Abstract
A series of novel, potent antagonists of the inhibitor of apoptosis proteins (IAPs) were synthesized in a highly convergent and rapid fashion (≤ 6 steps) using the Ugi four-component reaction as the key step, thus enabling rapid optimization of binding potency. These IAP antagonists compete with caspases 3, 7 and 9 for inhibition by X chromosome-linked IAP (XIAP) and bind strongly (nanomolar binding constants) to several crucial members of the IAP family of cancer pro-survival proteins to promote apoptosis, with a particularly unique selectivity for melanoma IAP (ML-IAP). Experiments in cell culture revealed powerful cancer cell growth inhibitory activity in multiple (breast, ovarian and prostate) cell lines with single agent toxicity at low nanomolar levels against SKOV-3 human ovarian carcinoma cells. Administration of the compounds to human foreskin fibroblast cells revealed no general toxicity to normal cells. Furthermore, computational modeling was performed, revealing key contacts between the IAP proteins and antagonists, suggesting a structural basis for the observed potency.
Although apoptosis (programmed cell death) is an essential part of normal homeostasis, the evasion of apoptosis by cells is one of the defining hallmarks of cancer.1 While advances in cancer chemotherapeutics over the last few years have improved life expectancy in many cases, the onset of intrinsic or acquired resistance remains a major barrier to effective treatment.2 Defective apoptotic signalling by caspases, a family of intracellular proteases, is an underlying cause of resistance to cell death.3 The activity of caspases is suppressed by a number of endogenous proteins, foremost among them being the inhibitor of apoptosis proteins (IAPs).4,5 In humans the IAP family consists of eight members, including X chromosome-linked IAP (XIAP), cellular IAP 1 (cIAP1), cellular IAP 2 (cIAP2), and melanoma IAP (ML-IAP). Each of the IAPs contains regions called baculoviral IAP repeat (BIR) domains which are 70–80 amino acids in length. In XIAP, the BIR2 domain and the linker preceding it inhibit the effector caspases 3 and 7, while BIR3 binds to, and antagonizes, the initiator caspase 9. The second mitochondria-derived activator of caspases (Smac) protein is an endogenous dimeric proapoptotic antagonist of XIAP. Acting through the intrinsic apoptotic pathway, Smac is released into the cytosol from the mitochondrial intermembrane space in response to cellular stress. Specifically, it is the Smac N-terminal AVPI sequence (Scheme 1B) that directly binds to a well, defined surface groove created by the BIR domains of each IAP family member and de-represses the actions of caspases, allowing apoptosis to proceed. Thus, the inhibition of IAPs by small molecules, or “inhibiting the inhibitors”, is a highly promising approach for the treatment of cancer.
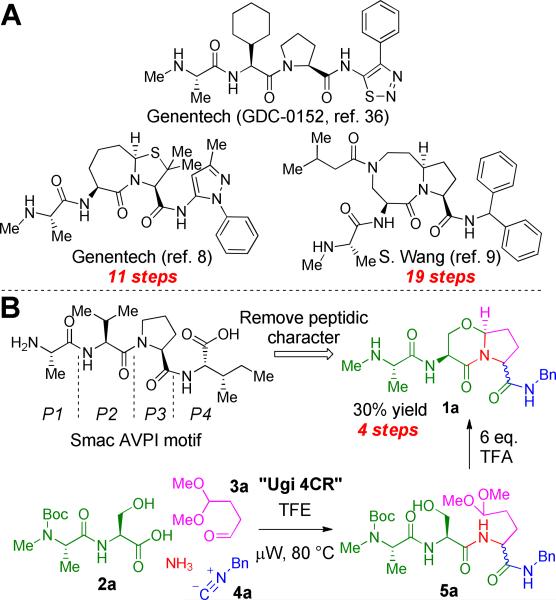
Scheme 1.
(A) Previous Smac mimetics from Genentech and S. Wang are based on the crucial N-terminal AVPI binding domain, but suffer from lengthy syntheses. (B) Novel Smac mimetic 1a can be accessed quickly in two chemical steps from dipeptide 2a.
Following initial studies by Fesik and co-workers,6,7 several research groups initiated programs directed at developing IAP antagonists that mimic the AVPI tetrapeptide sequence (Scheme 1A). For example, the synthesis and characterization of drug-like bicyclic peptidomimetics were reported separately by Genentech,8 S. Wang9 and P. Seneci.10 In addition to being potent IAP antagonists in vitro and in cells, these compounds exhibited promising drug-like properties, a logical consequence of their reduced peptidic nature compared with AVPI.
While such Smac peptidomimetics appear to be promising targets, previous syntheses of this framework have generally been laborious, requiring numerous (11–19) synthetic steps and purifications. A notable drawback to the reported procedures is their linear nature, in effect creating a bottleneck for rapid lead optimization. We therefore envisaged a scaffold that could mimic the pertinent interactions of AVPI with IAPs, avoid the typical issues associated with peptides as pharmaceutical agents, and yet could also be synthesized rapidly and efficiently in convergent fashion. This led us to hypothesize that peptidomimetic 1a and its derivatives might be both synthetically accessible and lead to potent, drug-like IAP antagonists.4 Although the [4,3,0]-bicyclic lactam core is known and has been studied for its propensity to adopt a reverse-turn conformation, application of previous methods to assemble 1a would require a lengthy linear synthesis or necessitate the use of specialized reaction conditions, such as anodic oxidation.11-18 We theorized that use of the Ugi four-component reaction (Ugi 4CR) had the potential to provide rapid access to the desired heterobicyclic structures.19 Utilization of this novel paradigm, if realized, would bring about the formation of six bonds and two stereocenters (one stereoselectively) over two steps. Herein we report the synthesis of novel, potent IAP antagonists via the highly efficient application of the Ugi 4CR.
