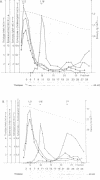
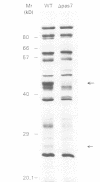
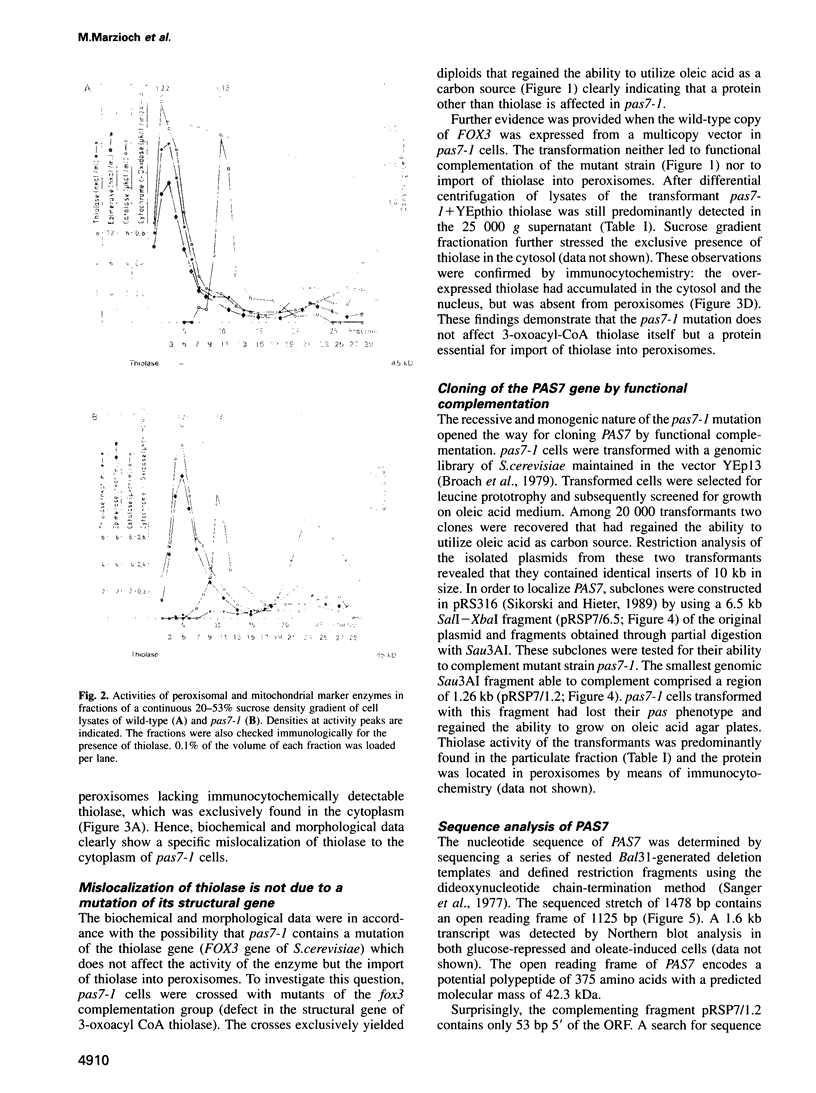
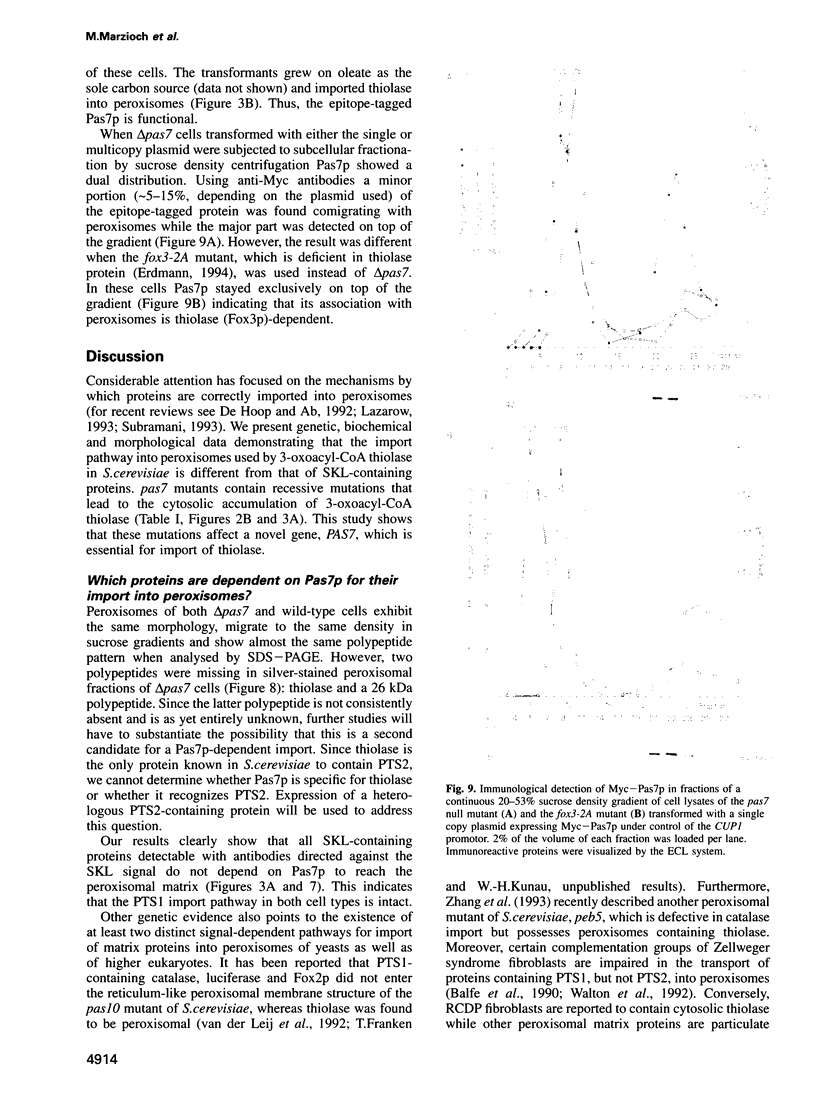
Abstract
To identify components of the peroxisomal import pathway in yeast, we have isolated pas mutants affected in peroxisome biogenesis. Two mutants assigned to complementation group 7 define a new gene, PAS7, whose product is necessary for import of thiolase, a PTS2-containing protein, but not for that of SKL (PTS1)-containing proteins, into peroxisomes. We have cloned PAS7 by complementation of the oleic acid non-utilizing phenotype of the pas7-1 strain. The DNA sequence predicts a 42.3 kDa polypeptide of 375 amino acids encoding a novel member of the beta-transducin related (WD-40) protein family. A Myc epitope-tagged Pas7p, expressed under the control of the CUP1 promotor, was functionally active. Subcellular localization studies revealed that in the presence of thiolase this epitope-tagged Pas7p in part associates with peroxisomes. However, in a thiolase-deficient mutant, Pas7p was entirely found in the cytoplasm. We suggest that Pas7p mediates the binding of thiolase to these organelles.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aitchison J. D., Murray W. W., Rachubinski R. A. The carboxyl-terminal tripeptide Ala-Lys-Ile is essential for targeting Candida tropicalis trifunctional enzyme to yeast peroxisomes. J Biol Chem. 1991 Dec 5;266(34):23197–23203. [PubMed] [Google Scholar]
- Balfe A., Hoefler G., Chen W. W., Watkins P. A. Aberrant subcellular localization of peroxisomal 3-ketoacyl-CoA thiolase in the Zellweger syndrome and rhizomelic chondrodysplasia punctata. Pediatr Res. 1990 Mar;27(3):304–310. doi: 10.1203/00006450-199003000-00023. [DOI] [PubMed] [Google Scholar]
- Berninger G., Schmidtchen R., Casel G., Knörr A., Rautenstrauss K., Kunau W. H., Schweizer E. Structure and metabolic control of the Yarrowia lipolytica peroxisomal 3-oxoacyl-CoA-thiolase gene. Eur J Biochem. 1993 Sep 1;216(2):607–613. doi: 10.1111/j.1432-1033.1993.tb18180.x. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Bodnar A. G., Rachubinski R. A. Cloning and sequence determination of cDNA encoding a second rat liver peroxisomal 3-ketoacyl-CoA thiolase. Gene. 1990 Jul 16;91(2):193–199. doi: 10.1016/0378-1119(90)90088-9. [DOI] [PubMed] [Google Scholar]
- Bout A., Teunissen Y., Hashimoto T., Benne R., Tager J. M. Nucleotide sequence of human peroxisomal 3-oxoacyl-CoA thiolase. Nucleic Acids Res. 1988 Nov 11;16(21):10369–10369. doi: 10.1093/nar/16.21.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Bruinenberg P. G., Evers M., Waterham H. R., Kuipers J., Arnberg A. C., AB G. Cloning and sequencing of the peroxisomal amine oxidase gene from Hansenula polymorpha. Biochim Biophys Acta. 1989 Jul 7;1008(2):157–167. doi: 10.1016/0167-4781(80)90003-2. [DOI] [PubMed] [Google Scholar]
