Aim44p undergoes septin-dependent localization to the actomyosin ring and regulates contractile ring closure and the abundance, phosphorylation, and dynamics of Hof1p, a regulator of actomyosin ring closure. It also interacts directly with Hof1p. Thus Aim44p is a novel regulator of contractile ring closure in budding yeast.
Abstract
Whereas actomyosin and septin ring organization and function in cytokinesis are thoroughly described, little is known regarding the mechanisms by which the actomyosin ring interacts with septins and associated proteins to coordinate cell division. Here we show that the protein product of YPL158C, Aim44p, undergoes septin-dependent recruitment to the site of cell division. Aim44p colocalizes with Myo1p, the type II myosin of the contractile ring, throughout most of the cell cycle. The Aim44p ring does not contract when the actomyosin ring closes. Instead, it forms a double ring that associates with septin rings on mother and daughter cells after cell separation. Deletion of AIM44 results in defects in contractile ring closure. Aim44p coimmunoprecipitates with Hof1p, a conserved F-BAR protein that binds both septins and type II myosins and promotes contractile ring closure. Deletion of AIM44 results in a delay in Hof1p phosphorylation and altered Hof1p localization. Finally, overexpression of Dbf2p, a kinase that phosphorylates Hof1p and is required for relocalization of Hof1p from septin rings to the contractile ring and for Hof1p-triggered contractile ring closure, rescues the cytokinesis defect observed in aim44∆ cells. Our studies reveal a novel role for Aim44p in regulating contractile ring closure through effects on Hof1p.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, cytokinesis involves actomyosin contractile ring closure and septum formation at the mother–bud neck (Bi et al., 1998). The actomyosin ring contains actin filaments formed by formin-dependent processes (Fang et al., 2010; Wloka and Bi, 2012). During actomyosin ring assembly, Myo1p, the only myosin II heavy chain in budding yeast, is recruited to the site of cell division by the septin-interacting protein Bni5p, the essential myosin light chain (Mlc1p), and an IQGAP family protein (Iqg1p/Cyk1p) in a highly regulated manner (Epp and Chant, 1997; Lippincott and Li, 1998b; Lee et al., 2002). Constriction of the actomyosin ring provides a force for ingression of the plasma membrane. During its constriction, Myo1p disassembles from the ring (Tully et al., 2009), and vesicles containing materials essential for the formation of new plasma membrane are deposited at the cell division site (Fang et al., 2010).
In yeast cells, cytokinesis also involves septation—the deposition of cell wall between mother and daughter cells. After actomyosin ring constriction begins, a specialized extension of the cell wall—the primary septum—begins to form at the bud neck through reactions catalyzed by chitin synthase II (Chs1p; Sburlati and Cabib, 1986). The primary septum is completed after contractile ring closure and separation of the plasma membranes of mother and daughter cells. Thereafter, a secondary septum is laid down on each side of the primary septum. Finally, hydrolytic enzymes are released from the daughter cell to degrade the primary septum and part of the secondary septum, allowing for cell separation (Kuranda and Robbins, 1991; Kovacech et al., 1996; Baladrón et al., 2002).
Septin proteins Cdc3p, Cdc10p, Cdc11p, Cdc12p, and Shs1p/Sep7p are also localized to the site of cell division. The septins, which were discovered in yeast, are conserved, GTP-binding proteins that form octameric filament-forming complexes (Pan et al., 2007; Bertin et al., 2008, 2012; Garcia et al., 2011). The septins provide structural support for assembly and maintenance of the actomyosin ring at the bud neck and play a role in primary septum formation (Longtine et al., 1996). Septins also function as a diffusion barrier during cell division, promoting asymmetric distribution of proteins critical for cytokinesis to the mother–bud neck (Longtine and Bi, 2003; Dobbelaere and Barral, 2004).
Before cell division, the septins localize to the presumptive bud site as a single cortical ring (Iwase et al., 2006). During bud growth, the septin ring forms an hourglass structure at the mother–bud neck, and at the onset of cytokinesis, the mitotic exit network (MEN) signals the septins to form a double ring that surrounds the actomyosin ring (Cid et al., 2001; Lippincott et al., 2001). The double-septin-ring structure is maintained throughout cytokinesis, after which the mother cell and daughter cell each retains a single septin ring at the cell division site (Cid et al., 2001).
Several proteins have been implicated in interactions between the septins and the actomyosin ring, including Hof1p, an F-BAR protein that regulates contractile ring closure. The localization of Hof1p during cell division is highly dynamic and tightly regulated. Hof1p binds to septins and type II myosins (Myo1p in yeast) and is recruited to septin rings at the mother–bud neck. Phosphorylation of Hof1p by the MEN kinase complex Mob1p/Dbf2p at the S313 residue results in release of Hof1p from septin rings and association of the protein with the actomyosin ring (Meitinger et al., 2011, 2013a). Actomyosin ring closure is also regulated by MEN (Vallen et al., 2000; Lippincott et al., 2001; Luca et al., 2001). Recent studies indicate that the SH3 domain of Hof1p is critical for maintaining the symmetry of Myo1p ring constriction during cytokinesis (Korinek et al., 2000; Vallen et al., 2000; Oh et al., 2013) and that phosphorylation of Hof1p by Mob1p/Dbf2p at S533 and S563 promotes its function in contractile ring closure (Meitinger et al., 2013a). Finally, Hof1p binds to Inn1p, a protein that localizes to the contractile ring and is required for coordinating contractile ring closure with plasma membrane ingression (Sanchez-Diaz et al., 2008). Hof1p begins to be degraded late in mitosis; however, whether its degradation is related to the regulation of actomyosin ring closure is controversial (Blondel et al., 2005; Stockstill et al., 2013).
Genome-wide screens revealed a role for Aim44p in the cell cycle (Doolin et al., 2001; Wang et al., 2011) and mitochondrial DNA inheritance (Hess et al., 2009). They also revealed that Aim44p can interact physically with Hof1p (Tonikian et al., 2009) and localizes to the mother–bud neck (Huh et al., 2003). While our article was in preparation, another group reported a role for Aim44p in cell polarity and septation during cell division and renamed the protein GTPase-mediated polarity switch 1 (Gps1p; Meitinger et al., 2013b). Here we show novel functions for Aim44p at cell division: namely, that Aim44p is recruited to the bud neck by the septins, where it regulates Hof1p phosphorylation and localization and promotes contractile ring closure.
RESULTS
aim44∆ cells exhibit defects in contractile ring closure during cytokinesis
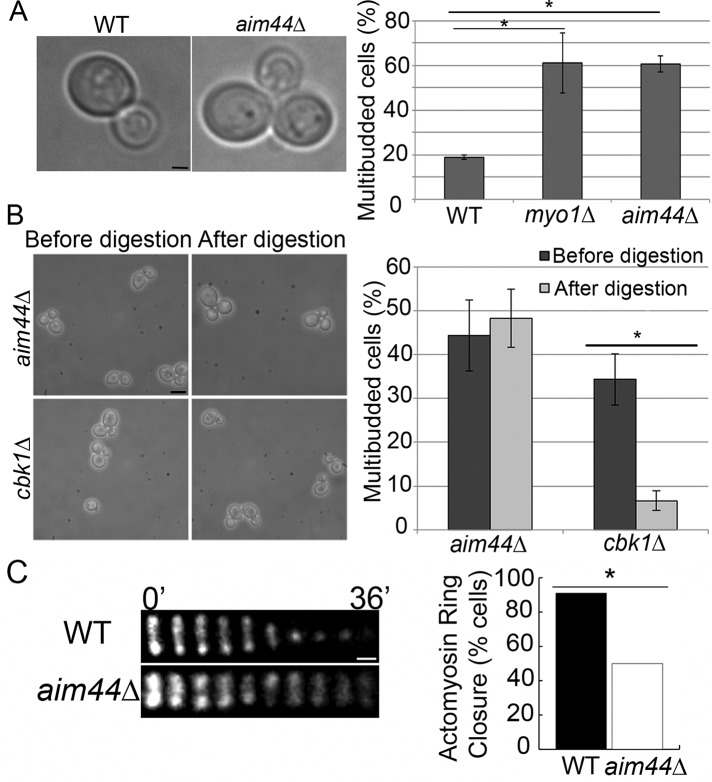
We find that deletion of AIM44 results in a multibudded phenotype, which is a hallmark of cytokinesis failure. In the wild-type cells used for these studies, 18.8 ± 1.0% cells examined are multibudded (García-Rodríguez et al., 2009). Deletion of MYO1, a known mediator of cytokinesis, results in 69.2% increase in multibudded cells compared with wild-type cells (61.2 ± 13.5%, p = 0.006). Similarly, in aim44∆ cells, we detect 69% increase in multibudded cells compared with wild-type cells (60.7 ± 3.6%, p = 4 × 10−5; Figure 1A). From these data, we conclude that Aim44p contributes to mother–daughter separation in S. cerevisiae.