RESULTS AND DISCUSSION
Synthetic proof of concept
Our initial test of the feasibility of using the Ugi 4CR as the key step in the construction of compound 1a is shown in Scheme 1B. Dipeptide 2a,20 ammonia, butanedial monoacetal (3a)21 and commercially available benzyl isocyanide (4a, R=Bn) were stirred in 2,2,2-trifluoroethanol (TFE)22 under microwave irradiation at 80 °C for 20 min. We were delighted to find that the Ugi 4CR product 5a (R=Bn) was produced cleanly as a 1:1 mixture of diastereomers. Next, to test the stereoselective formation of the 6,5-heterobicycle 1a (R=Bn), a six-fold molar excess of trifluoroacetic acid (TFA) was added to the crude product 5a from the previous step. As a result, several transformations were accomplished in one pot: acid-induced oxocarbenium ion formation and capture by the amide nitrogen to form the five-membered ring, loss of methanol from the resulting N,O-acetal to give a transient N-acyliminium ion which was stereoselectively trapped by the pendant serine hydroxyl group to generate the bicycle, and finally cleavage of the N-terminal amine Boc-protecting group. The resulting stereochemistry at the N,O-acetal stereocenter was exclusively as shown and was controlled by the configuration at L-serine.11-18 The product was then purified by flash chromatography over basic alumina to yield peptidomimetic 1a in 30% overall yield from 2a.
Expansion of the reaction scope
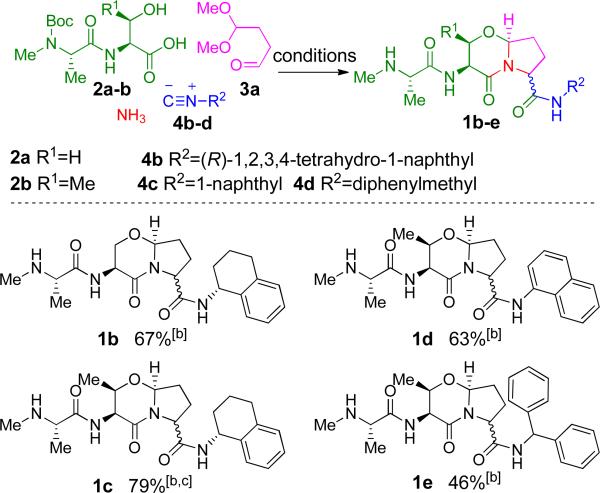
With a robust method in place, we then sought to expand the substrate scope by varying the Ugi 4CR carboxylic acid and isocyanide components. As shown in Figure 1, the methodology was extended to various readily available starting materials, together with aldehyde 3a and ammonia. The Ugi 4CR was conducted as before (Scheme 1B), followed by treatment with TFA. Pleasingly, two-step yields were good despite the entropic disadvantage of constructing comparatively large scaffolds in a single step. Both aliphatic and aromatic isocyanides performed well in the reaction, generally giving overall yields above 60% for the two step process.
Figure 1.
Ugi 4CR results: expansion of the substrate scope. [a] Conditions: 1 eq. 2a or 2b, 1.05 eq. 3a, 1 eq. 4b, 4c or 4d in TFE, μW at 80 °C, 20 min; 6 eq. TFA, DCM, 23 °C; [b] 1:1 d.r. [c] Yield is of semi-pure 1c, which was further purified by reverse-phase HPLC for analysis.
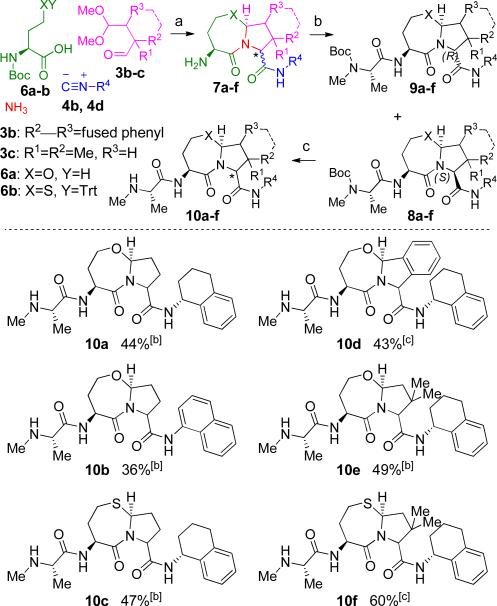
The ambit of the reaction sequence was further increased by varying the carboxylic acid and aldehyde components to generate a more diverse set of heterocycles. In place of the dipeptides, the protected amino acids 6a (X=O) and 6b (X=S) were utilized, as shown in Figure 2. This modification served two purposes: first, preparation of the homoserine analog of 2a (Scheme 1B) was complicated by formation of an inseparable γ-lactone byproduct.23 Second, the change allowed for chromatographic separation of the two diastereomers 8 and 9. The reaction sequence was conducted as shown in Figure 2 with the Ugi 4CR followed by the aforementioned acid-induced reaction cascade. However, primary amine 7 was isolated and used without purification in the coupling reaction with Boc-N-Me-Ala-OH to give a 1:1 mixture of diastereomers 8 and 9. For the majority of entries (10a–c, 10e), the two diastereomers were fully or partially separated by a single flash chromatography on silica gel. A final TFA-induced removal of the amine protecting group gave compounds 10a–f in impressive overall yields (36–60%) for the four step sequence with only a single purification. As may be expected, the homocysteine-derived compounds (10c and 10f, 47% and 60%, respectively) were delivered in higher yields compared to their oxygen-containing counterparts (10a and 10e, respectively), with 10e and 10f benefiting additionally from the Thorpe-Ingold (gem-dialkyl) effect24 during ring closure.
Figure 2.
Further expansion of the Ugi 4CR product output. [a] Conditions: a) 1 eq. 6a (XY=OH) or 6b (XY=STrt), 1.05 eq. 3a, 3b or 3c, 1 eq. 4b or 4d in TFE, μW at 80 °C, 20 min; 8–10 eq. TFA, DCM, 32 °C; b) Boc-N-Me-Ala-OH, HOBT, EDC, NMM, THF; c) 8 eq. TFA, DCM 23 °C; [b] Total yield given; 10a–c and 10e were separated as enriched diastereomers (≥3:1 d.r., see SI for details) after third step for analysis; [c] Diastereomers not separated.
