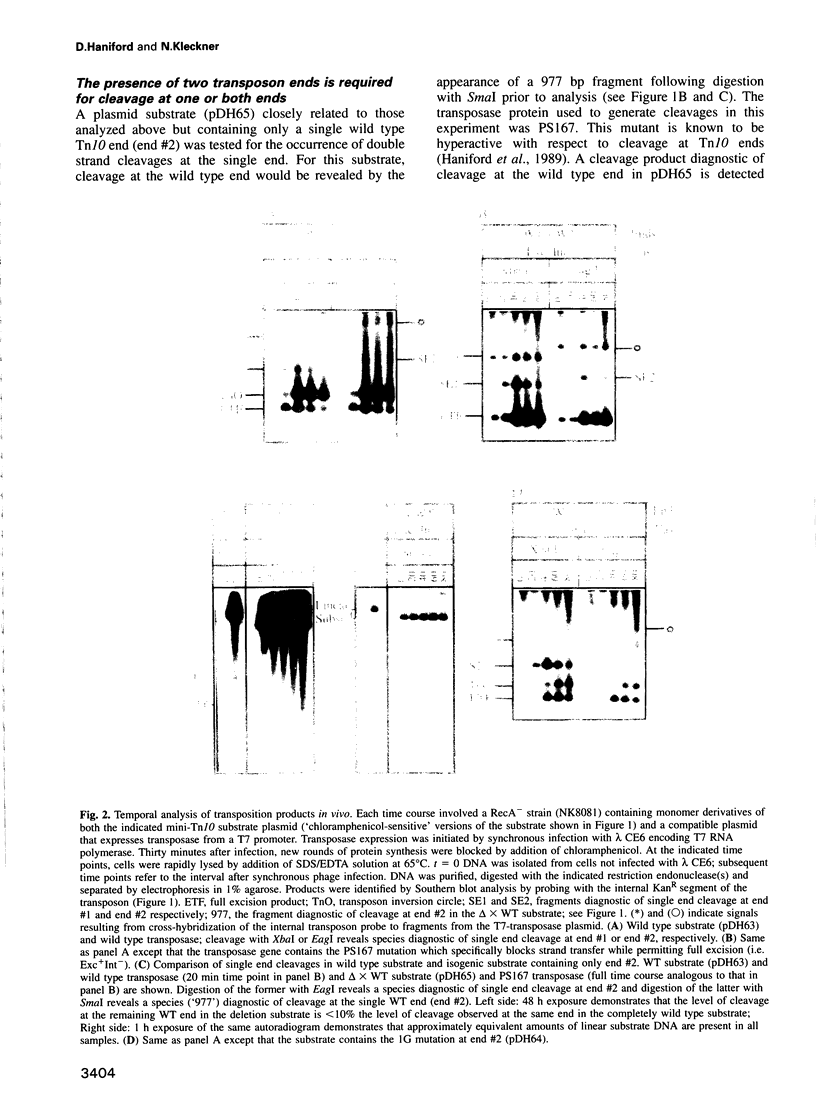
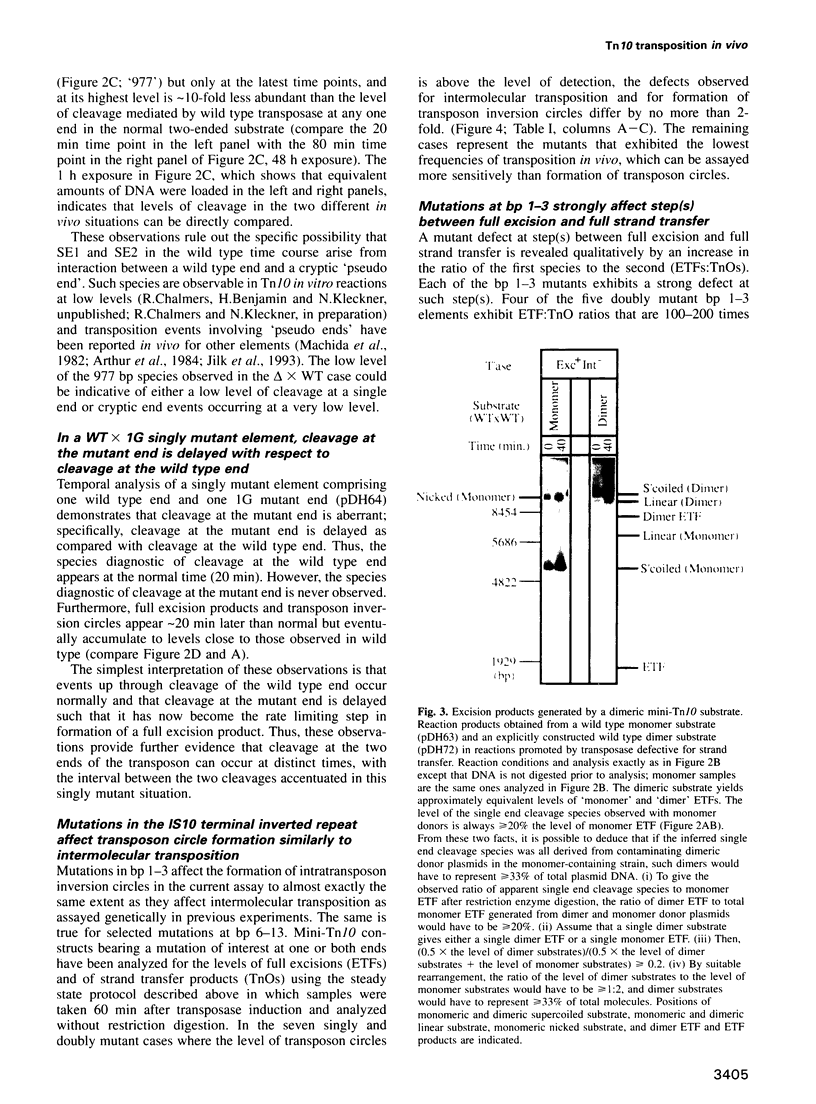
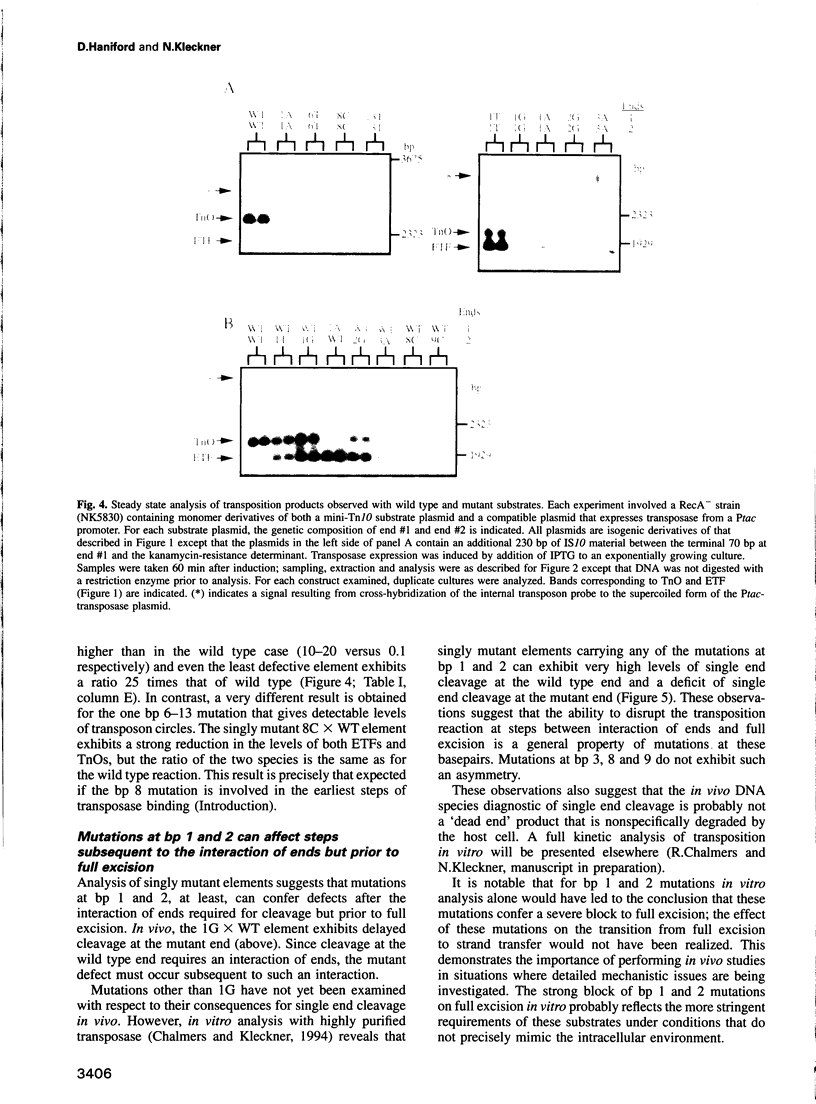
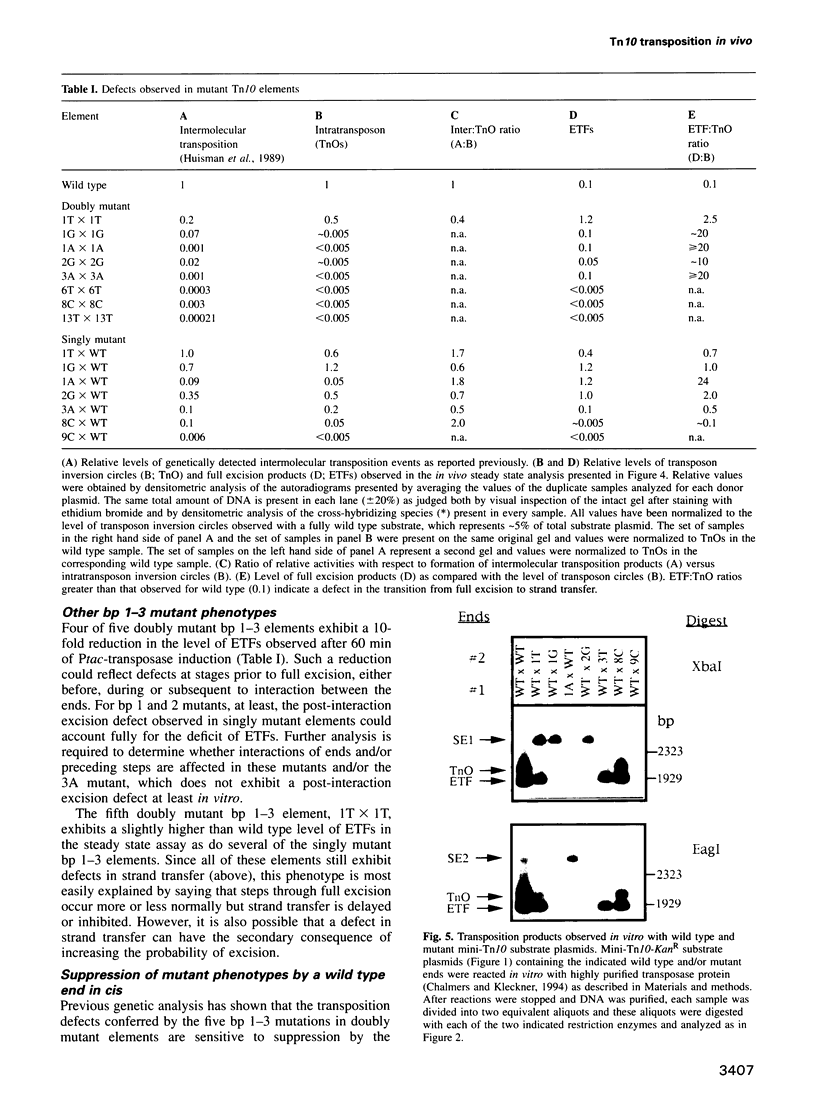




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arciszewska L. K., Craig N. L. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res. 1991 Sep 25;19(18):5021–5029. doi: 10.1093/nar/19.18.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur A., Nimmo E., Hettle S., Sherratt D. Transposition and transposition immunity of transposon Tn3 derivatives having different ends. EMBO J. 1984 Aug;3(8):1723–1729. doi: 10.1002/j.1460-2075.1984.tb02038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton R. J., Kubo K. M., Feng J. N., Craig N. L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993 Mar 26;72(6):931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- Bainton R., Gamas P., Craig N. L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991 May 31;65(5):805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- Bender J., Kleckner N. Genetic evidence that Tn10 transposes by a nonreplicative mechanism. Cell. 1986 Jun 20;45(6):801–815. doi: 10.1016/0092-8674(86)90555-6. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Kleckner N. Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4648–4652. doi: 10.1073/pnas.89.10.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H. W., Kleckner N. Intramolecular transposition by Tn10. Cell. 1989 Oct 20;59(2):373–383. doi: 10.1016/0092-8674(89)90298-5. [DOI] [PubMed] [Google Scholar]
