Abstract
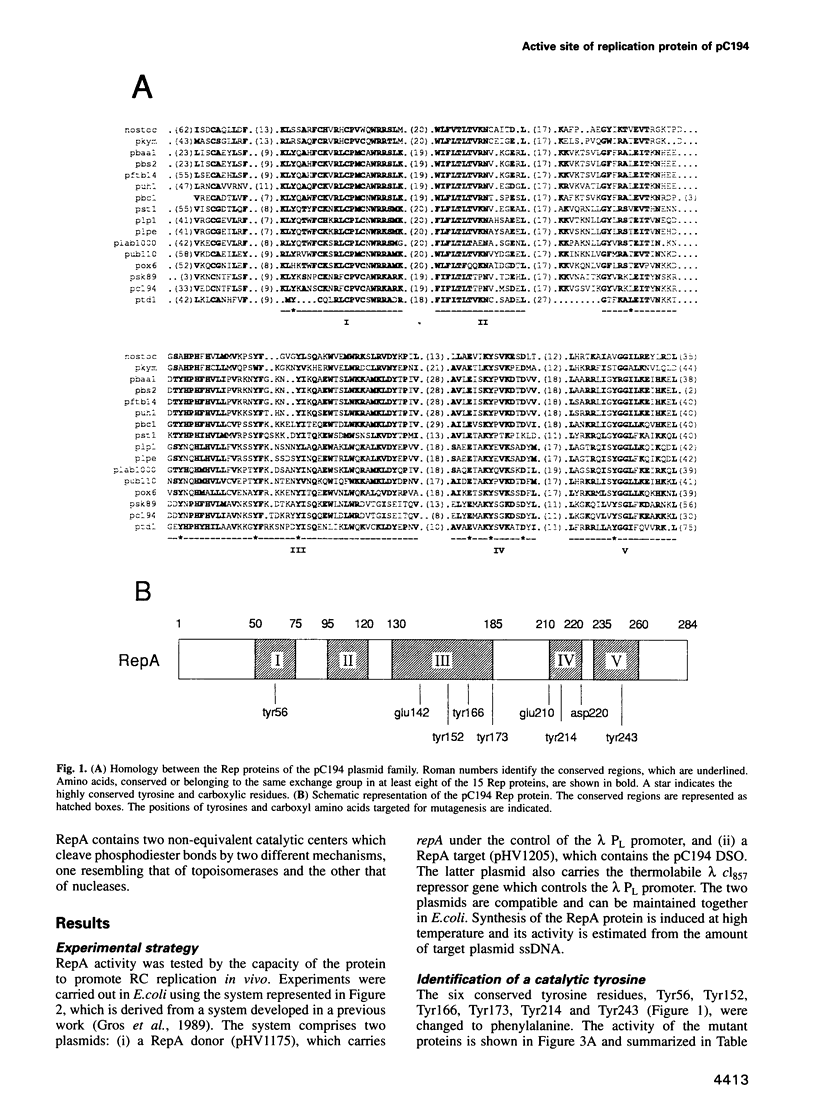
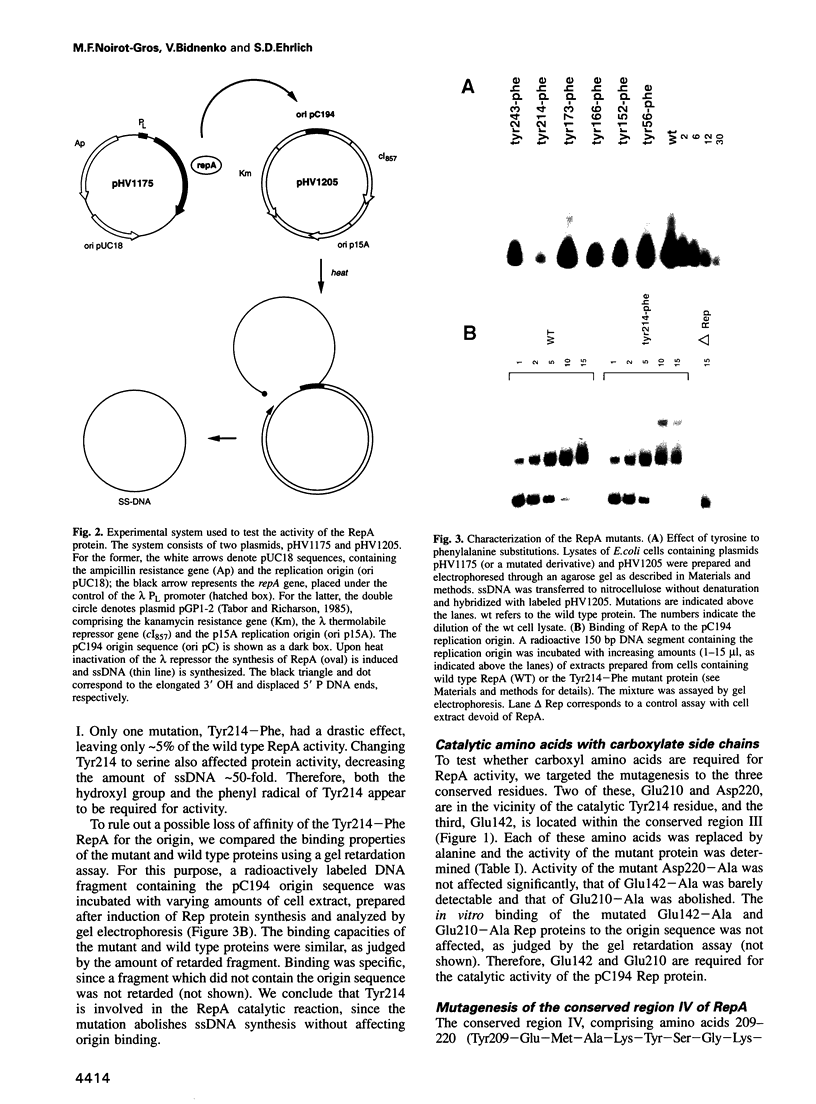
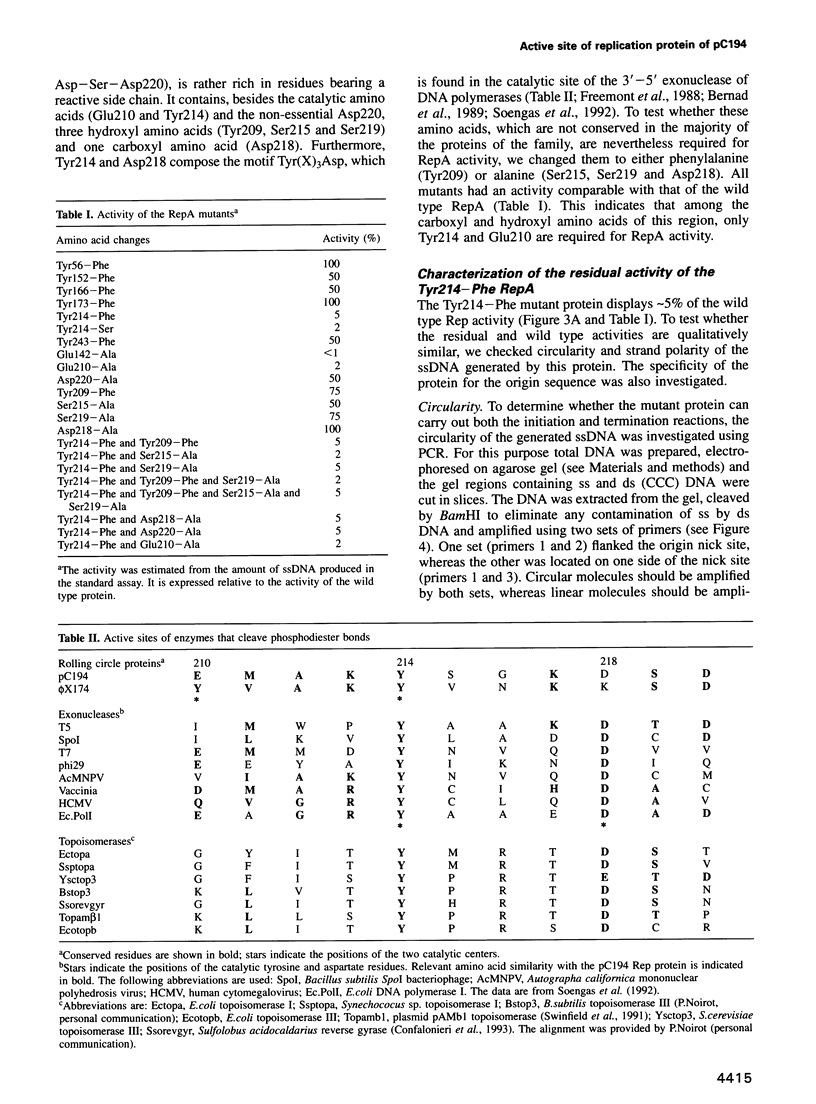
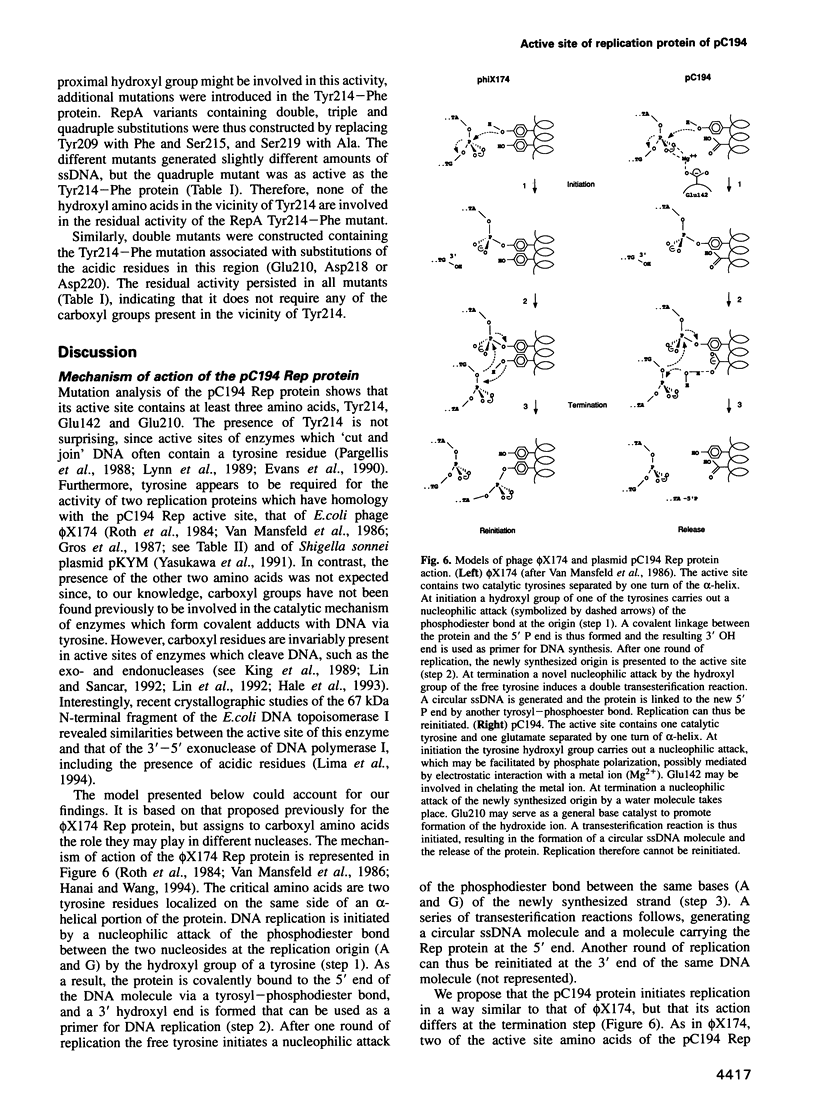
Mutation analysis of the rolling circle (RC) replication initiator protein RepA of plasmid pC194 was targeted to tyrosine and acidic amino acids (glutamate and aspartate) which are well conserved among numerous related plasmids. The effect of mutations was examined by an in vivo activity test. Mutations of one tyrosine and two glutamate residues were found to greatly impair or abolish activity, without affecting affinity for the origin, as deduced from in vitro gel mobility assays. We conclude that all three amino acids have a catalytic role. Tyrosine residues were found previously in active sites of different RC plasmid Rep proteins and topoisomerases, but not in association with acidic residues, which are a hallmark of the active sites of DNA hydrolyzing enzymes, such as the exo- and endonucleases. We propose that the active site of RepA contains two different catalytic centers, corresponding to a tyrosine and a glutamate. The former may be involved in the formation of the covalent DNA-protein intermediate at the initiation step of RC replication, and the latter may catalyze the release of the protein from the intermediate at the termination step.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Boe L., Gros M. F., te Riele H., Ehrlich S. D., Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989 Jun;171(6):3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri F., Elie C., Nadal M., de La Tour C., Forterre P., Duguet M. Reverse gyrase: a helicase-like domain and a type I topoisomerase in the same polypeptide. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4753–4757. doi: 10.1073/pnas.90.10.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. R., Chen J. W., Parsons R. L., Bauer T. K., Teplow D. B., Jayaram M. Identification of the active site tyrosine of Flp recombinase. Possible relevance of its location to the mechanism of recombination. J Biol Chem. 1990 Oct 25;265(30):18504–18510. [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Replication origin of a single-stranded DNA plasmid pC194. EMBO J. 1989 Sep;8(9):2711–2716. doi: 10.1002/j.1460-2075.1989.tb08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale S. P., Poole L. B., Gerlt J. A. Mechanism of the reaction catalyzed by staphylococcal nuclease: identification of the rate-determining step. Biochemistry. 1993 Jul 27;32(29):7479–7487. doi: 10.1021/bi00080a020. [DOI] [PubMed] [Google Scholar]
- Hanai R., Wang J. C. The mechanism of sequence-specific DNA cleavage and strand transfer by phi X174 gene A* protein. J Biol Chem. 1993 Nov 15;268(32):23830–23836. [PubMed] [Google Scholar]
- Heitman J., Model P. Mutants of the EcoRI endonuclease with promiscuous substrate specificity implicate residues involved in substrate recognition. EMBO J. 1990 Oct;9(10):3369–3378. doi: 10.1002/j.1460-2075.1990.tb07538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Maxwell A. Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11232–11236. doi: 10.1073/pnas.90.23.11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee J., Jayaram M. Mechanism of site-specific recombination. Logic of assembling recombinase catalytic site from fractional active sites. J Biol Chem. 1993 Aug 15;268(23):17564–17570. [PubMed] [Google Scholar]