Utility of the convertible isocyanide
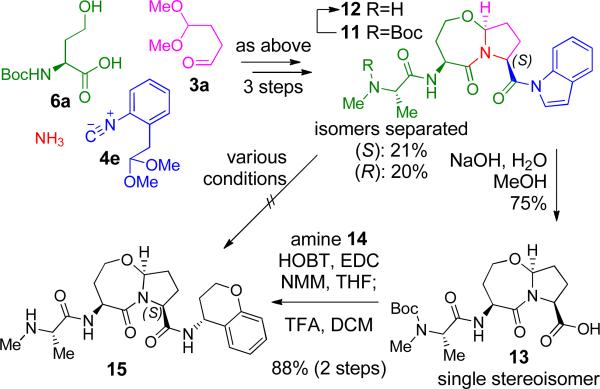
In order to expand the potential product scope, application of a convertible isocyanide, an activated carboxylate surrogate which allows for selective functionalization of the C, terminal amide, was investigated (Scheme 2). In theory, isocyanide 4e25 fulfilled our needs well; its conversion from the acetanilide after the Ugi 4CR to N-acylindole is achieved under moderately acidic conditions, similar to those used for conversion from 5a to 1a (Scheme 1B), thus requiring no additional steps.26
Scheme 2.
N-Acylindole 11 as a handle for the synthesis of stereochemically pure C-terminal carboxylate derivatives.
Thus, isocyanide 4e was used with 6a, 3a and ammonia in the 3 step reaction sequence to yield pure 11 (21% 3-step yield + 20% R-isomer, separated isomers) as shown in Scheme 2. In an attempt to directly displace the indole moiety, 11 was subjected to treatment with several nucleophiles to no avail; degradation was predominant to product formation in these cases. This resistance to nucleophilic attack and substitution is surprising given the well, established propensity of N-acylindoles to undergo this type of reactivity, as demonstrated by Fukuyama.25 Extrapolation of the three-dimensional structure of 11 from the X-ray crystal structure of the similar naphthyl amide 8b (Figure 3A) indicates that steric hindrance at the reactive carbonyl should not be a significant barrier to reaction. Fortunately, hydrolysis of 11 proceeded well and carboxylic acid 13 was isolated in 75% yield. A subsequent coupling reaction with (R)-chroman-4-ylamine (14) followed by deprotection gave the final compound 15 in 88% yield. Thus, the synthetic strategy described in Scheme 2 adds versatility by allowing for the synthesis of a variety of C-terminal carboxylate derivatives in stereochemically pure form.
Figure 3.
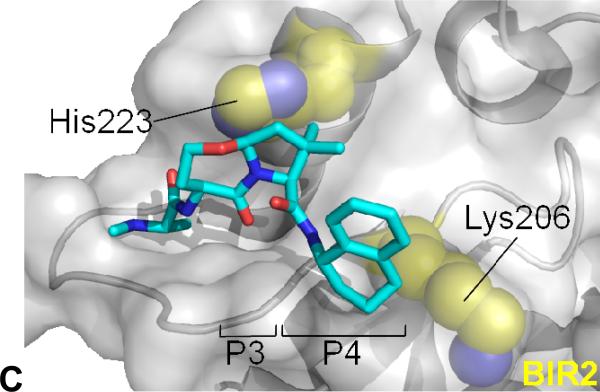
(A) Compound 8b (ellipsoids at 50% probability level) as determined by single crystal X-ray analysis. (B) Compound 10e (cyan) modelled onto the surface groove occupied by Smac in complex with cIAP1 BIR3 (grey surface) and (C) cIAP1 BIR2. (B) 10e was superimposed on PBD entry 3MUP followed by a conformational search over rotatable bonds. Key contacts Trp323, Phe324 and Gly306 indicated (brown). (C) BIR2 from 1I3O was overlayed on the model and is displayed with compound 10e. Key contacts His223 and Lys206 (equivalent to Trp 323 and Gly306 of cIAP1 BIR3) indicated (yellow).
Compound 8b was further analyzed by single crystal X-ray diffraction analysis (Figure 3A).27 The refined structure revealed that the more polar of the two diastereomers possesses the desired stereochemistry, corresponding to the AVPI stereoconfiguration at P3 (Scheme 1B). The stereochemistry of the separated isomers of the remaining compounds in Figure 2 was similarly and tentatively correlated based on polarity due to their structural similarity.28
Biological activity of IAP antagonists
The new compounds were evaluated in a panel of relevant biochemical assays to determine their activity against IAP family proteins. To measure binding potency in vitro, a series of fluorescence polarization (FP) competition assays were employed independently for the BIR domains of XIAP, cIAP1, cIAP2 and ML-IAP (Table 1).29 Most of the compounds synthesized and tested showed only low to moderate levels of binding to the BIR2 domain of XIAP, with 10e showing the best potency (Ki=9.64 μM). We previously showed that IAP antagonists possessing a C-terminal 1-naphthyl amide exhibited more potent binding to the BIR2 domain relative to BIR3 of XIAP;30 however, this effect was not apparent for the present compounds (e.g. 1d, 10b).
Table 1.
Fluorescence polarization competition assay data.