- Butt T. R., Sternberg E. J., Gorman J. A., Clark P., Hamer D., Rosenberg M., Crooke S. T. Copper metallothionein of yeast, structure of the gene, and regulation of expression. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3332–3336. doi: 10.1073/pnas.81.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y., van den Berg M., Tabak H. F., Distel B. An efficient positive selection procedure for the isolation of peroxisomal import and peroxisome assembly mutants of Saccharomyces cerevisiae. Genetics. 1993 Nov;135(3):731–740. doi: 10.1093/genetics/135.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M. J., Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991 Nov 5;266(31):21150–21157. [PubMed] [Google Scholar]
- Erdmann R., Kunau W. H. A genetic approach to the biogenesis of peroxisomes in the yeast Saccharomyces cerevisiae. Cell Biochem Funct. 1992 Sep;10(3):167–174. doi: 10.1002/cbf.290100306. [DOI] [PubMed] [Google Scholar]
- Erdmann R. The peroxisomal targeting signal of 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. Yeast. 1994 Jul;10(7):935–944. doi: 10.1002/yea.320100708. [DOI] [PubMed] [Google Scholar]
- Evan G. I., Lewis G. K., Ramsay G., Bishop J. M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985 Dec;5(12):3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong H. K., Hurley J. B., Hopkins R. S., Miake-Lye R., Johnson M. S., Doolittle R. F., Simon M. I. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989 Feb 7;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Gietl C., Faber K. N., van der Klei I. J., Veenhuis M. Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3151–3155. doi: 10.1073/pnas.91.8.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C. Glyoxysomal malate dehydrogenase from watermelon is synthesized with an amino-terminal transit peptide. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5773–5777. doi: 10.1073/pnas.87.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Glover J. R., Andrews D. W., Subramani S., Rachubinski R. A. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J Biol Chem. 1994 Mar 11;269(10):7558–7563. [PubMed] [Google Scholar]
- Goebl M., Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991 May;16(5):173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Gould S. G., Keller G. A., Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987 Dec;105(6 Pt 2):2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Keller G. A., Schneider M., Howell S. H., Garrard L. J., Goodman J. M., Distel B., Tabak H., Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990 Jan;9(1):85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., Krisans S., Keller G. A., Subramani S. Antibodies directed against the peroxisomal targeting signal of firefly luciferase recognize multiple mammalian peroxisomal proteins. J Cell Biol. 1990 Jan;110(1):27–34. doi: 10.1083/jcb.110.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Ishii N., Kagamiyama H., Osumi T., Hashimoto T. Structural analysis of cDNA for rat peroxisomal 3-ketoacyl-CoA thiolase. J Biol Chem. 1987 Jun 15;262(17):8151–8158. [PubMed] [Google Scholar]
- Hijikata M., Wen J. K., Osumi T., Hashimoto T. Rat peroxisomal 3-ketoacyl-CoA thiolase gene. Occurrence of two closely related but differentially regulated genes. J Biol Chem. 1990 Mar 15;265(8):4600–4606. [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hiltunen J. K., Wenzel B., Beyer A., Erdmann R., Fosså A., Kunau W. H. Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J Biol Chem. 1992 Apr 5;267(10):6646–6653. [PubMed] [Google Scholar]