FIGURE 1:
aim44∆ cells have a defect in contractile ring closure during cytokinesis. (A) Wild-type and aim44∆ cells were grown to late log phase (OD600 = 1.5) in SC glucose-based medium at 30°C, and the percentage of cells in multibudded clusters was determined. Left, transmitted-light image of wild-type cell with a single bud and an aim44∆ cell with multiple buds. Right, quantitation of the multibudded phenotype in wild-type, myo1∆, and aim44∆ cells. myo1Δ cells, which have defects in contractile ring constriction, exhibit higher levels of multibudded cells than do wild-type cells (p = 0.006). The aim44Δ cells show a statistically significant increase in the level of multibudded cells over wild type (p = 4.0 × 10−5). Error bars show SDs from three independent experiments. n ≥ 100 cells/strain per experiment. Scale bar, 1 μm. (B) The percentage of multibudded cells in aim44∆ and cbk1∆ cells was determined before and after treatment with Zymolyase 20T (0.1 mg/ml for 10 min at room temperature). Left, phase-contrast images of aim44∆ and cbk1∆ cells before and after Zymolyase treatment. Right, quantitation of multibudded phenotype before and after treatment. Zymolyase treatment results in cell separation in the cbk1∆ septation mutant (p = 0.002) but not in aim44∆ cells (p = 0.5). Errors bars show SDs from n > 800 cells/strain. Scale bar, 5 μm. (C, D) Wild-type and aim44∆ cells expressing MYO1 C-terminally tagged at its chromosomal locus with GFP were grown to mid log phase and synchronized in G1 phase by incubation with pheromone (10 μM α-factor) for 2 h at 30°C. Cells were then washed and placed in fresh media. The contractile ring was visualized beginning 60 min after release from G1 arrest by time-lapse imaging at 4-min intervals over a 40-min period. (C) Montage of the contractile ring in single cells over time. In wild-type cells (top), contractile ring closure is complete within 10 min. In contrast, in the aim44∆ cell shown, the contractile ring does not close during the 40-min imaging period (bottom). Scale bar, 0.3 μm. (D) Quantitation of the number of wild-type and aim44∆ cells that exhibit contractile ring closure (n = 52 and 66 for wild-type and aim44∆ cells, respectively; p = 7 × 10−7, chi-squared test). Results are pooled from three independent time-lapse imaging experiments.
Failure of yeast cells to separate can indicate a defect in contractile ring closure or septation. To determine whether aim44∆ cells have septation defects, we treated cells with Zymolyase, an enzyme isolated from Arthrobacter luteus that digests cell wall polymers (Lippincott and Li, 1998a). Zymolyase treatment results in cell separation in strains with septation defects, such as cbk1∆ cells, but not in yeast with defects in contractile ring closure (Hartwell, 1971). The proportion of cbk1∆ cells in multibudded clusters significantly decreases after digestion with Zymolyase (before digestion, 34 ± 6%; after digestion, 7 ± 2%; p = 0.002; Figure 1B). In contrast, Zymolyase treatment has no significant effect on the multibudded phenotype of aim44∆ cells (before digestion, 44 ± 8%; after digestion, 48 ± 7%; p = 0.5; Figure 1B). Thus the multibudded phenotype of aim44∆ cells is not due to defects in septation that are sensitive to Zymolyase treatment.
In light of this, we studied the effect of deletion of AIM44 on contractile ring closure. Myo1p, the type II myosin of the actomyosin ring, was tagged at its chromosomal locus using green fluorescent protein (GFP) in wild-type and aim44∆ cells. Previous studies indicate that this tag has no effect on Myo1p function (Lippincott and Li, 1998b). Deletion of AIM44 does not affect assembly of Myo1p into a ring at the bud neck (Figure 1C). However, deletion of AIM44 results in defects in contractile ring closure. In synchronized wild-type cells, contractile ring closure, visualized as a decrease in the diameter of the Myo1p-GFP–labeled contractile ring, occurs in 91% of the cells analyzed (n = 66) and is completed within 10 min from the onset of contraction (Figure 1C). In contrast, the contractile ring fails to close in 50% of the aim44∆ cells examined (n = 52; Figure 1). Of note, in the aim44∆ cells that do undergo contractile ring constriction, the rate of ring closure is not statistically different from that in wild-type cells (p = 0.96). However, in aim44Δ cells with contractile ring closure defects, Myo1p-GFP persists as a ring that does not contract and ultimately dissipates (Figure 1C). These aim44∆ cells eventually develop another bud and become multibudded. These findings are consistent with a role for Aim44p in regulating contractile ring closure.
Aim44p-GFP assembles into a ring structure at the bud neck that transiently colocalizes with the septins and the contractile ring
In yeast and other cell types, proteins that mediate or regulate cytokinesis localize to the site of cell division and often associate with either the actomyosin contractile ring or septin rings. High-throughput subcellular localization experiments revealed that Aim44p localizes to the bud neck (Huh et al., 2003). We tagged endogenous AIM44 at its C-terminus with GFP and found that Aim44p-GFP is fully functional (Supplemental Figure S1). Aim44p-GFP localizes to a ring at the incipient bud site (Figure 2). During early stages of bud development, Aim44p-GFP forms a single ring extending around the bud neck, consistent with the identification of Aim44p as a bud-neck protein (Huh et al., 2003). After contractile ring closure, the Aim44p-GFP ring thickens and separates into two rings; each ring remains at the site of cell division on the mother and daughter cells after cell separation (Figure 2).
FIGURE 2:
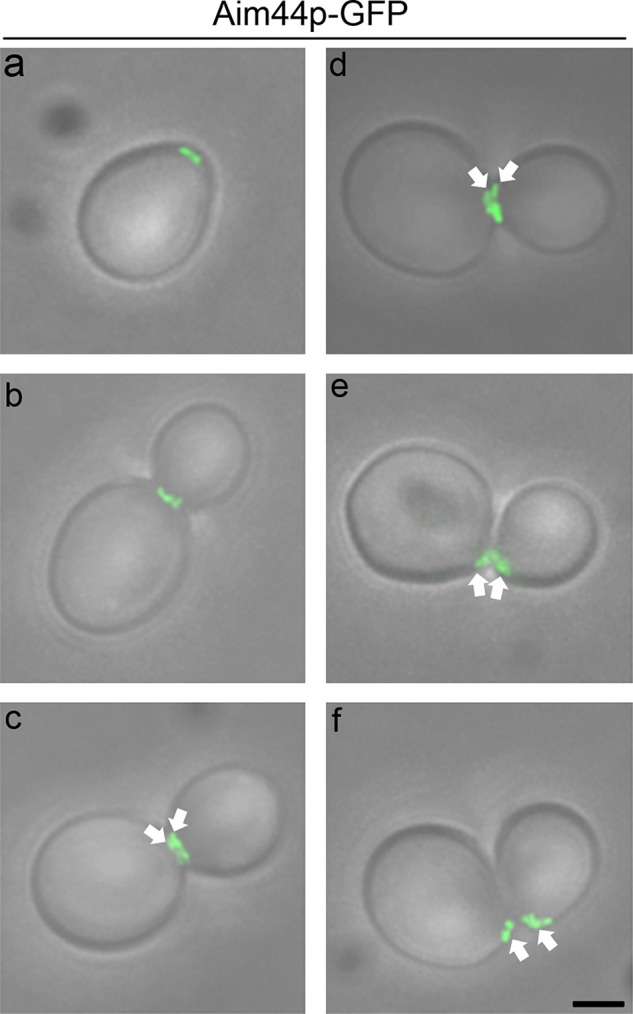
Aim44p-GFP localization throughout the cell cycle. Cells expressing AIM44 tagged at its chromosomal locus with GFP (green) were grown as for Figure 1 and imaged by fluorescence and phase contrast microscopy. Micrographs of the fluorescence images superimposed on transmitted-light images depict representative cells at different stages in the cell division cycle. Aim44p-GFP is recruited to the selected bud site, where it forms a ring structure (a). As the bud emerges and grows, Aim44p-GFP localizes to a single ring (b) and later to a double ring (c and d, highlighted with white arrows) at the bud neck. Finally, Aim44p persists as a ring on newly separated mother and daughter cells (e and f, highlighted with white arrows). Scale bar, 1 μm (bottom right).
