Abstract
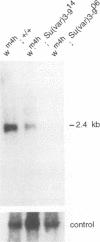
Modifier mutations of position-effect variegation (PEV) represent a useful tool for a genetic and molecular dissection of genes connected with chromatin regulation in Drosophila. The Su(var)3-9 gene belongs to the group of haplo suppressor loci which manifest a triplo enhancer effect. Mutations show a strong suppressor effect even in the presence of PEV enhancer mutations, indicating a central role of this gene in the regulation of PEV. By molecular analysis, Su(var)3-9 could be correlated with a 2.4 kb transcript which encodes a putative protein of 635 amino acids containing a chromo domain and a region of homology to Enhancer of zeste and trithorax, two antagonistic regulators of the Antennapedia and Bithorax gene complexes, as well as to the human protein ALL-1/Hrx which is implicated in acute leukemias. This region of homology is found in all four proteins at the C-terminus. The homology of Su(var)3-9 to both negative (Polycomb and Enhancer of zeste) and positive (trithorax) regulators of the Antennapedia and Bithorax complexes also suggests similarities in the molecular processes connected with stable transmission of a determined state and the clonal propagation of heterochromatinization.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anborgh P. H., Parmeggiani A., Jonák J. Site-directed mutagenesis of elongation factor Tu. The functional and structural role of residue Cys81. Eur J Biochem. 1992 Sep 1;208(2):251–257. doi: 10.1111/j.1432-1033.1992.tb17180.x. [DOI] [PubMed] [Google Scholar]
- Arai K., Clark B. F., Duffy L., Jones M. D., Kaziro Y., Laursen R. A., L'Italien J., Miller D. L., Nagarkatti S., Nakamura S. Primary structure of elongation factor Tu from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Brands J. H., Maassen J. A., van Hemert F. J., Amons R., Möller W. The primary structure of the alpha subunit of human elongation factor 1. Structural aspects of guanine-nucleotide-binding sites. Eur J Biochem. 1986 Feb 17;155(1):167–171. doi: 10.1111/j.1432-1033.1986.tb09472.x. [DOI] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. F., Elgin S. C. Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res. 1992 Nov 25;20(22):6067–6074. doi: 10.1093/nar/20.22.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V., Stokes D. G., Perry R. P. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., Krauss V., Reuter G., Saumweber H. The enhancer of position-effect variegation of Drosophila, E(var)3-93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., Szidonya J., Korge G., Sehnert M., Taubert H., Archoukieh E., Tschiersch B., Morawietz H., Wustmann G., Hoffmann G. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics. 1993 Feb;133(2):279–290. doi: 10.1093/genetics/133.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., James T. C., Foster-Hartnett D. M., Hartnett T., Ngan V., Elgin S. C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Morris G. D., Reuter G., Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992 Jun;131(2):345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C. Position effect variegation in Drosophila: towards a genetics of chromatin assembly. Bioessays. 1989 Jul;11(1):14–17. doi: 10.1002/bies.950110105. [DOI] [PubMed] [Google Scholar]
- Epstein H., James T. C., Singh P. B. Cloning and expression of Drosophila HP1 homologs from a mealybug, Planococcus citri. J Cell Sci. 1992 Feb;101(Pt 2):463–474. doi: 10.1242/jcs.101.2.463. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gatti M., Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annu Rev Genet. 1992;26:239–275. doi: 10.1146/annurev.ge.26.120192.001323. [DOI] [PubMed] [Google Scholar]
- Grigliatti T. Position-effect variegation--an assay for nonhistone chromosomal proteins and chromatin assembly and modifying factors. Methods Cell Biol. 1991;35:587–627. [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Gu Y., Nakamura T., Alder H., Prasad R., Canaani O., Cimino G., Croce C. M., Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell. 1992 Nov 13;71(4):701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hartmann-Goldstein I. J. On the relationship between heterochromatization and variegation in Drosophila, with special reference to temperature-sensitive periods. Genet Res. 1967 Oct;10(2):143–159. doi: 10.1017/s0016672300010880. [DOI] [PubMed] [Google Scholar]