| XIAP | cIAP1 | cIAP2 | ML-IAP | ||
|---|---|---|---|---|---|
| Cmpd[a] | BIR2 Ki (μM) | BIR3 Ki (μM) | BIR3 Ki (μM) | BIR3 Ki (μM) | Ki (μM) |
| 1a | >56.0 | >39.1 | 8.98 | 28.9 | >25 |
| 1b | >56.0 | 13.3 | 1.18 | 3.31 | 8.33 |
| 1c | >56.0 | 6.87 | 1.05 | 2.70 | 5.07 |
| 1d | 51.0 | >39.1 | 5.36 | 9.71 | >7.5 |
| 1e | >56.0 | 28.3 | 1.49 | 3.81 | >7.5 |
| (S)-10a | >56.0 | 0.44 | 0.061 | 0.149 | 0.344 |
| (S)-10b | 17.2 | 9.23 | 0.411 | 0.799 | 4.71 |
| (S)-10c | >56.0 | 0.61 | 0.130 | 0.260 | 0.043 |
| 10d | >56.0 | 0.93 | 0.034 | 0.056 | 0.314 |
| (S)-10e | 9.64 | 0.10 | 0.022 | 0.047 | 0.002 |
| 10f | 15.6 | 0.27 | 0.085 | 0.116 | 0.004 |
| (S)-12 | >56.0 | 32.6 | 6.97 | 9.72 | >25 |
| (S)-15 | >56.0 | 0.35 | <0.010 | 0.071 | 0.294 |
| (R)-15 | >56.0 | >39.1 | 12.7 | 27.2 | ND |
| GDC- | 29.8 | 0.258 | 0.102 | 0.144 | 0.041 |
| 0152[b] | (0.112) | (0.028) | (0.017) | (0.043) | (0.014) |
More promising results were observed for the XIAP BIR3 domain binding data. Initial assay results obtained for the 6,5-bicyclic compounds (1a–1e), while modest, were important in verifying the scaffold as a potential lead structure as well as providing some vital preliminary SAR data. The most potent XIAP BIR3 ligand of the 6,5-bicyclics, 1c (P2 derived from threonine), gave a Ki of 6.87 μM. This is compared to the two-fold lower binding affinity for 1b (P2 derived from serine) against XIAP BIR3, suggesting that the area of the pocket occupied by the P2 region of 1c may be receptive to substituents. The 6,5-bicyclic derivatives (1a–1e) also exhibited micromolar binding affinities for the BIR3 domains of cIAP1 and cIAP2, results which helped to confirm the multi-IAP antagonist activity of the series. A significant breakthrough in potency was made, however, in the 7,5-bicyclic series. Compound 10a exhibited a thirty-fold increase in binding affinity for XIAP BIR3 (Ki=440 nM) and was similarly potent against cIAP1 and cIAP2 BIR3, with Ki values of 61 and 149 nM, respectively. In addition, the analog of 10a possessing a chroman-4-ylamide [(S)-15] in place of the tetrahydronaphth-1-ylamide displayed a similar potency profile to 10a. Substitution of oxygen for sulfur within the seven-membered heterocyclic ring (P2: homoserine→homocysteine), as in 10a→10c, resulted in a slight reduction in potency against XIAP, cIAP1 and cIAP2. On the other hand, the sulfur-containing 10c showed a ten-fold improvement in binding affinity for the BIR domain of ML-IAP (Ki=43 nM) compared with 10a (Ki=344 nM). Furthermore, introduction of a gem-dimethyl group to the P3 pyrrolidine ring, as in 10e and 10f, resulted in a significant improvement in binding affinity, especially against ML-IAP. Thus, the Ki values for 10e and 10f against ML-IAP are 2 nM and 4 nM, respectively. These results are significant given that ML-IAP/Livin31-33 is a member of the IAP family of proteins that has been shown to be a potent Smac ligand and validated as a target for anticancer therapy.34,35 Interestingly, the phenyl-fused tricyclic derivative 10d was more potent against cIAP1 and cIAP2 than XIAP BIR3 and ML-IAP. Overall, the SAR data indicate that there are exploitable hydrophobic regions within the protein active site on both sides of the core heterobicyclic scaffold. Finally, the clinical candidate GDC-0152 recently reported by Genentech (see Scheme 1A) was tested side-by-side in the FP assays for comparative purposes. Interestingly, we found the compound to be less potent in our hands than the reported values.36
Unique selectivity for ML-IAP
The exceptional level of ML-IAP binding of 10e and 10f merits further comment. Previously reported IAP antagonists typically exhibit a higher level of binding to this IAP family member relative to XIAP, but generally no more than a range of 2- to 10-fold (see Table 1, GDC-0152).8,37,38 The 50- and 67-fold increase seen with 10e and 10f, respectively, is certainly unique for small molecule IAP antagonists and could explain the significant increase (relative to the XIAP BIR2/3 binding values) in induction of cell death in certain cancer cell lines. In addition, it has been shown that ML-IAP is not widely expressed in normal tissues, and so specificity for this IAP member could be important and exploitable in a therapeutic context.39
Molecular modelling provides insights to binding
We next investigated the putative interactions of compound 10e with the BIR3 domain of cIAP1 and the BIR2 domain of XIAP using modelling in silico.10,40 Our studies implicate several factors that could account for the observed BIR selectivity of this compound (Figures 3B and 3C). Firstly, binding of 10e to the BIR2 domain of XIAP appears to be disfavored due to a less adept interaction between the 7,5-bicyclic structure (P2–P3) and His223 as well as clashes between the C-terminal substituent in P4 and Lys206 (Figure 3C), consistent with findings on peptide selectivity differences between BIR2 and BIR3 domains.41 The BIR3 domain of cIAP1 (Figure 3B), as well as XIAP and ML-IAP, contain strictly conserved Trp- (Trp323 in cIAP1) and Gly-residues (Gly306 in cIAP1) in these positions that are more suitable to accommodate compound 10e and related scaffolds relative to BIR2. The model further illustrates that the gem-dimethyl group in the P3 pyrrolidine ring of 10e aids binding to the BIR3 domain of cIAP1 by efficiently occupying a hydrophobic space between Trp323 and Phe324 of cIAP1 BIR3 (Figure 3B).