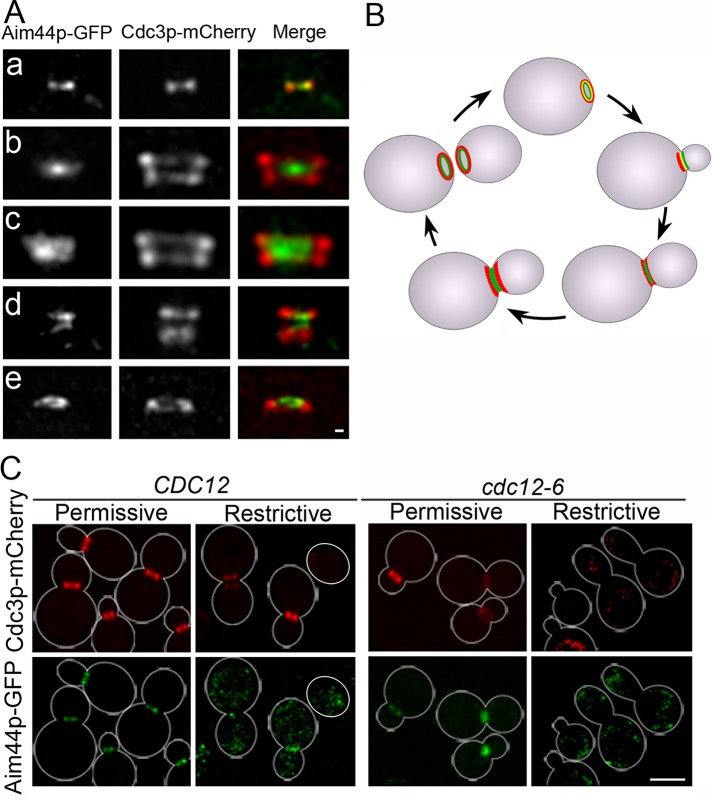
Because the localization pattern of Aim44p-GFP during mitosis resembles that of the septins, we studied the localization of Aim44p-GFP relative to the septin Cdc3p, tagged at its C-terminus with mCherry, in wild-type yeast cells (Figure 3). At the beginning of the cell division cycle, Aim44p-GFP and Cdc3p-mCherry are recruited to concentric rings at the selected bud site. During early bud development, the single Aim44p-GFP and septin rings partially colocalize. Later in the cell cycle, when septins localize to double rings, Aim44p-GFP localizes to a single ring between the septin double rings. Thereafter, when septin rings decrease in size, Aim44p-GFP localizes to two rings that are within and partially overlap with the double septin rings. Finally, after cell separation, the double septin and Aim44p rings separate such that the mother cell and daughter cell each has an Aim44p ring within a septin ring.
FIGURE 3:
Aim44p undergoes septin-dependent assembly into rings that are encircled by septin rings. Wild-type yeast expressing AIM44 tagged at its chromosomal locus with GFP (green) and plasmid-borne Cdc3p-mCherry (red) were grown to mid log phase (OD600 = 0.1–0.5) in SC and imaged by fluorescence microscopy. Representative images show the localization of Aim44p-GFP and the septin ring marked by Cdc3p-mCherry at the bud neck at various points throughout the cell division cycle. (A) Projections of deconvolved z-series showing side views of Aim44p-GFP (green) and Cdc3p-mCherry (red) at the mother–bud neck. Concentric single rings of Aim44p-GFP and septins during early bud development (a). A single Aim44p ring between two double septin rings in a cell with a medium to large bud (b). After contractile ring closure, Aim44p-GFP forms a thick single ring that is concentric and partially overlaps the septin rings (c). After cell division, both Aim44p-GFP and Cdc3p-mCherry are present as rings on both the mother and daughter cells, marking the former site of cell division (d). Concentric rings of Cdc3p-mCherry and Aim44p-GFP in a cell after cell separation (e). Scale bar, 0.3 μm. (B) Schematic of the localizations of Aim44p-GFP (green) and the septin rings (red) throughout the cell division cycle. (C) A temperature-sensitive septin strain cdc12-6 and the wild-type parent CDC12 strain expressing Aim44p-GFP (green) and a plasmid-borne septin Cdc3p-mCherry (red) were grown to mid log phase at permissive temperature (room temperature). An aliquot of each strain was shifted to restrictive temperature (34°C) for 2 h, and all cells were imaged by fluorescence microscopy. Shift of cdc12-6 mutant cells to restrictive temperature results in localization of Aim44p-GFP and Cdc3p-mCherry to abnormal diffuse and punctate cytosolic structures. Cell outlines based on transmitted-light images are shown in white. Scale bars, 5 μm.
Because Aim44p colocalizes with the single septin ring early in the cell division cycle and septins serve as a scaffold for assembly of the contractile ring, we tested whether Aim44p ring assembly is septin dependent. We detect both the single and double septin rings in aim44Δ cells (Supplemental Figure S3A). Thus septin ring assembly does not require Aim44p. In contrast, Aim44p-GFP ring assembly depends on septin ring formation. Specifically, we find that shift of the temperature-sensitive septin mutant cdc12-6 to restrictive temperatures disrupts Aim44p localization (Figure 3C). Incubation of wild-type CDC12 cells at 34°C results in a modest (10.6%) decrease in Aim44p localization to rings. In contrast, shift of the cdc12-6 mutant from room temperature to 34°C results in complete loss of septin rings, as described previously (Cid et al., 2001), and complete loss of detectable Aim44p rings (rings present in 98.5% of cells at permissive temperature and 0% at restrictive temperature; n ≥ 200 for each sample). In cdc12-6 cells at 34°C, Aim44p-GFP and Cdc3p-mCherry localize to punctate cytosolic structures. From these data, we conclude that septin ring formation is required for Aim44p ring formation.