- Bushman F. D., Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Craigie R. Integration of human immunodeficiency virus DNA: adduct interference analysis of required DNA sites. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3458–3462. doi: 10.1073/pnas.89.8.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L., Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990 Sep 7;62(5):967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- Chalmers R. M., Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J Biol Chem. 1994 Mar 18;269(11):8029–8035. [PubMed] [Google Scholar]
- Craigie R., Fujiwara T., Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990 Aug 24;62(4):829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987 Nov 6;51(3):493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi M., Mizuuchi K. Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell. 1984 Dec;39(2 Pt 1):387–394. doi: 10.1016/0092-8674(84)90017-5. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Binding of the IS903 transposase to its inverted repeat in vitro. EMBO J. 1992 Sep;11(9):3449–3455. doi: 10.1002/j.1460-2075.1992.tb05424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Hwang L., Grindley N. D. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V., Abrams H., Roe T., Lifson J., Brown P. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990 Jun;64(6):2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991 Dec 20;67(6):1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Farnet C. M., Haseltine W. A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl A., Lütticke S., Saedler H. TnpA product encoded by the transposable element En-1 of Zea mays is a DNA binding protein. EMBO J. 1988 Dec 20;7(13):4045–4053. doi: 10.1002/j.1460-2075.1988.tb03298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford D. B., Benjamin H. W., Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991 Jan 11;64(1):171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Chelouche A. R., Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989 Oct 20;59(2):385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa H., Ikeda K., Wishart W. L., Ohtsubo E. Specific binding of transposase to terminal inverted repeats of transposable element Tn3. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8220–8224. doi: 10.1073/pnas.84.23.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilk R. A., Makris J. C., Borchardt L., Reznikoff W. S. Implications of Tn5-associated adjacent deletions. J Bacteriol. 1993 Mar;175(5):1264–1271. doi: 10.1128/jb.175.5.1264-1271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990 Oct 5;63(1):87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. A novel type of transposon generated by insertion element IS102 present in a pSC101 derivative. Cell. 1982 Aug;30(1):29–36. doi: 10.1016/0092-8674(82)90008-3. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991 Jul 12;66(1):129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker T. A., Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992 Jul 24;70(2):303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- Morisato D., Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987 Oct 9;51(1):101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- Murphy J. E., Goff S. P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992 Aug;66(8):5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonti R. V., Keating D. H., Roth J. R. Lethal transposition of Mud phages in Rec- strains of Salmonella typhimurium. Genetics. 1993 Jan;133(1):17–28. doi: 10.1093/genetics/133.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Harkness T., Chaconas G. Stimulation of the Mu A protein-mediated strand cleavage reaction by the Mu B protein, and the requirement of DNA nicking for stable type 1 transpososome formation. In vitro transposition characteristics of mini-Mu plasmids carrying terminal base pair mutations. J Biol Chem. 1991 Feb 15;266(5):3118–3124. [PubMed] [Google Scholar]
- Vink C., van Gent D. C., Elgersma Y., Plasterk R. H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991 Sep;65(9):4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiater L. A., Grindley N. D. Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. EMBO J. 1988 Jun;7(6):1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]