- Lima C. D., Wang J. C., Mondragón A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994 Jan 13;367(6459):138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- Lin J. J., Phillips A. M., Hearst J. E., Sancar A. Active site of (A)BC excinuclease. II. Binding, bending, and catalysis mutants of UvrB reveal a direct role in 3' and an indirect role in 5' incision. J Biol Chem. 1992 Sep 5;267(25):17693–17700. [PubMed] [Google Scholar]
- Lin J. J., Sancar A. Active site of (A)BC excinuclease. I. Evidence for 5' incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J Biol Chem. 1992 Sep 5;267(25):17688–17692. [PubMed] [Google Scholar]
- Lynn R. M., Bjornsti M. A., Caron P. R., Wang J. C. Peptide sequencing and site-directed mutagenesis identify tyrosine-727 as the active site tyrosine of Saccharomyces cerevisiae DNA topoisomerase I. Proc Natl Acad Sci U S A. 1989 May;86(10):3559–3563. doi: 10.1073/pnas.86.10.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Oda Y., Iwai S., Inoue H., Ohtsuka E., Kanaya S., Kimura S., Katsuda C., Katayanagi K., Morikawa K. How does RNase H recognize a DNA.RNA hybrid? Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11535–11539. doi: 10.1073/pnas.88.24.11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman B. J., Grindley N. D. Mutants of the gamma delta resolvase: a genetic analysis of the recombination function. Cell. 1984 Sep;38(2):463–469. doi: 10.1016/0092-8674(84)90501-4. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Parsons R. L., Evans B. R., Zheng L., Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. Possible role of Arg-308 in coupling substrate binding to catalysis. J Biol Chem. 1990 Mar 15;265(8):4527–4533. [PubMed] [Google Scholar]