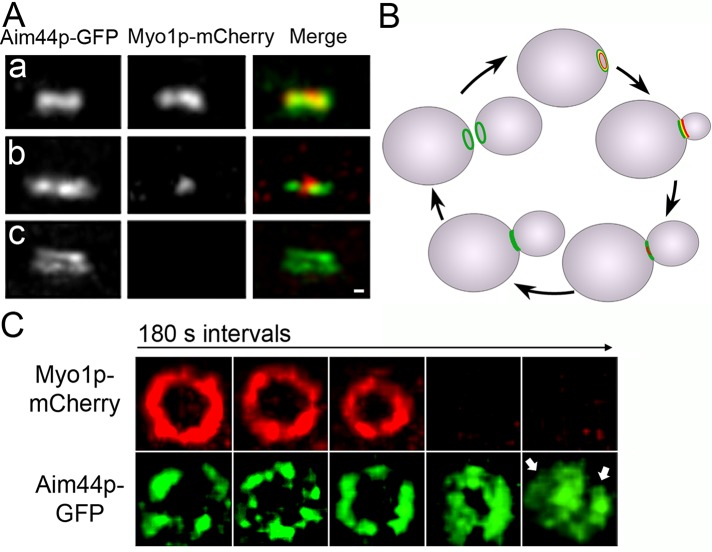
We also examined the localization of Aim44p-GFP relative to the contractile ring component Myo1p, which was tagged at its C-terminus with mCherry. Deletion of MYO1 does not affect Aim44p-GFP ring formation (Supplemental Figure S3B). However, Aim44p-GFP colocalizes with the Myo1p-mCherry–labeled contractile ring when both proteins form rings at the presumptive bud site and through most of the cell division cycle (Figure 4, A and B). Of interest, the ring formed by Aim44p-GFP does not contract when the contractile ring contracts. Instead, the Aim44p ring thickens and forms a double ring, which separates concomitant with cell separation such that mother cell and daughter cell each retains one Aim44p ring (Figure 4). The relative localization of Cdc3p-mCherry and Myo1p-GFP during the cell cycle is shown in Supplemental Figure S2.
FIGURE 4:
Localization of Aim44p-GFP and Myo1p-mCherry during cell division in wild-type cells. AIM44 and MYO1 were tagged with GFP and mCherry, respectively, at their chromosomal loci. Representative images show the bud neck of mid-log-phase cells imaged by fluorescence microscopy at different points throughout the cell division cycle. (A) Projections of deconvolved z-series showing side views of Aim44p-GFP (green) and Myo1p-mCherry (red). Single rings formed by Aim44p-GFP and Myo1p-mCherry characteristic of early bud growth (a). Aim44p-GFP remains a single ring that colocalizes with the actomyosin ring through most of the cell division cycle. During contractile ring closure, the diameter of the actomyosin ring decreases (b). However, the Aim44p ring does not contract. Aim44p forms a double ring after contractile ring closure (c). Scale bar, 0.3 μm. (B) Schematic of the localizations of Aim44p-GFP (green) and the actomyosin contractile ring (red) throughout the cell division cycle. (C) Cells expressing Myo1p-mCherry and Aim44p-GFP were treated with pheromone as for Figure 1, immobilized in a microfluidic chamber such that the mitotic rings were localized parallel to the microscope stage, and imaged 60–90 min after release from pheromone-induced G1 arrest. The z-series were obtained every 3 min to observe ring dynamics before, during, and after contractile ring closure. The Myo1p-mCherry ring contracts, but the Aim44p-GFP ring does not. After completion of contractile ring closure, Aim44p-GFP forms two rings (marked by two white arrows).
Aim44p coimmunoprecipitates with Hof1p and regulates Hof1p phosphorylation and localization
We investigated another regulator of contractile ring closure, Hof1p, and obtained evidence for a role for Aim44p in regulating Hof1p phosphorylation and localization. First, we confirmed that Hof1p undergoes cell cycle–dependent changes in abundance and phosphorylation. Specifically, we detect changes in the electrophoretic mobility of Hof1p over time after release from G1 arrest. The mobility shifts are sensitive to treatment with calf intestinal alkaline phosphatase and are therefore due to Hof1p phosphorylation (Supplemental Figure S4).
Next we assessed the effect of deletion of AIM44 on the level and phosphorylation state of Hof1p. Hof1p-13Myc is phosphorylated 75 min after release from G1 arrest in wild-type cells and 90 min after release from G1 arrest in aim44∆ cells (Figure 5A). Moreover, in wild-type cells, Hof1p undergoes dephosphorylation 105 min after release from G1 arrest, but this does not occur until 120 min after release from G1 arrest in aim44∆ cells. Thus deletion of AIM44 results in a delay in cell cycle–regulated phosphorylation and dephosphorylation of Hof1p.
FIGURE 5:
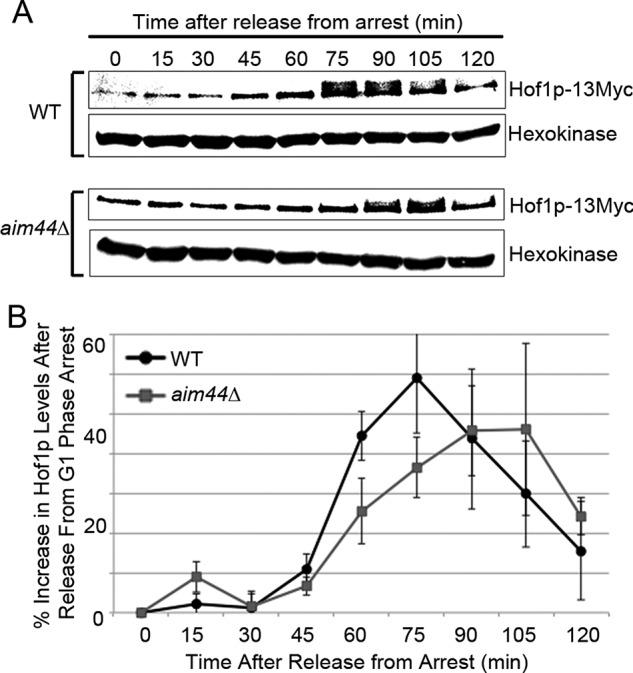
Deletion of AIM44 results in defect in cell cycle–linked changes in Hof1p abundance and phosphorylation. Wild-type and aim44Δ cells expressing Hof1p tagged with 13 copies of Myc at its chromosomal locus were synchronized as for Figure 6. Aliquots were removed from cell cultures for protein extraction every 15 min for 120 min after release from pheromone-induced G1 arrest. (A) Western blot decorated with antibodies against the Myc epitope on Hof1p and the loading control, hexokinase. The shift in electrophoretic mobility of Hof1p detected at 75–105 min in wild-type cells and 90–105 min in aim44∆ cells represents phosphorylation, as determined by sensitivity to treatment with calf intestinal alkaline phosphatase (Supplemental Figure S4). (B) Relative levels of Hof1p-13Myc were quantified by scanning densitometry. Quantifications are averages from Western blots from three independent experiments. Error bars represent SEM.
Total levels of Hof1p are also affected by deletion of AIM44 (Figure 5B). In wild-type cells, we detect an increase in the level of Hof1p from 30 to 75 min after release from pheromone-induced G1 arrest and a decline in Hof1p levels from 75 to 120 min after release from G1 arrest. Deletion of AIM44 alters cell cycle–linked changes in Hof1p levels. Specifically, we observe a decrease in the rate of cell cycle-linked increases in Hof1p levels, a delay in the time when maximum Hof1p levels occurs (90–105 min for aim44∆ cells vs. 75 min for wild-type cells), and a 22% decrease in the maximum level of Hof1p. Using spindle length as an indicator of mitotic progression, we find that deletion of AIM44 has no effect on cell cycle progression (Supplemental Figure S5). Thus deletion of AIM44 affects the abundance and phosphorylation of Hof1p without affecting overall rates cell cycle progression before the point of cell division.
Because deletion of AIM44 results in defects in phosphorylation of Hof1p and Dbf2p catalyzes phosphorylation of Hof1p, we assessed the effect of overexpression of Dbf2p on cytokinesis in aim44∆ cells. We confirmed that incubation in galactose-based media results in overexpression of Dbf2p in wild-type cells and aim44∆ cells. We also found that the level of overexpression of Dbf2p is similar in both cell types (Figure 6A). Next we monitored cytokinesis, by quantitation of the number of multibudded cells, in cells with wild-type or elevated levels of Dbf2p. Overexpression of Dbf2p has no effect on the level of multibudded cells observed in wild-type cells (p = 0.52). In contrast, overexpression of Dbf2p results in a decrease in the number of multibudded cells in aim44∆ from 55% in aim44∆ cells to 24% in aim44∆ cells that overexpress Dbf2p (p = 0.002; Figure 6B). Thus the defect in cytokinesis observed upon deletion of AIM44 can by rescued by overexpression of a kinase that catalyzes phosphorylation of Hof1p.