- Höhfeld J., Veenhuis M., Kunau W. H. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991 Sep;114(6):1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igual J. C., Matallaná E., Gonzalez-Bosch C., Franco L., Pérez-Ortin J. E. A new glucose-repressible gene identified from the analysis of chromatin structure in deletion mutants of yeast SUC2 locus. Yeast. 1991 May-Jun;7(4):379–389. doi: 10.1002/yea.320070408. [DOI] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990 Sep-Oct;6(5):363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Karin M., Najarian R., Haslinger A., Valenzuela P., Welch J., Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc Natl Acad Sci U S A. 1984 Jan;81(2):337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. A., Krisans S., Gould S. J., Sommer J. M., Wang C. C., Schliebs W., Kunau W., Brody S., Subramani S. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J Cell Biol. 1991 Sep;114(5):893–904. doi: 10.1083/jcb.114.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F., Langeder A., Raupachova J., Binder M., Hartig A. Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. J Cell Biol. 1993 Feb;120(3):665–673. doi: 10.1083/jcb.120.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunau W. H., Beyer A., Franken T., Götte K., Marzioch M., Saidowsky J., Skaletz-Rorowski A., Wiebel F. F. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;75(3-4):209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- Kunau W. H., Hartig A. Peroxisome biogenesis in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1992 Aug;62(1-2):63–78. doi: 10.1007/BF00584463. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lazarow P. B. Genetic approaches to studying peroxisome biogenesis. Trends Cell Biol. 1993 Mar;3(3):89–93. doi: 10.1016/0962-8924(93)90079-g. [DOI] [PubMed] [Google Scholar]
- Lithgow T., Høj P. B., Hoogenraad N. J. Do cytosolic factors prevent promiscuity at the membrane surface? FEBS Lett. 1993 Aug 23;329(1-2):1–4. doi: 10.1016/0014-5793(93)80179-x. [DOI] [PubMed] [Google Scholar]
- Luirink J., Dobberstein B. Mammalian and Escherichia coli signal recognition particles. Mol Microbiol. 1994 Jan;11(1):9–13. doi: 10.1111/j.1365-2958.1994.tb00284.x. [DOI] [PubMed] [Google Scholar]
- McCollum D., Monosov E., Subramani S. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells--the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol. 1993 May;121(4):761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno de la Garza M., Schultz-Borchard U., Crabb J. W., Kunau W. H. Peroxisomal beta-oxidation system of Candida tropicalis. Purification of a multifunctional protein possessing enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase and 3-hydroxyacyl-CoA epimerase activities. Eur J Biochem. 1985 Apr 15;148(2):285–291. doi: 10.1111/j.1432-1033.1985.tb08837.x. [DOI] [PubMed] [Google Scholar]