- Pourmotabbed T., Dell'Acqua M., Gerlt J. A., Stanczyk S. M., Bolton P. H. Kinetic and conformational effects of lysine substitutions for arginines 35 and 87 in the active site of staphylococcal nuclease. Biochemistry. 1990 Apr 17;29(15):3677–3683. doi: 10.1021/bi00467a013. [DOI] [PubMed] [Google Scholar]
- Rasooly A., Novick R. P. Replication-specific inactivation of the pT181 plasmid initiator protein. Science. 1993 Nov 12;262(5136):1048–1050. doi: 10.1126/science.8235621. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Roth M. J., Brown D. R., Hurwitz J. Analysis of bacteriophage phi X174 gene A protein-mediated termination and reinitiation of phi X DNA synthesis. II. Structural characterization of the covalent phi X A protein-DNA complex. J Biol Chem. 1984 Aug 25;259(16):10556–10568. [PubMed] [Google Scholar]
- Seery L. T., Nolan N. C., Sharp P. M., Devine K. M. Comparative analysis of the pC194 group of rolling circle plasmids. Plasmid. 1993 Nov;30(3):185–196. doi: 10.1006/plas.1993.1051. [DOI] [PubMed] [Google Scholar]
- Serre M. C., Zheng L., Jayaram M. DNA splicing by an active site mutant of Flp recombinase. Possible catalytic cooperativity between the inactive protein and its DNA substrate. J Biol Chem. 1993 Jan 5;268(1):455–463. [PubMed] [Google Scholar]
- Sherman P. A., Fyfe J. A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas M. S., Esteban J. A., Lázaro J. M., Bernad A., Blasco M. A., Salas M., Blanco L. Site-directed mutagenesis at the Exo III motif of phi 29 DNA polymerase; overlapping structural domains for the 3'-5' exonuclease and strand-displacement activities. EMBO J. 1992 Nov;11(11):4227–4237. doi: 10.1002/j.1460-2075.1992.tb05517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozhamannan S., Dabert P., Moretto V., Ehrlich S. D., Gruss A. Plus-origin mapping of single-stranded DNA plasmid pE194 and nick site homologies with other plasmids. J Bacteriol. 