FIGURE 6:
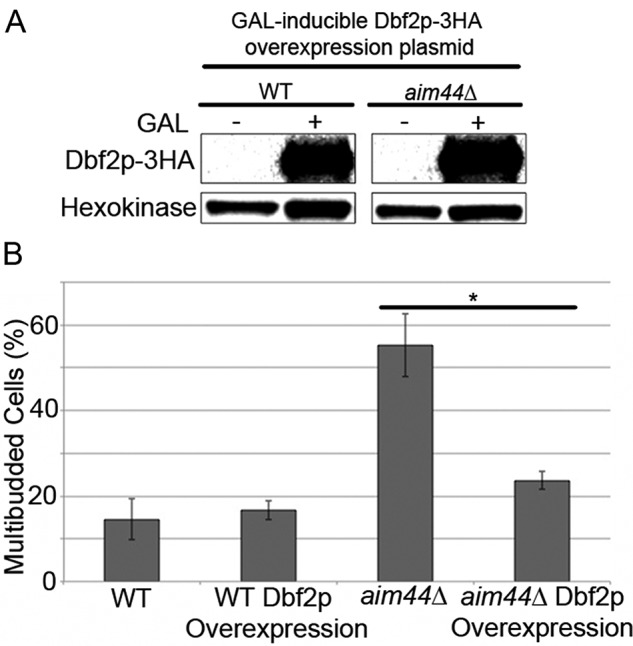
The cell division defect of aim44Δ cells is alleviated with overexpression of the kinase, Dbf2p. Wild-type and aim44Δ cells were transformed with a plasmid bearing the DBF2 gene that was tagged with HA and expressed under control of the GAL-inducible promoter. The cells were grown to late log phase (OD600 = 1.5) in SC glucose-based medium or galactose-based medium at 30°C. (A) Western blots showing the level of Dbf2p-HA detected using anti-HA antibodies and the load control, hexokinase, detected with anti–hexokinase 1p antibody in whole-cell extracts of glucose- or galactose-grown cells. (B) Percentage of multibudded cells in wild-type and aim44∆ cells that express either endogenous Dbf2p or overexpress Dbf2p determined as for Figure 1. Error bars show SDs from three independent experiments. n ≥ 100 cells/strain per experiment.
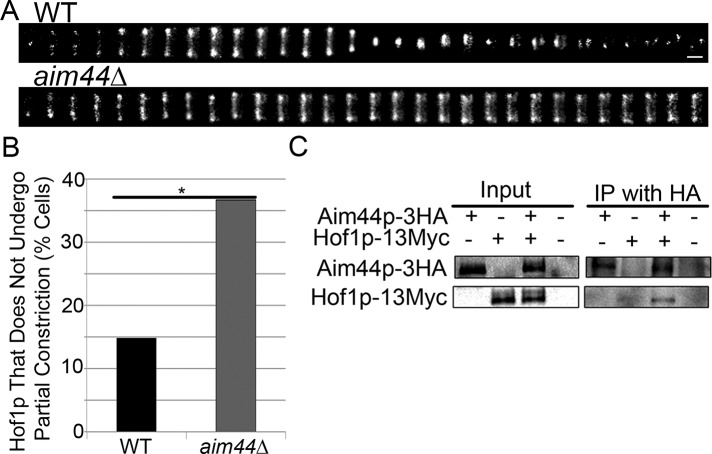
Because phosphorylation of Hof1p affects its localization, we carried out time-lapse imaging to study the effect of deletion of AIM44 on Hof1p dynamics. We confirmed that Hof1p-GFP localizes to a double ring at the bud neck in a wild-type strain expressing Hof1p C-terminally tagged at its chromosomal locus with GFP (Figure 7). Later in the cell cycle, Hof1p localizes to a single, medial ring that undergoes partial constriction as the actomyosin ring contracts. It then separates into two rings that localize to both mother and daughter cells after cell division. Eighty-five percent of wild-type cells examined display this pattern of Hof1p-GFP dynamics (Figure 7B). In contrast, 40% of aim44Δ cells exhibit defects in Hof1p dynamics (p = 0.0003). In these cells, Hof1p localizes to a double ring at the bud neck that does not undergo partial contraction during the 3-h imaging period (Figure 7B). aim44∆ cells that exhibit defects in Hof1p dynamics also do not undergo cell separation. Thus deletion of AIM44 results in defects in Hof1p abundance, phosphorylation, dynamics, and function in promoting contractile ring closure.
FIGURE 7:
Aim44p regulates Hof1p localization and dynamics at the bud neck and coimmunoprecipitates with Hof1p. (A) Wild-type and aim44Δ cells expressing Hof1p-GFP were synchronized as for Figure 6. Hof1p-GFP were imaged every 5 min beginning 60 min after release from G1 arrest for a total time of 3 h. Montages showing changes in Hof1p localization and dynamics at the bud neck as a function of time after release from pheromone-induced G1 arrest in wild-type cells (top) and aim44∆ mutants (bottom). Scale bar, 0.3 μm. (B) Quantitation of wild-type and aim44∆ cells that exhibit Hof1p-GFP ring contraction (p = 0.0003, chi-squared test). n = 101 and 120 for wild-type and aim44∆ cells, respectively. Data are pooled from two independent experiments. (C) Yeast cells expressing no tagged proteins, Aim44p-3HA, Hof1p-13Myc, or both Aim44p-3HA and Hof1-13Myc were grown to mid log phase and treated with pheromone as for Figure 1. Whole-cell extracts were prepared from each sample 75 min after release from pheromone-induced G1 arrest, and samples were immunoprecipitated using anti-HA antibodies. Western blots show whole-cell extracts and immunoprecipitated proteins detected using anti-HA and anti-Myc antibodies. Hof1p-Myc coimmunoprecipitates with Aim44p-HA.
To further characterize the mechanism of Aim44p function in regulation of Hof1p, we tested whether the two proteins interact. Previous genome-wide two-hybrid screens revealed that Hof1p and Aim44p have the capacity to bind to each other (Tonikian et al., 2009). We tested whether this interaction is physiologically significant. To do so, we tagged Aim44p and Hof1p at their chromosomal loci in wild-type cells with hemagglutinin (HA) and Myc epitopes, respectively. We then synchronized cells and carried out immunoprecipitation with whole-cell extracts prepared 75 min after release from pheromone-induced G1 arrest. We found that Aim44p coimmunoprecipitates with Hof1p (Figure 7C). Hof1p detected in the immunoprecipitate is the unphosphorylated form of the protein. Thus Aim44p binds to unphosphorylated Hof1p. There is no detectable phosphorylated Hof1p in the immunoprecipitate. Therefore, it is not clear whether Aim44p can bind to the phosphorylated form of the protein.
DISCUSSION
Our studies support a role for Aim44p in actomyosin contractile ring closure during cytokinesis. First, the localization of Aim44p is consistent with a role in this process. Specifically, we find that Aim44p forms a single ring within the single septin ring at the bud neck early in the cell cycle. The Aim44p ring colocalizes with the actomyosin ring throughout the cell cycle until contractile ring closure. During contractile ring closure, the Aim44p ring does not contract. After contractile ring closure, the Aim44p ring thickens and forms two rings that are within and closely opposed to the double septin rings. Finally, during cell separation, the double rings of septins and Aim44p also separate such that a single Aim44p ring remains within a single septin ring on both mother and daughter cells, marking the site of division. The localization of Aim44p to the actomyosin ring at the bud neck depends on the septins but not Myo1p, the type II myosin of the contractile ring in yeast.
Aim44p is not required for assembly of septin or actomyosin rings at the bud neck. However, it does regulate actomyosin ring closure. Deletion of AIM44 results in a statistically significant increase in multibudded cells and a decrease in the frequency of contractile ring closure. In aim44∆ cells that do undergo contractile ring constriction, the rate of ring closure is not significantly different from that observed in wild-type cells. Thus Aim44p is not required for formation of structures underlying the contractile ring, but it regulates the efficiency of contractile ring closure.