- Motley A., Hettema E., Distel B., Tabak H. Differential protein import deficiencies in human peroxisome assembly disorders. J Cell Biol. 1994 May;125(4):755–767. doi: 10.1083/jcb.125.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T., Tsukamoto T., Hata S., Yokota S., Miura S., Fujiki Y., Hijikata M., Miyazawa S., Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991 Dec 31;181(3):947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Söllner T., Neupert W. Mitochondrial import receptors for precursor proteins. Trends Biochem Sci. 1991 Feb;16(2):63–67. doi: 10.1016/0968-0004(91)90026-r. [DOI] [PubMed] [Google Scholar]
- Preisig-Müller R., Kindl H. Thiolase mRNA translated in vitro yields a peptide with a putative N-terminal presequence. Plant Mol Biol. 1993 Apr;22(1):59–66. doi: 10.1007/BF00038995. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozawa N., Suzuki Y., Orii T., Moser A., Moser H. W., Wanders R. J. Standardization of complementation grouping of peroxisome-deficient disorders and the second Zellweger patient with peroxisomal assembly factor-1 (PAF-1) defect. Am J Hum Genet. 1993 Apr;52(4):843–844. [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small G. M., Szabo L. J., Lazarow P. B. Acyl-CoA oxidase contains two targeting sequences each of which can mediate protein import into peroxisomes. EMBO J. 1988 Apr;7(4):1167–1173. doi: 10.1002/j.1460-2075.1988.tb02927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991 Nov;10(11):3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leij I., Franse M. M., Elgersma Y., Distel B., Tabak H. F. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11782–11786. doi: 10.1073/pnas.90.24.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leij I., Van den Berg M., Boot R., Franse M., Distel B., Tabak H. F. Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J Cell Biol. 1992 Oct;119(1):153–162. doi: 10.1083/jcb.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenhuis M., Mateblowski M., Kunau W. H., Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987 Jun;3(2):77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- Walton P. A., Gould S. J., Feramisco J. R., Subramani S. Transport of microinjected proteins into peroxisomes of mammalian cells: inability of Zellweger cell lines to import proteins with the SKL tripeptide peroxisomal targeting signal. Mol Cell Biol. 1992 Feb;12(2):531–541. doi: 10.1128/mcb.12.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., Driessen A. J., Hartl F. U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu Rev Biochem. 1991;60:101–124. doi: 10.1146/annurev.bi.60.070191.000533. [DOI] [PubMed] [Google Scholar]
- Zhang J. W., Han Y., Lazarow P. B. Novel peroxisome clustering mutants and peroxisome biogenesis mutants of Saccharomyces cerevisiae. J Cell Biol. 1993 Dec;123(5):1133–1147. doi: 10.1083/jcb.123.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoop M. J., Ab G. Import of proteins into peroxisomes and other microbodies. Biochem J. 1992 Sep 15;286(Pt 3):657–669. doi: 10.1042/bj2860657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voorn L., Ploegh H. L. The WD-40 repeat. FEBS Lett. 1992 Jul 28;307(2):131–134. doi: 10.1016/0014-5793(92)80751-2. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Protein targeting signals. Curr Opin Cell Biol. 1990 Aug;2(4):604–608. doi: 10.1016/0955-0674(90)90100-s. [DOI] [PubMed] [Google Scholar]