1990 Aug;172(8):4543–4548. doi: 10.1128/jb.172.8.4543-4548.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinfield T. J., Jannière L., Ehrlich S. D., Minton N. P. Characterization of a region of the Enterococcus faecalis plasmid pAM beta 1 which enhances the segregational stability of pAM beta 1-derived cloning vectors in Bacillus subtilis. Plasmid. 1991 Nov;26(3):209–221. doi: 10.1016/0147-619x(91)90044-w. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Yeheskiely E., van der Marel G. A., van Boom J. H., Plasterk R. H. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991 Dec 25;19(24):6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H., Hase T., Sakai A., Masamune Y. Rolling-circle replication of the plasmid pKYM isolated from a gram-negative bacterium. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10282–10286. doi: 10.1073/pnas.88.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Moscoso M., Espinosa M. Rolling circle-replicating plasmids from gram-positive and gram-negative bacteria: a wall falls. Mol Microbiol. 1993 May;8(5):789–796. doi: 10.1111/j.1365-2958.1993.tb01625.x. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Oude Groeneger A. A., Plasterk R. H. Identification of amino acids in HIV-2 integrase involved in site-specific hydrolysis and alcoholysis of viral DNA termini. Nucleic Acids Res. 1993 Jul 25;21(15):3373–3377. doi: 10.1093/nar/21.15.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., van Teeffelen H. A., Baas P. D., Jansz H. S. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986 May 27;14(10):4229–4238. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]