Potent anticancer effects of IAP antagonists
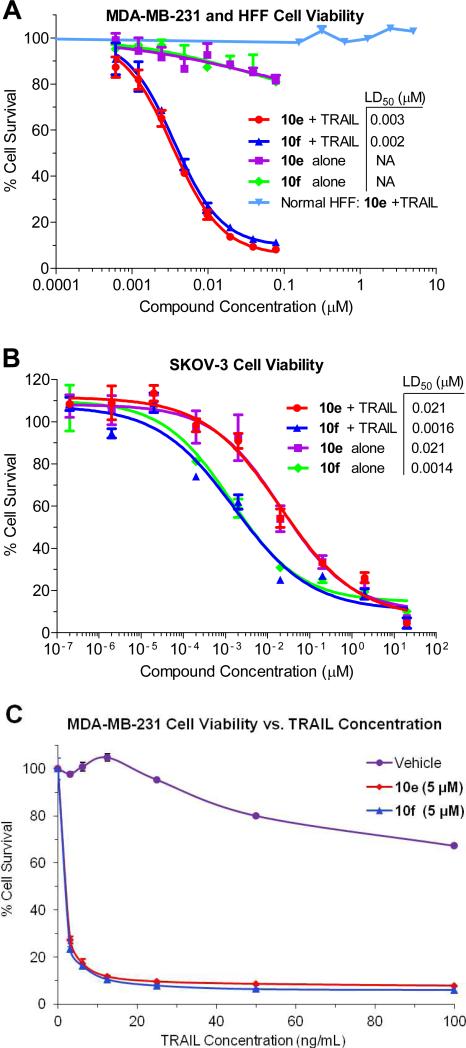
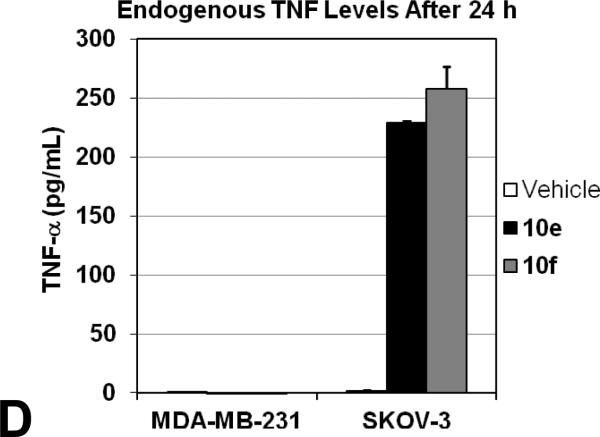
Based on the impressive in vitro data, IAP antagonists 10e and 10f were tested to determine their effects on cancer cell viability in a relevant cellular context. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a member of the TNF family of cytokines which triggers apoptosis via binding to the cell surface death receptors DR4 and DR5.42 TRAIL has been shown to act in combination with other therapeutic agents, including IAP antagonists, to induce tumor cell death.4 We therefore tested the ability of 10e and 10f to sensitize TRAIL-resistant MDA-MB-231 breast adenocarcinoma cells to apoptosis.23 As shown in Figure 4A, 10e and 10f each potently sensitized MDA-MB-231 cells to a concentration of TRAIL (5 ng/mL) that did not induce cell death alone, with LD50 values of 3 and 2 nM, respectively. Compounds 10e and 10f also induced cell death in SKOV-3 ovarian cancer cells as single agents (no TRAIL) at low nanomolar concentrations (Figure 4B). Interestingly, when 10e and 10f were co-administered with TRAIL (100 ng/mL), no significant difference in potency was observed.43 SKOV-3 cells may be sensitive to these Smac mimetics in the absence of exogenous TRAIL due to endogenous production of TNF family cytokines, as has been observed with other IAP antagonists and confirmed in our hands with 10e and 10f (Figure 5D).44 This is consistent with 10e and 10f being potent inhibitors of cIAP1 and cIAP2, proteins which suppress apoptosis in the context of TNF receptor signalling. Finally, 10e was tested against human foreskin fibroblasts (HFF) to evaluate its effects in normal cells (Figure 4A). No toxicity was observed, even in the presence of TRAIL (100 ng/mL). Taken together, these data suggest the potential utility of our novel IAP antagonists as therapeutic agents to treat certain forms of cancer with minimal adverse effects.
Figure 4.
Cell death induction by IAP antagonists. (A) MDA,MB,231 cells were treated with compounds 10e or 10f and 0 or 5 ng/mL TRAIL. The ability of the IAP antagonists to promote apoptosis is in stark contrast to their effect on non-cancerous HFF cells (100 ng/mL TRAIL). (B) SKOV-3 cells were treated with compounds 10e or 10f and 0 or 5 ng/mL TRAIL. (C) MDA-MB-231 cell viability dose-response curves for TRAIL-induced apoptosis (20 h) in the presence of vehicle or 5 M of 10e or 10f.
Figure 5.
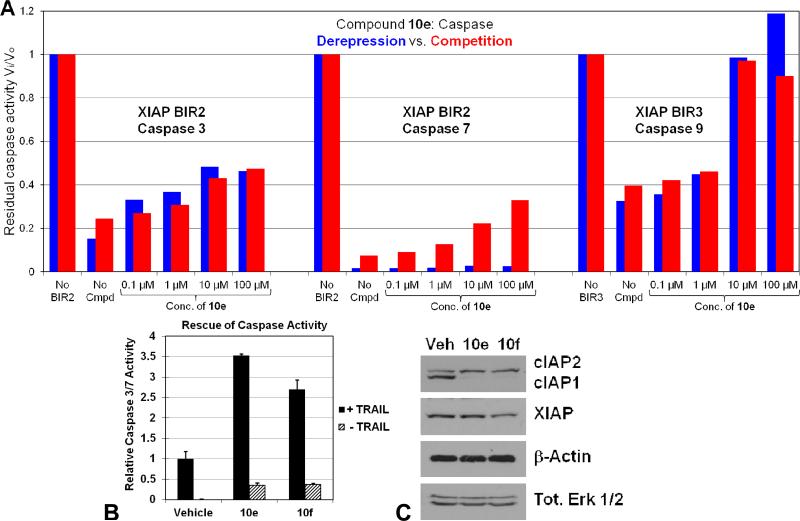
Caspase rescue ability of Smac mimetics. (A) Purified recombinant human caspases 3 [0.1 nM], -7 [1 nM] or -9 [2.2 TM] were used in combination with XIAP BIR2 [1 nM] and BIR3 [2.1 TM]. During the derepression assay (blue), caspases and the BIRs were allowed to form a complex for 30 minutes before the addition of increasing concentrations of 10e. For the competition assay (red), increasing concentrations of 10e were added at the same time as the enzyme and inhibitor. The ratio of inhibited to uninhibited caspase activity (Vi/Vo) was obtained by monitoring the release of fluorescence from the fluorogenic substrates Ac-DEVD-afc for caspases 3 and 7 or Ac-LEHD-afc for caspase 9. (B) Caspase 3/7 activity assays in MDA-MB-231 cells pretreated with vehicle (0.1% DMSO) or 5 M of 10e or 10f before treatment with 100 or 0 ng/mL TRAIL for 4 h. Activity is normalized to that of vehicle + or – TRAIL values, respectively, and all assays are carried out in triplicate as least twice. (C) Immunoblot analysis of cIAP1/2 and XIAP expression in MDA-MB-231 cells treated with vehicle (0.1% DMSO) or 5 M of 10e or 10f for 24 h. Tot. Erk 1/2 and β-actin are shown as equal loading controls. (D) Treatment of SKOV-3 cells with 5 M of 10e or 10f for 24 h resulted in robust production of TNF while the same treatment of MDA-MB-231 cells failed to produce any detectable TNF.