Consistent with this, we find that deletion of AIM44 affects the phosphorylation and localization of Hof1p, a protein that localizes to the bud neck, binds to septins and Myo1p, regulates contractile ring closure, and is detected as an Aim44p binding partner in a genome-wide protein interaction screen (Tonikian et al., 2009). We find that cell cycle–regulated changes in Hof1p abundance and phosphorylation are altered in aim44∆ cells. We also detect defects in Hof1p localization in aim44∆ cells. In wild-type cells, Hof1p assembles as two rings on septin rings at the bud neck early in the cell division cycle. In response to a series of phosphorylations, Hof1p then moves from septin rings to the actomyosin ring. The Hof1p ring undergoes partial contraction with the actomyosin ring, ultimately associating with septin rings on mother and daughter cells upon cell separation.
Recent studies indicate that phosphorylation of Hof1p affects its binding to the septins, which in turn affects septin stability, in addition to its function in regulating actomyosin ring closure (Meitinger et al., 2013a). Although we find that assembly of the septin ring at the bud neck does not depend on Aim44p, it is possible that the maintenance of a stable septin scaffold may also be affected by Aim44p regulation of Hof1p phosphorylation.
However, we find that deletion of AIM44 results in defects in Hof1p localization. In aim44∆ cells, Hof1p localizes to two large rings. Therefore it is likely that Hof1p can associate with septin rings in aim44∆ cells. However, in aim44∆ cells with contractile ring closure defects, Hof1p does not undergo cell cycle–linked movement from double rings to the single actomyosin ring. We also find that Aim44p coimmunoprecipitates with Hof1p. It is not clear whether Aim44p has the capacity to bind to phospho-Hof1p. Nonetheless, our studies indicate that it can bind to the unphosphorylated form of the protein. Finally, we find that overexpression of Dbf2p, the kinase that catalyzes phosphorylation of Hof1p, which leads to movement of Hof1p from the septin rings to the actomyosin ring, rescues the contractile ring closure defect in aim44∆ cells.
These findings support a model for Aim44p regulation of contractile ring closure. According to this model, Aim44p assembles into a ring at the interface between septin and actomyosin rings early in the cell division cycle. Later in the cell cycle, as Hof1p levels increase and it assembles into a ring within the septin ring, Aim44p interacts with the unphosphorylated form Hof1p. This interaction then promotes cell cycle–regulated phosphorylation of Hof1p by Mob1p/Dbf2p, which leads to movement of Hof1p from the septin rings to the contractile ring and Hof1p-triggerred contractile ring closure. Aim44p remains associated with septin rings during contractile ring closure, septation, and cell–cell separation as rings that are encircled by septin rings.
During the preparation of this article, Meitinger et al. (2013b) found that wild-type Aim44p (referred to in that work as Gps1p) localizes to structures at the bud neck that are similar to those that we report and can bind directly to two signal transduction GTPase proteins—Cdc42p and Rho1p—which also localize to the bud neck during cytokinesis. They identified a role for Aim44p in regulating secondary septum formation through effects on Rho1p. They also observed a role for Aim44p in preventing the activation of Cdc42p at the bud neck after cytokinesis and found that this mechanism ensures that new daughter cells are not produced at the site of cytokinesis from the previous round of cell division.
Meitinger et al. (2013b) argue that Aim44p does not function in contractile ring closure, based on findings that contractile ring closure occurs at wild-type rates in aim44∆ cells. We also find that actomyosin ring closure occurs at the wild-type rate in aim44Δ cells that do undergo this process. Of importance, however, we find that the actomyosin ring fails to close in 50% of aim44Δ cells. Thus we find that Aim44p regulates the efficiency of contractile ring closure rather than the rate of closure.
Moreover, Meitinger et al. (2013b) find that there is a defect in secondary septum formation in aim44∆ cells. We find that Zymolyase-dependent cell wall degradation under conditions that allow for cell separation in a known septation mutant, cbk1∆, does not result in separation of multibudded aim44∆ cells. These findings indicate that aim44∆ cells have either a cytokinesis defect that appears before the septation defect or septation defects that are not similar to those of cbk1∆ cells.
Finally, recent studies indicate that Rho1p and Cdc42p are activated during actomyosin ring assembly, inactivated during actomyosin ring contraction, and activated again during septum formation (Atkins et al., 2013; Onishi et al., 2013). Therefore it is possible that defects in Rho1p and/or Cdc42p regulation may contribute to the defects in contractile ring closure in aim44∆ cells.
Taken together, this and other recent work indicates that Aim44p has multiple functions late in the yeast budding process. It functions in contractile ring closure through effects on Hof1p phosphorylation and localization in addition to its role in septation through effects on Cdc42p. Indeed, Meitinger et al. (2013b) find that an aim44∆ hof1∆ mutant exhibits more-severe growth defects than either aim44∆ or hof1∆ single-mutant yeast. This is genetic evidence that Aim44p has Hof1p-independent functions in addition to the Aim44p- and Hof1p-dependent function in contractile ring closure described here. In summary, our studies reveal a novel role for Aim44p in contractile ring closure through effects on Hof1p phosphorylation and localization to the actomyosin ring. Because Hof1p, septins, and contractile ring components are conserved, it is possible that there is an orthologue of Aim44p in other cell types. Further studies will expand our understanding of how Aim44p contributes to the coordinated regulation of the septins and actomyosin ring closure.
MATERIALS AND METHODS
Strains, plasmids, and genetic manipulation
S. cerevisiae strains used in this study are summarized in Supplemental Table S1. All yeast strains were created in the BY4741 wild-type background, except for the temperature-sensitive septin strain (YEF743 cdc12-6) and the wild-type control (YEF6866 CDC12). The two YEF strains were kindly provided by Erfei Bi (University of Pennsylvania, Philadelphia, PA).
AIM44 was tagged at its C-terminus with GFP by transformation with a PCR-amplified GFP tagging cassette from the pFA6a-GFP system (Addgene, Cambridge, MA) containing HIS3 and 40–base pair homology to the endogenous locus. The protein Hof1p was fused with 13 copies of Myc at its C-terminus tag by transformation with a PCR-amplified pFA6a-13Myc plasmid containing a His3Mx6 gene for selection and 40–base pair homology to HOF1. The AIM44 genomic locus was disrupted in cells expressing Myo1p-GFP and Hof1p-GFP by transformation with a PCR-amplified cassette from pFA6a-KanMX6 containing the kanMX selectable marker and 40–base pair homology to the endogenous locus. The AIM44 genomic locus was disrupted in Hof1-13Myc:His3MX6 cells by transformation with a PCR-amplified sequence containing LEU2 derived from the pOM cassette system (Tamm, 2009) and 40–base pair homology to the endogenous locus. Similarly, the MYO1 genomic locus was disrupted by transformation with a PCR-amplified sequence containing LEU2 derived from the pOM cassette system and 40–base pair homology to the endogenous locus. To tag Myo1p with mCherry, cells were transformed with a PCR-amplified pcY3090-02 plasmid containing a hphMX4 gene for selection with hygromycin B and 40–base pair homology to MYO1 (Young et al., 2012). TUB1 was tagged at its C-terminus with GFP by transformation with DNA from the plasmid pTS988 digested with the restriction enzyme XbaI (New England BioLabs, Ipswich, MA) in wild-type and aim44Δ cells. AIM44 was tagged with 3HA at its C-terminus in wild-type and Hof1p-13Myc–expressing cells by transformation with a PCR-amplified cassette from the pFA6a-3HA system containing kanMX6 and 40–base pair homology to the endogenous locus. Construction of the cbk1Δ strain (García-Rodríguez et al., 2009), Myo1p-GFP expression strain (Huckaba et al., 2006), and myo1Δ strain (Higuchi et al., 2013) are described in previous work. In the YEF6866 CDC12 and YEF743 cdc12-6 strains, a C-terminus GFP tag was fused to AIM44 by transformation with a PCR-amplified, kanMX-encoding cassette from pFA6a-GFP(S65T)-kanMX6 containing 40–base pair homology to the endogenous locus. These strains were then transformed with the YIP128-CDC3-mCherry plasmid, kindly provided by John Pringle (Stanford University, Stanford, CA). aim44Δ cells and cells expressing Aim44p-GFP and Myo1p-GFP were also transformed with the YIP128-CDC3-mCherry plasmid. After transformation using the lithium acetate method, cells were selected on solid media with the appropriate selective solid media (Gietz et al., 1995). For Dbf2p overexpression studies, wild-type and aim44Δ cells were transformed with an open reading frame expression vector plasmid, pGB1805, with a GAL-inducible promoter controlling the overexpression of Dbf2p tagged with an HA epitope (Thermo Scientific, Pittsburgh, PA). In-frame tagging or disruption of a gene was verified using PCR amplification. Escherichia coli DH5-α–competent cells (Stratagene, Santa Clara, CA) were used for amplification of all plasmids used in this study. Plasmid clones were recovered from bacteria with a MiniPrep kit (Qiagen, Valencia, CA).
