Abstract
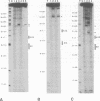
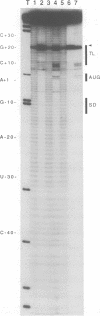
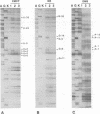
We footprinted the interaction of model mRNAs with 30S ribosomal subunits in the presence or absence of tRNA(fMet) or tRNA(Phe) using chemical probes directed at the sugar-phosphate backbone or bases of the mRNAs. When bound to the 30S subunits in the presence of tRNA(fMet), the sugar-phosphate backbones of gene 32 mRNA and 022 mRNA are protected from hydroxyl radical attack within a region of about 54 nucleotides bounded by positions -35 (+/- 2) and +19, extending to position +22 when tRNA(Phe) is used. In 70S ribosomes, protection is extended in the 5' direction to about position -39 (+/- 2). In the absence of tRNA, the 30S subunit protects only nucleotides -35 (+/- 2) to +5. Introduction of a stable tetraloop hairpin between positions +10 and +11 of gene 32 mRNA does not interfere with tRNA(fMet)-dependent binding of the mRNA to 30S subunits, but results in loss of protection of the sugar-phosphate backbone of the mRNA downstream of position +5. Using base-specific probes, we find that the Shine-Dalgarno sequence (A-12, A-11, G-10 and G-9) and the initiation codon (A+1, U+2 and G+3) of gene 32 mRNA are strongly protected by 30S subunits in the presence of initiator tRNA. In the presence of tRNA(Phe), the same Shine-Dalgarno bases are protected, as are U+4, U+5 and U+6 of the phenylalanine codon. Interestingly, A-1, immediately preceding the initiation codon, is protected in the complex with 30S subunits and initiator tRNA, while U+2 and G+3 are protected in the complex with tRNA(Phe) in the absence of initiator tRNA. Additionally, specific bases upstream from the Shine-Dalgarno region (U-33, G-32 and U-22) as well as 3' to the initiation codon (G+11) are protected by 30S subunits in the presence of either tRNA. These results imply that the mRNA binding site of the 30S subunit covers about 54-57 nucleotides and are consistent with the possibility that the ribosome interacts with mRNA along its sugar-phosphate backbone.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balakin A., Skripkin E., Shatsky I., Bogdanov A. Transition of the mRNA sequence downstream from the initiation codon into a single-stranded conformation is strongly promoted by binding of the initiator tRNA. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):119–123. doi: 10.1016/0167-4781(90)90151-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangu R., Wollenzien P. The mRNA binding track in the Escherichia coli ribosome for mRNAs of different sequences. Biochemistry. 1992 Jun 30;31(25):5937–5944. doi: 10.1021/bi00140a033. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontsova O., Dokudovskaya S., Kopylov A., Bogdanov A., Rinke-Appel J., Jünke N., Brimacombe R. Three widely separated positions in the 16S RNA lie in or close to the ribosomal decoding region; a site-directed cross-linking study with mRNA analogues. EMBO J. 1992 Aug;11(8):3105–3116. doi: 10.1002/j.1460-2075.1992.tb05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989 Dec;3(12A):1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A., Noller H. F. Hydroxyl radical cleavage of tRNA in the ribosomal P site. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):7851–7855. doi: 10.1073/pnas.89.17.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. W., Cantor C. R. Structure of ribosome-bound messenger RNA as revealed by enzymatic accessibility studies. J Mol Biol. 1985 Jan 20;181(2):241–251. doi: 10.1016/0022-2836(85)90088-9. [DOI] [PubMed] [Google Scholar]
- La Teana A., Pon C. L., Gualerzi C. O. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagunez-Otero J., Trifonov E. N. mRNA periodical infrastructure complementary to the proof-reading site in the ribosome. J Biomol Struct Dyn. 1992 Dec;10(3):455–464. doi: 10.1080/07391102.1992.10508662. [DOI] [PubMed] [Google Scholar]
- Langer J. A., Jurnak F., Lake J. A. Elongation factor Tu ternary complex binds to small ribosomal subunits in a functionally active state. Biochemistry. 1984 Dec 4;23(25):6171–6178. doi: 10.1021/bi00320a043. [DOI] [PubMed] [Google Scholar]
- Lill R., Wintermeyer W. Destabilization of codon-anticodon interaction in the ribosomal exit site. J Mol Biol. 1987 Jul 5;196(1):137–148. doi: 10.1016/0022-2836(87)90516-x. [DOI] [PubMed] [Google Scholar]
- McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. mRNA degradation by processive 3'-5' exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991 Sep 5;221(1):81–95. [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Murakawa G. J., Nierlich D. P. Mapping the lacZ ribosome binding site by RNA footprinting. Biochemistry. 1989 Oct 3;28(20):8067–8072. doi: 10.1021/bi00446a015. [DOI] [PubMed] [Google Scholar]
- Platt T., Yanofsky C. An intercistronic region and ribosome-binding site in bacterial messenger RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2399–2403. doi: 10.1073/pnas.72.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringquist S., MacDonald M., Gibson T., Gold L. Nature of the ribosomal mRNA track: analysis of ribosome-binding sites containing different sequences and secondary structures. Biochemistry. 1993 Sep 28;32(38):10254–10262. doi: 10.1021/bi00089a048. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel J., Jünke N., Brimacombe R., Dukudovskaya S., Dontsova O., Bogdanov A. Site-directed cross-linking of mRNA analogues to 16S ribosomal RNA; a complete scan of cross-links from all positions between '+1' and '+16' on the mRNA, downstream from the decoding site. Nucleic Acids Res. 1993 Jun 25;21(12):2853–2859. doi: 10.1093/nar/21.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengart M. L., Fatscher H. P., Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990 Apr 11;18(7):1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. N. Translation framing code and frame-monitoring mechanism as suggested by the analysis of mRNA and 16 S rRNA nucleotide sequences. J Mol Biol. 1987 Apr 20;194(4):643–652. doi: 10.1016/0022-2836(87)90241-5. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. Tetraloops and RNA folding. Nature. 1990 Aug 16;346(6285):613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- Wollenzien P., Expert-Bezançon A., Favre A. Sites of contact of mRNA with 16S rRNA and 23S rRNA in the Escherichia coli ribosome. Biochemistry. 1991 Feb 19;30(7):1788–1795. doi: 10.1021/bi00221a009. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Kozarich J. W., Stubbe J. The mechanism of free base formation from DNA by bleomycin. A proposal based on site specific tritium release from Poly(dA.dU). J Biol Chem. 1983 Apr 25;258(8):4694–4697. [PubMed] [Google Scholar]
- Zamir A., Miskin R., Elson D. Inactivation and reactivation of ribosomal subunits: amino acyl-transfer RNA binding activity of the 30 s subunit of Escherichia coli. J Mol Biol. 1971 Sep 14;60(2):347–364. doi: 10.1016/0022-2836(71)90299-3. [DOI] [PubMed] [Google Scholar]