The resistance of MDA-MB-231 cells to TRAIL-induced apoptosis and the potent ability of 10e and 10f to rescue this condition are demonstrated in Figure 4C. The impact of TRAIL alone was only first observed at a concentration of 30 ng/mL, and at the maximum dose of 100 ng/mL only 30% of the cells had undergone cell death. This contrasts sharply with MDA-MB-231 cell viability in the presence of 10e and 10f (5 M) which demonstrated maximum cell death induction at around 10 ng/mL TRAIL in both cases.
Single agent toxicity, although indicative of the power of the small molecule to illicit a cellular response, may not be a desirable feature for a therapeutic agent, in the case of IAP antagonists. This kind of activity invokes activation of the non-canonical NF-κB inflammatory pathway for the production of TNF-related death receptor ligands which, although attractive on a cellular level, may be less desirable in a clinical setting.5 Thus, this point of distinction between our compounds and previously reported small- molecule Smac mimetics, at least in regards to the MDA-MB-231 cell line, could be advantageous.
Abrogation of IAP inhibition of caspases
To demonstrate that the observed binding of our IAP antagonists to IAP family members results in concomitant enhanced caspase activity, several experiments were performed (Figure 5). As an initial test, the IAP antagonists were evaluated in competition and derepression (caspase and XIAP BIR domain premixed) dose-response assays with the appropriate caspase-XIAP BIR domain combination (Figure 5A). Consistent with its relative affinity for the BIR2 and BIR3 domains of XIAP (Table 1), the Smac mimetic 10e showed good rescue of caspase 9 activity (BIR3), even to levels equal to that involving no BIR3 at higher concentrations. Caspase 3 and 7 (BIR2) rescue ability was somewhat attenuated, especially in the case of caspase 7. Interestingly, 10e was in effect equally capable of outcompeting and derepressing the caspase-IAP complex for caspases 3 and 9, but only showed moderate competition and negligible derepression for caspase 7, likely because caspase 7 is more strongly inhibited by XIAP than is caspase 3, and thus more difficult to derepress. This finding is fully consistent with the two-site binding mode of inhibition of caspases by XIAP, and that mutation of the caspase, interacting groove on BIR2 has a larger impact on caspase 7 inhibition than on caspase 3.45
Caspase activity was then investigated in MDA-MB-231 cells treated with TRAIL in conjunction with 5 μM 10e or 10f (Figure 5B). Both compounds showed marked ability to promote caspase 3/7 activity, with 10e eliciting a ≥3.5-fold increase over vehicle. A more modest impact on caspase activity was observed when 10e and 10f were added in the absence of TRAIL (Figure 5B), consistent with the MDA-MB-231 cell viability data (Figure 4A). In addition, immunoblot analysis of MDA-MB-231 cell lysates revealed that 10e and 10f induce degradation of cIAP1 within minutes (only 24 h time point shown), consistent with other previously described Smac mimetics in the same cell line (Figure 5C). Taken together, these data show a direct correlation between the IAP inhibitory activity of the compounds and a caspase-dependent mechanism.
CONCLUSION
In conclusion, we have developed a highly efficient synthesis of novel IAP antagonists with potent activity both in binding and cellular assays. In addition, the high selectivity of the present Smac mimetics for ML-IAP over XIAP is unique. The defining features of our approach include its brevity, operational simplicity with relatively few purifications, and convergent nature of the synthesis allowing access to diverse structures. These new compounds should prove to be valuable tools that can be used to explore the role of ML-IAP in normal and cancerous cells, which is the topic of current research. Expansion of the methodology for the synthesis of additional scaffolds and their biological evaluation is underway and will be reported in due course.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants U54 HG00503, U01 CA113318, R03 MH081277 and CA081534. We thank A. Rheingold and Y. Su of UCSD for X-ray and MS analysis, respectively, and J. Badger (DeltaG technologies) for structural modelling software.
ABBREVIATIONS
- IAPs
Inhibitor of apoptosis proteins
- XIAP
X chromosome-linked IAP
- cIAP
cellular IAP
- ML-IAP
melanoma IAP
- BIR
baculoviral IAP repeat
- Smac
second mitochondria-derived activator of caspases
- Ugi 4CR
Ugi four-component reaction
- TFE
2,2,2-trifluoroethanol
- TFA
trifluoroacetic acid
- Bn
benzyl
- FP
fluorescence polarization
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- HFF
human foreskin fibroblasts
Footnotes
Author Contributions
M.V. designed the synthetic strategy, performed all of the chemistry and co-wrote the paper. K.W. performed the XIAP and cIAP FP assays along with the MDA-MB-231 PPC-1 cell viability assays. D.F. conducted caspase 3/7 activity assays, immunoblot analysis and TNF ELISA assay, along with TRAIL dose-response and normal HFF cell viability assays. P.S.L. conducted the ML-IAP FP assays along with the SKOV-3 cell viability assays. P.D.M. performed the modelling computations. G.S.S. designed and S.J.S. and M.L.G. performed the caspase derepression studies. S.R.G. and R.J.A. synthesized GDC-0152. N.D.P.C. co-wrote the paper and conceived the Smac mimetic scaffold. S.J.R., K.V., G.S.S., J.C.R. and N.D.P.C. supervised scientific work.