Media and culture conditions
E. coli was cultivated in Luria–Bertani (LB) medium (1% peptone, 0.5% yeast extract, 1% sodium chloride, pH 7.0) at 37°C with ampicillin (100 g/l) to select for plasmid-carrying cells. S. cerevisiae strains were grown in synthetic complete (SC) medium at 30°C on a rotary shaker at 225 rpm, as previously described (Fehrenbacher et al., 2004). Solid medium was prepared as described with the addition of agar (2% wt/vol). For selection of genetically manipulated cells, SC medium was prepared without histidine for selection of cells auxotrophic for HIS3, without leucine for selection of cells auxotrophic for LEU2, or with the addition of Geneticin (G418) sulfate (200 μg/ml; Invitrogen, Carlsbad, CA) for cells expressing the kanMX6 gene. Media were prepared with galactose instead of glucose as the carbon source for experiments requiring media containing galactose. For experiments with temperature-sensitive YEF6866 and YEF743 strains expressing Aim44p-GFP and Cdc3p-mCherry, cells were grown as described but using 20 and 34°C as permissive and restrictive temperature, respectively. Other yeast methods were performed as previously described (Sherman, 2002).
Quantification of multibudded cells
To quantify the frequency of multibudded cells, cultures were grown to late log phase in SC medium to OD600 of at least 1.5. Cells were sonicated and visualized using a light microscope. Data shown were pooled from three independent experiments. To check for a cytokinesis defect, we incubated cells with Zymolyase 20T (0.1 mg/ml) at room temperature for 10 min.
Synchronization of cells
To induce cell-cycle arrest in G1 phase, we treated mid-log-phase cultures with 10 μM α-factor (Genemed Synthesis, San Antonio, TX) in fresh SC for 2 h at 30°C. Arrest was verified by light microscopy, indicated by at least 70% of cells exhibiting a schmoo morphology. Cells were released from arrest by washing three times in cold (4°C) water before resuspending in fresh SC medium.
Preparation of cells for microscopy, microscopy, and image analysis
For the localization studies in fixed cells, we used an AxioObserver.Z1 microscope equipped with a Colibri LED excitation source, a wide-field camera (Orca ER, Hamamatsu Photonics, Bridgewater, NJ), and AxioVision acquisition software. We used a 100×/1.3 numerical aperture (NA) EC PlanNeofluar objective (Carl Zeiss, Jena, Germany) and set the camera to 1 × 1 binning to optimize spatial resolution. To visualize mCherry, we used an excitation of 570 nm and a standard rhodamine/RGP filter set with an exposure time of 300 ms for Cdc3p-mCherry and 100 ms for Myo1p-mCherry. To visualize GFP, we used excitation at 470 nm (100% LED power) and a standard GFP filter set with an exposure time of 500 ms for Aim44p-GFP and 300 ms for Myo1p-GFP. We acquired z-stacks consisting of 13 slices with 0.5-μm spacing. Time-lapse imaging of Hof1p-GFP–expressing cells was also performed using this microscope and the 470-nm LED for excitation with an exposure time of 125 ms. For this experiment, cells were imaged using the CellASIC (EMD Millipore, Billerica, MA) microfluidic flow chamber (Y04C plate) controlled by the ONIX Control System and software. Cells were imaged every 5 min for 3 h beginning 1 h after release from arrest in G1 phase.
For time-lapse imaging of wild-type cells expressing Myo1-mCherry and Aim44p-GFP, cells were held upright in microfluidic chambers composed of polydimethylsiloxane with a depth of 20 μm and a diameter of 6 μm, such that actomyosin and Aim44p rings were positioned parallel to the microscope state. Imaging was carried out using a spinning disk confocal microscope consisting of the CSU-X1 spinning disk attachment (Yokogawa Electronic Corporation, Tokyo, Japan) on a Nikon Ti Eclipse inverted microscope (Nikon, Melville, NY) equipped with an electron-multiplying charge-coupled device (CCD) camera (DU-897; Andor Technologies, Belfast, United Kingdom; Evolve, Photometrics, Tucson, AZ), 488- and 561-nm, 50-mW lasers, and a CFI Plan Apo 100×/1.45 NA oil objective (Nikon). Aim44p-GFP was imaged using 488-nm excitation illumination with 500-ms exposure times and 20% laser power. Myo1p-mCherry was imaged using 561-nm excitation illumination with 300-ms exposure times and 30% laser power.
Experiments with temperature-sensitive strains were performed using a Nikon Eclipse Ti microscope with a 60×/1.4NA oil PlanApo Nikon objective and Perfect Focus System equipped with a Hamamatsu 10-600 Orca R2 camera, an ASI Nano-Drive piezoelectric focus drive (Applied Scientific Instruments, Eugene, OR), a Lambda smart shutter controller and emission filter wheel (Sutter Instruments, Novato, CA), spectral LMM5 100-mW lasers, and a Yokogawa (Sugar Land, TX) CSU 10 spinning-disk confocal attachment upgraded with a Borealis system (Spectral Applied Research, Natick, MA). The 491- and 561-nm lasers were used for excitation at 100% with exposure times of 800 and 225 ms for green and red, respectively.
Live-cell imaging for the actomyosin contractile ring studies was performed using agarose pads as previously described (Fehrenbacher et al., 2004). Imaging was performed on an Axioskop 2 microscope (Zeiss) with a 100×/1.4 NA Plan-Apochromat objective and an Orca 1 cooled CCD camera (Hamamatsu) or an E600 microscope (Nikon) with a Plan-Apo 100×/1.4 NA objective and an Orca-ER cooled CCD camera with fluorescein isothiocyanate filter sets. Hardware was controlled by Openlab software. Three-dimensional images over time were acquired by obtaining optical sections in 0.5-μm spacing using a piezoelectric focus motor mounted on the objective lens (Polytech PI, Auburn, MA). An exposure time of 250 ms was used to visualize Myo1p-GFP. Cells were imaged every 4 min for ∼48 min beginning 60 min after release from arrest in G1 phase.
For the cell cycle progression analysis in wild-type and aim44Δ cells expressing Tub1-GFP, cells were fixed using 3.6% paraformaldehyde and stained with the DNA-binding dye 4′,6-diamidino-2-phenylindole (Life Technologies, Grand Island, NY) mixed with Mounting Solution, as described previously (Swayne et al., 2010). Fixed cells were then imaged on an Axioskop 2 microscope with a 100×/1.4 NA Plan-Apochromat objective and an Orca 1 cooled CCD camera. We acquired z-sections with 0.5-μm spacing.
Image processing and analysis
Images were deconvolved using a constrained iterative restoration algorithm in Volocity software (Perkin Elmer-Cetus, Waltham, MA). Maximum projections were reconstructed using Volocity software or ImageJ (Schneider et al., 2012). Volocity was also used for measurement of spindle length. For statistical analysis, p values were calculated with the Student's t test (two-tailed distribution), the chi-squared test, or Kruskal–Wallis analysis.