- Hovemann B., Richter S., Walldorf U., Cziepluch C. Two genes encode related cytoplasmic elongation factors 1 alpha (EF-1 alpha) in Drosophila melanogaster with continuous and stage specific expression. Nucleic Acids Res. 1988 Apr 25;16(8):3175–3194. doi: 10.1093/nar/16.8.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. C., Elgin S. C. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986 Nov;6(11):3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. S., Gelbart W. M. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993 Oct;13(10):6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990 Oct 5;63(1):97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Locke J., Kotarski M. A., Tartof K. D. Dosage-dependent modifiers of position effect variegation in Drosophila and a mass action model that explains their effect. Genetics. 1988 Sep;120(1):181–198. doi: 10.1093/genetics/120.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo A. M., Huang D. H., Mozer B. A., Dawid I. B. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer S., Franke A., Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992 Jul;6(7):1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- Möller W., Amons R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 1985 Jul 1;186(1):1–7. doi: 10.1016/0014-5793(85)81326-0. [DOI] [PubMed] [Google Scholar]
- Nagata S., Nagashima K., Tsunetsugu-Yokota Y., Fujimura K., Miyazaki M., Kaziro Y. Polypeptide chain elongation factor 1 alpha (EF-1 alpha) from yeast: nucleotide sequence of one of the two genes for EF-1 alpha from Saccharomyces cerevisiae. EMBO J. 1984 Aug;3(8):1825–1830. doi: 10.1002/j.1460-2075.1984.tb02053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Orlando V., Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993 Dec 17;75(6):1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Nucleic acid hybridization to the DNA of cytological preparations. Methods Cell Biol. 1975;10:1–16. doi: 10.1016/s0091-679x(08)60727-x. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Hennig W. Heterochromatin: junk or collectors item? Chromosoma. 1990 Dec;100(1):3–7. doi: 10.1007/BF00337597. [DOI] [PubMed] [Google Scholar]
- Paro R., Hogness D. S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990 Dec;6(12):416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Parry P., Djabali M., Bower M., Khristich J., Waterman M., Gibbons B., Young B. D., Evans G. Structure and expression of the human trithorax-like gene 1 involved in acute leukemias. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4738–4742. doi: 10.1073/pnas.90.10.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce J. J., Singh P. B., Gaunt S. J. The mouse has a Polycomb-like chromobox gene. Development. 1992 Apr;114(4):921–929. doi: 10.1242/dev.114.4.921. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Powers J. A., Eissenberg J. C. Overlapping domains of the heterochromatin-associated protein HP1 mediate nuclear localization and heterochromatin binding. J Cell Biol. 1993 Jan;120(2):291–299. doi: 10.1083/jcb.120.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Giarre M., Farah J., Gausz J., Spierer A., Spierer P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990 Mar 15;344(6263):219–223. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- Reuter G., Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992 Sep;14(9):605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Reuter G., Szidonya J. Cytogenetic analysis of variegation suppressors and a dominant temperature-sensitive lethal in region 23-26 of chromosome 2L in Drosophila melanogaster. Chromosoma. 1983;88(4):277–285. doi: 10.1007/BF00292904. [DOI] [PubMed] [Google Scholar]
- Reuter G., Werner W., Hoffmann H. J. Mutants affecting position-effect heterochromatinization in Drosophila melanogaster. Chromosoma. 1982;85(4):539–551. doi: 10.1007/BF00327349. [DOI] [PubMed] [Google Scholar]
- Reuter G., Wolff I. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol Gen Genet. 1981;182(3):516–519. doi: 10.1007/BF00293947. [DOI] [PubMed] [Google Scholar]
- Roseman R. R., Pirrotta V., Geyer P. K. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993 Feb;12(2):435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Spradling A. C. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983 Sep 24;11(18):6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Chiang A., Bender W., Shimell M. J., O'Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993 Jul;158(1):131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- Singh P. B., Miller J. R., Pearce J., Kothary R., Burton R. D., Paro R., James T. C., Gaunt S. J. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991 Feb 25;19(4):789–794. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Karpen G. H. Sixty years of mystery. Genetics. 1990 Dec;126(4):779–784. doi: 10.1093/genetics/126.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982 Oct 22;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Tkachuk D. C., Kohler S., Cleary M. L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992 Nov 13;71(4):691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- Wedeen C., Harding K., Levine M. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell. 1986 Mar 14;44(5):739–748. doi: 10.1016/0092-8674(86)90840-8. [DOI] [PubMed] [Google Scholar]
- Wustmann G., Szidonya J., Taubert H., Reuter G. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol Gen Genet. 1989 Jun;217(2-3):520–527. doi: 10.1007/BF02464926. [DOI] [PubMed] [Google Scholar]
- Zink B., Paro R. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature. 1989 Feb 2;337(6206):468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]