Supporting Information
Experimental procedures and data can be obtained free of charge via the internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman MM. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 3.Pop C, Salvesen GS. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S. Design of small-molecule Smac mimetics as IAP antagonists. In: Vassilev L, Fry D, editors. Small-Molecule Inhibitors of Protein-Protein Interactions. Springer; Berlin: 2011. pp. 89–113. [DOI] [PubMed] [Google Scholar]
- 5.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discovery. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 6.Oost TK, Sun C, Armstrong RC, Al-Assaad A-S, Betz SF, Deckwerth TL, Ding H, Elmore SW, Meadows RP, Olejniczak ET, Oleksijew A, Oltersdorf T, Rosenberg SH, Shoemaker AR, Tomaselli KJ, Zou H, Fesik SW. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J. Med. Chem. 2004;47:4417–4426. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 8.Zobel K, Wang L, Varfolomeev E, Franklin MC, Elliott LO, Wallweber HJA, Okawa DC, Flygare JA, Vucic D, Fairbrother WJ, Deshayes K. Design, synthesis, and biological activity of a potent Smac mimetic that sensitizes cancer cells to apoptosis by antagonizing IAPs. ACS Chem. Biol. 2006;1:525–533. doi: 10.1021/cb600276q. [DOI] [PubMed] [Google Scholar]
- 9.Cai Q, Sun H, Peng Y, Lu J, Nikolovska-Coleska Z, McEachern D, Liu L, Qiu S, Yang C-Y, Miller R, Yi H, Zhang T, Sun D, Kang S, Guo M, Leopold L, Yang D, Wang S. A potent and orally active antagonist (SM, 406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J. Med. Chem. 2011;54:2714–2726. doi: 10.1021/jm101505d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cossu F, Malvezzi F, Canevari G, Mastrangelo E, Lecis D, Delia D, Seneci P, Scolastico C, Bolognesi M, Milani M. Recognition of Smac-mimetic compounds by the BIR domain of cIAP1. Protein Sci. 2010;19:2418–2429. doi: 10.1002/pro.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun H, Martin C, Kesselring D, Keller R, Moeller KD. Building functionalized peptidomimetics: use of electroauxiliaries for introducing N-acyliminium ions into peptides. J. Am. Chem. Soc. 2006;128:13761–13771. doi: 10.1021/ja064737l. [DOI] [PubMed] [Google Scholar]
- 12.Slomczynska U, Chalmers DK, Cornille F, Smythe ML, Beusen DD, Moeller KD, Marshall GR. Electrochemical cyclization of dipeptides to form novel bicyclic, reverse,turn peptidomimetics. 2. Synthesis and conformational analysis of 6,5-bicyclic systems. J. Org. Chem. 1996;61:1198–1204. [Google Scholar]
- 13.Baldwin JE, Hulme C, Schofield CJ, Edwards AJ. Synthesis of potential β-turn bicyclic dipeptide mimetics. J. Chem. Soc., Chem. Commun. 1993:935–936. [Google Scholar]
- 14.Claridge TDW, Hulme C, Kelly RJ, Lee V, Nash IA, Schofield CJ. Synthesis and analysis of Leu-enkephalin analogues containing reverse turn peptidomimetics. Bioorg. Med. Chem. Lett. 1996;6:485–490. [Google Scholar]
- 15.Robl JA, Sun C-Q, Stevenson J, Ryono DE, Simpkins LM, Cimarusti MP, Dejneka T, Slusarchyk WA, Chao S, Stratton L, Misra RN, Bednarz MS, Asaad MM, Cheung HS, Abboa-Offei BE, Smith PL, Mathers PD, Fox M, Schaeffer TR, Seymour AA, Trippodo NC. Dual metalloprotease inhibitors: mercaptoacetyl-based fused heterocyclic dipeptide mimetics as inhibitors of angiotensin-converting enzyme and neutral endopeptidase. J. Med. Chem. 1997;40:1570–1577. doi: 10.1021/jm970041e. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Jiang W, Schmitt AC. A convenient and versatile synthesis of 6,5- and 7,5-fused bicyclic lactams as peptidomimetics. Tet. Lett. 2001;42:4943–4945. [Google Scholar]
- 17.Chiou W-H, Mizutani N, Ojima I. Highly efficient synthesis of azabicyclo[x.y.0]alkane amino acids and congeners by means of Rh-catalyzed cyclohydrocarbonylation. J. Org. Chem. 2006;72:1871–1882. doi: 10.1021/jo061692y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornille F, Slomczynska U, Smythe ML, Beusen DD, Moeller KD, Marshall GR. Electrochemical cyclization of dipeptides toward novel bicyclic, reverse-turn peptidomimetics. 1. Synthesis and conformational analysis of 7,5- bicyclic systems. J. Am. Chem. Soc. 1995;117:909–917. [Google Scholar]
- 19.Dömling A, Ugi I. Multicomponent reactions with isocyanides. Angew. Chem. Int. Ed. 2000;39:3168–3210. doi: 10.1002/1521-3773(20000915)39:18<3168::aid-anie3168>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 20.See Supporting Information for details on the two step synthesis of 2a. Although the Smac AVPI tetrapeptide possesses normal alanine (not N-methylated), it has been observed that antagonists containing N-methyl alanine show better activity; see reference 4.
- 21.Takeishi K, Sugishima K, Sasaki K, Tanaka K. Rhodium-catalyzed intramolecular hydroacylation of 5- and 6-alkynals: convenient synthesis of α-alkylidenecycloalkanones and cycloalkenones. Chem. Eur. J. 2004;10:5681–5688. doi: 10.1002/chem.200400679. [DOI] [PubMed] [Google Scholar]
- 22.Kazmaier U, Hebach C. Peptide syntheses via Ugi reactions with ammonia. Synlett. 2003:1591–1594. [Google Scholar]
- 23.See Supporting Information (Fig. SI-1) for details.
- 24.Jung ME, Piizzi G. gem-disubstituent effect: theoretical basis and synthetic applications. Chem. Rev. 2005;105:1735–1766. doi: 10.1021/cr940337h. [DOI] [PubMed] [Google Scholar]
- 25.Arai E, Tokuyama H, Linsell MS, Fukuyama T. Aminophenyl)-acetaldehyde dimethyl acetal: a novel reagent for the protection of carboxylic acids. Tet. Lett. 1998;39:71–74. [Google Scholar]
- 26.See Supporting Information for details on synthesis and intermediate structures.