Protein methods
Protein was isolated from yeast cells as previously described (Boldogh et al., 1998). Protein concentration was determined using the bicinchoninic acid assay (Pierce, Rockford, IL). Calf intestinal alkaline phosphatase (New England BioLabs) treatment was carried out using 10 U of enzyme for 1 h at 37°C. 13Myc-tagged proteins were probed on nitrocellulose membrane using a monoclonal anti-Myc (9E10 1-1) primary antibody (Evan et al., 1985) and a horseradish peroxidase–conjugated secondary antibody (Promega, Madison, WI). HA-tagged proteins were detected using a monoclonal mouse anti-HA antibody (12CA5; Roche, Nutley, NJ). Hexokinase was detected using a rabbit polyclonal anti-hexokinase antibody (LS-C59302) purchased from LifeSpan Biosciences (Seattle, WA). Signal was detected using the SuperSignal West Pico chemiluminescent substrate (Pierce). Luminescence was recorded using a ChemiDoc MP imaging system and analyzed using Image Lab software (Bio-Rad, Hercules, CA). For the coimmunoprecipitation experiments, protein extracts were added to Dynabeads Protein G (Life Technologies) according to the manufacturer's instructions and incubated with a monoclonal mouse anti-HA antibody (clone 12CA5; 11 583 816 001; Roche) to mediate the pull down of Aim44p-3HA. The antibodies used for probing the nitrocellulose membrane were a monoclonal rabbit anti-Myc antibody (71D10; Cell Signaling, Danvers, MA) and a monoclonal mouse anti-HA antibody (clone 16B12; MMS-101P; Covance, Princeton, NJ).
Supplementary Material
Acknowledgments
We thank the members of the Pon laboratory for technical assistance and valuable discussions, John Pringle for yeast strains, Erfei Bi for plasmids, Zhou Zhou and Fred Chang (Columbia University, New York, NY) for microfluidic chambers, Julie Canman (Columbia University) for valuable guidance and use of her spinning disk confocal microscope, and Theresa C. Swayne for assistance during preparation of the manuscript. This work was supported by awards from the National Institutes of Health 2 TL1 RR 24158-6 to D.M.A.W. and GM045735, GM045735S1, and GM096445 to L.P. GM045735S1 was issued from the National Institutes of Health under the American Recovery and Reinvestment Act of 2009. Imaging in the Confocal and Specialized Microscopy Shared Resource was supported by National Institutes of Health P30 CA13696 to the Herbert Irving Comprehensive Cancer Center at Columbia University.
Abbreviations used:
- GFP
green fluorescent protein
- HA
hemagglutinin
- MEN
mitotic exit network
- SC
synthetic complete
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-06-0317) on January 22, 2014.
REFERENCES
- Atkins BD, Yoshida S, Saito K, Wu C-F, Lew DJ, Pellman D. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol. 2013;202:231–240. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baladrón V, Ufano S, Dueñas E, Martín-Cuadrado AB, del Rey F, Vázquez de Aldana CR. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot Cell. 2002;1:774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park S-S, Garcia G, Patanwala I, Ng H-L, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Pierson J, Thai L, McDonald KL, Zehr EA, García G, Peters P, Thorner J, Nogales E. Three-dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae. Mol Biol Cell. 2012;23:423–432. doi: 10.1091/mbc.E11-10-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M, Bach S, Bamps S, Dobbelaere J, Wiget P, Longaretti C, Barral Y, Meijer L, Peter M. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 2005;24:1440–1452. doi: 10.1038/sj.emboj.7600627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Adamiková L, Sánchez M, Molina M, Nombela C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 2001;147:1437–1450. doi: 10.1099/00221287-147-6-1437. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Doolin MT, Johnson AL, Johnston LH, Butler G. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol Microbiol. 2001;40:422–432. doi: 10.1046/j.1365-2958.2001.02388.x. [DOI] [PubMed] [Google Scholar]
- Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191:1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher KL, Yang H-C, Gay AC, Huckaba TM, Pon LA. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Garcia G, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011;195:993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez LJ, Crider DG, Gay AC, Salanueva IJ, Boldogh IR, Pon LA. Mitochondrial inheritance is required for MEN-regulated cytokinesis in budding yeast. Curr Biol. 2009;19:1730–1735. doi: 10.1016/j.cub.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hess DC, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Vevea JD, Swayne TC, Chojnowski R, Hill V, Boldogh IR, Pon LA. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr Biol. 2013;23:2417–2422. doi: 10.1016/j.cub.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba TM, Lipkin T, Pon LA. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J Cell Biol. 2006;175:957–969. doi: 10.1083/jcb.200609155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Bi E, Epp JA, Wang L, Ho J, Chant J. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr Biol. 2000;10:947–950. doi: 10.1016/s0960-9822(00)00626-6. [DOI] [PubMed] [Google Scholar]
- Kovacech B, Nasmyth K, Schuster T. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol Cell Biol. 1996;16:3264–3274. doi: 10.1128/mcb.16.7.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranda MJ, Robbins PW. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- Lee PR, Song S, Ro H-S, Park CJ, Lippincott J, Li R, Pringle JR, De Virgilio C, Longtine MS, Lee KS. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6906–6920. doi: 10.1128/MCB.22.19.6906-6920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998a;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998b;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–1386. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13:403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol. 2001;21:6972–6983. doi: 10.1128/MCB.21.20.6972-6983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Palani S, Hub B, Pereira G. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol Biol Cell. 2013a;24:1290–1304. doi: 10.1091/mbc.E12-08-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Richter H, Heisel S, Hub B, Seufert W, Pereira G. A safeguard mechanism regulates Rho GTPases to coordinate cytokinesis with the establishment of cell polarity. PLoS Biol. 2013b;11:e1001495. doi: 10.1371/journal.pbio.1001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell. 2013;24:1305–1320. doi: 10.1091/mbc.E12-11-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol. 2013;202:311–329. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Malmberg RL, Momany M. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol Biol. 2007;7:103–120. doi: 10.1186/1471-2148-7-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A, Marchesi V, Murray S, Jones R, Pereira G, Edmondson R, Allen T, Labib K. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast. Nat Cell Biol. 2008;10:395–406. doi: 10.1038/ncb1701. [DOI] [PubMed] [Google Scholar]
- Sburlati A, Cabib E. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J Biol Chem. 1986;261:15147–15152. [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Stockstill KE, Park J, Wille R, Bay G, Joseph A, Shannon KB. Mutation of Hof1 PEST motif phosphorylation sites leads to retention of Hof1 at the bud neck and a decrease in the rate of myosin contraction. Cell Biol Int. 2013;37:314–325. doi: 10.1002/cbin.10042. [DOI] [PubMed] [Google Scholar]
- Swayne TC, Boldogh IR, Pon LA. Imaging of the cytoskeleton and mitochondria in fixed budding yeast cells. In: Gavin RH, editor. Cytoskeleton Methods and Protocols. Totowa, NJ: Humana Press; 2010. pp. 171–184. [DOI] [PubMed] [Google Scholar]
- Tamm T. Plasmids with E2 epitope tags: tagging modules for N- and C-terminal PCR-based gene targeting in both budding and fission yeast, and inducible expression vectors for fission yeast. Yeast. 2009;26:55–66. doi: 10.1002/yea.1650. [DOI] [PubMed] [Google Scholar]
- Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully GH, Nishihama R, Pringle JR, Morgan DO. The anaphase-promoting complex promotes actomyosin-ring disassembly during cytokinesis in yeast. Mol Biol Cell. 2009;20:1201–1212. doi: 10.1091/mbc.E08-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hou L, Qian M, Li F, Deng M. Integrating multiple types of data to predict novel cell cycle-related genes. BMC Syst Biol. 2011;5(Suppl 1):S9. doi: 10.1186/1752-0509-5-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Young CL, Raden DL, Caplan JL, Czymmek KJ, Robinson AS. Cassette series designed for live-cell imaging of proteins and high-resolution techniques in yeast. Yeast. 2012;29:119–136. doi: 10.1002/yea.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.