- 27.CCDC 879698 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
- 28.The more polar diastereomer possesses S-stereochemistry. This hypothesis was verified by the biological assay data shown in Table 1.
- 29.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal. Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 30.González-Lopez M, Welsh K, Finlay D, Ardecky RJ, Ganji SR, Su Y, Yuan H, Teriete P, Mace PD, Riedl SJ, Vuori K, Reed JC, Cosford NDP. Design, synthesis and evaluation of monovalent Smac mimetics that bind to the BIR2 domain of the anti-apoptotic protein XIAP. Bioorg. Med. Chem. Lett. 2011;21:4332–4336. doi: 10.1016/j.bmcl.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin J-H, Deng G, Huang Q, Morser J. KIAP, a novel member of the inhibitor of apoptosis protein family. Biochem. Biophys. Res. Commun. 2000;279:820–831. doi: 10.1006/bbrc.2000.4027. [DOI] [PubMed] [Google Scholar]
- 32.Vucic D, Stennicke HR, Pisabarro MT, Salvesen GS, Dixit VM. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr. Biol. 2000;10:1359–1366. doi: 10.1016/s0960-9822(00)00781-8. [DOI] [PubMed] [Google Scholar]
- 33.Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J. Biol. Chem. 2001;276:3238–3246. doi: 10.1074/jbc.M003670200. [DOI] [PubMed] [Google Scholar]
- 34.Abd-Elrahman I, Hershko K, Neuman T, Nachmias B, Perlman R, Ben-Yehuda D. The inhibitor of apoptosis protein Livin (ML-IAP) plays a dual role in tumorigenicity. Cancer Res. 2009;69:5475–5480. doi: 10.1158/0008-5472.CAN-09-0424. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Zhang Q, Liu B, Han M, Shan B. Challenge and promise: roles for Livin in progression and therapy of cancer. Mol. Cancer. Ther. 2008;7:3661–3669. doi: 10.1158/1535-7163.MCT-08-0480. [DOI] [PubMed] [Google Scholar]
- 36.The authors used 5-carboxyflourescein (5-FAM)-conjugated AVP-diphenylalanine-AKK (AVP-diPhe-FAM) for the MLXBIR3SG, XIAP-BIR3, XIAP-BIR2, cIAP1-BIR3 and cIAP2-BIR3 assays and AVPFAK(5-FAM)K (Hid-FAM) for cIAP1, BIR2 and cIAP2-BIR2 assays, whereas AVPIAQK-rhodamine was the probe used in our XIAP, cIAP and ML-IAP assays. See: Flygare JA, Beresini M, Budha N, Chan H, Chan IT, Cheeti S, Cohen F, Deshayes K, Doerner K, Eckhardt SG, Elliott LO, Feng B, Franklin MC, Reisner SF, Gazzard L, Halladay J, Hymowitz SG, La H, LoRusso P, Maurer B, Murray L, Plise E, Quan C, Stephan J-P, Young SG, Tom J, Tsui V, Um J, Varfolomeev E, Vucic D, Wagner AJ, Wallweber HJA, Wang L, Ware J, Wen Z, Wong H, Wong JM, Wong M, Wong S, Yu R, Zobel K, Fairbrother WJ. Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (IAP) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152) J. Med. Chem. 2012;55:4101–4113. doi: 10.1021/jm300060k.
- 37.Cohen F, Alicke B, Elliott LO, Flygare JA, Goncharov T, Keteltas SF, Franklin MC, Frankovitz S, Stephan J-P, Tsui V, Vucic D, Wong H, Fairbrother WJ. Orally Bioavailable Antagonists of Inhibitor of Apoptosis Proteins Based on an Azabicyclooctane Scaffold. J. Med. Chem. 2009;52:1723–1730. doi: 10.1021/jm801450c. [DOI] [PubMed] [Google Scholar]
- 38.Cohen F, Koehler MFT, Bergeron P, Elliott LO, Flygare JA, Franklin MC, Gazzard L, Keteltas SF, Lau K, Ly CQ, Tsui V, Fairbrother WJ. Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres. Bioorg. Med. Chem. Lett. 2010;20:2229–2233. doi: 10.1016/j.bmcl.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 39.Varfolomeev E, Moradi E, Dynek JN, Zha J, Fedorova AV, Deshayes K, Fairbrother WJ, Newton K, Le Couter J, Vucic D. Characterization of ML,IAP protein stability and physiological role in vivo. Biochem. J. 2012;447:427–436. doi: 10.1042/BJ20121103. [DOI] [PubMed] [Google Scholar]
- 40.See Supporting Information for modelling details.
- 41.Eckelman BP, Drag M, Snipas SJ, Salvesen GS. The mechanism of peptide-binding specificity of IAP BIR domains. Cell Death Differ. 2008;15:920–928. doi: 10.1038/cdd.2008.6. [DOI] [PubMed] [Google Scholar]
- 42.Hellwig CT, Rehm M. TRAIL signaling and synergy mechanisms used in TRAIL-based combination therapies. Mol. Cancer. Ther. 2012;11:3–13. doi: 10.1158/1535-7163.MCT-11-0434. [DOI] [PubMed] [Google Scholar]
- 43.The authors show that a 2- to 3-fold increase in LBW242-induced cell death was observed with the addition of 50 ng/mL TRAIL, see: Petrucci E, Pasquini L, Bernabei M, Saulle E, Biffoni M, Accarpio F, Sibio S, Di Giorgio A, Di Donato V, Casorelli A, Benedetti-Panici P, Testa U. A Small Molecule SMAC Mimic LBW242 Potentiates TRAIL- and Anticancer Drug-Mediated Cell Death of Ovarian Cancer Cells. PLoS ONE. 2012;7:e35073. doi: 10.1371/journal.pone.0035073.
- 44.Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X. Autocrine TNFα Signaling Renders Human Cancer Cells Susceptible to Smac-Mimetic-Induced Apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott FL, Denault J-B, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.