Abstract
The spatial organization of the nucleus results in a compartmentalized structure that affects all aspects of nuclear function. This compartmentalization involves genome organization as well as the formation of nuclear bodies and plays a role in many functions, including gene regulation, genome stability, replication, and RNA processing. Here we review the recent findings associated with the spatial organization of the nucleus and reveal that a common theme for nuclear proteins is their ability to participate in a variety of functions and pathways. We consider this multiplicity of function in terms of Crowdsourcing, a recent phenomenon in the world of information technology, and suggest that this model provides a novel way to synthesize the many intersections between nuclear organization and function.
Keywords: Nuclear organization, genome organization, nuclear bodies
1. Introduction
The nucleus is functionally compartmentalized. For example, there is ample evidence for the deterministic positioning of gene loci, chromatin, and even chromosomes according to physical and functional characteristics. In particular, the radial distribution of genes and chromosomes has been shown to have a defined pattern, with active transcription and gene rich chromosomes occupying the nuclear interior, and gene poor chromosomes and silenced, developmentally regulated gene loci positioned at the nuclear periphery. Moreover, the nucleus is unique as an organelle in that many of its functions are compartmentalized in non-membranous assemblies, referred to as nuclear bodies (NBs). These NBs are themselves spatially localized non-randomly within the nucleus. The mechanistic basis for the arrangement of the genome and its functional outcome remain to be fully understood.
The idea of self-organization has been implemented as a means to conceive the dynamic organization of the genome. Self-organization in the context of nuclear cell biology has been understood as a non-hierarchical association of factors that result in functionally competent stable-state structures [1]. For example, double stranded DNA repair foci have been generated in the absence of DNA breaks simply by tethering individual components of the pathway to a lac operator array integrated into the genome [2]. Similarly, NBs have been created de novo using the same strategy [3]. We have shown that the principles of self-organization describe the cell-specific chromosomal topologies that arise through coordinate gene regulation during cellular differentiation [4]. Thus, the nucleus is an open system not at equilibrium, and rather than the dynamic association of nuclear proteins and modified chromatin devolving into ever greater entropy, they form functional centers and characteristic organizational patterns.
There is an inherent promiscuity of nuclear proteins with many being involved in a wide range of networks and functions. A prime example of this is the nuclear intermediate filament protein lamin A/C (encoded by LMNA), a central component of the nuclear lamina. Mutations in LMNA have been implicated in a wide variety of human disease phenotypes, collectively referred to as laminopathies [5]. The basis for a single protein being involved in myriad functions, from replication, to transcription, to cell signaling, remains a perplexing problem. Of course, lamin A/C’s multi-functionality can at least partially be attributed to its role in the nucleoskeleton, a network of laminar and other proteins that is thought to provide a substrate for nuclear activities. As reviewed below, however, there are many such examples of a nuclear protein intersecting numerous and varied functional pathways. While self-organization provides a useful model to describe the functional dynamics of the genome, it may be inadequate to address the multi-functionality of regulatory and structural nuclear proteins. In particular, self-organizing systems are comprised of defined components, albeit non-hierarchical in their association. As we review extensively below, the dynamic organization of genome function more closely resembles a multi-agent system, with the factors involved in a particular function originating from diverse and often unexpected sources.
For many decades the genetic code and its central dogma have provided a colorful metaphor for understanding computer technology. In a structural sense, the hard disk drive has been considered something of a genome (storage of information); the central processing unit the machinery that replicates and transcribes (processes information); and random access memory the proteins that carry out cellular function (running programs). Beyond the clear parallels of information storage, processing, and function, modeling computer technology from the standpoint of molecular biology provides a comparison to a deeply complex system, in a sense predicting the potential of information technology (IT). Intriguingly, the relatively recent emergence of the Web 2.0, which comprises the myriad social uses of the internet that harness the possibilities of the billions of World Wide Web (W3) users, may turn the tables and provide models for biological insight. The behaviors that emerge from the interactions of ‘agents’ in the W3 provide a laboratory to explore dynamic biological systems in ways that we are not currently capable. In particular, what can the Web 2.0 inform us about the multiplicity of functional associations of nuclear proteins and the genome?
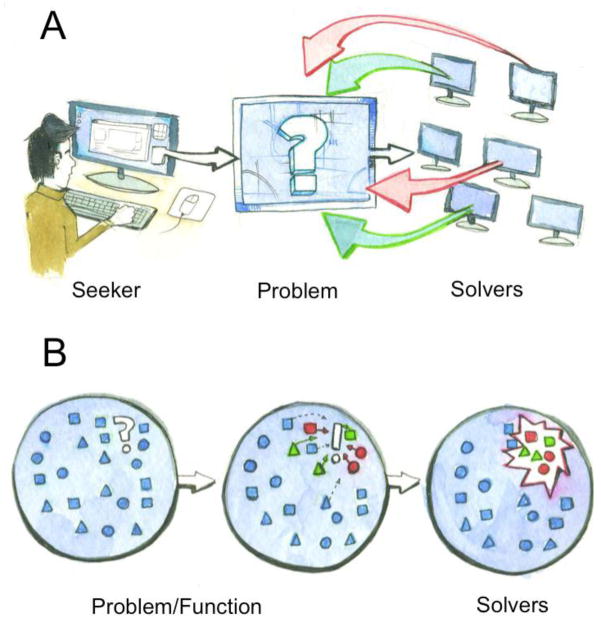
We offer that the nascent Web 2.0 phenomenon of Crowdsourcing may provide a useful analytical model to address the multi-functionality of nuclear proteins. Crowdsourcing, in the context of the W3, is comprised of three central elements: seeker, problem, and solvers. The seeker is a company or agency that is involved in a given purpose (from commercial to non-profit), which is in need of a solution to a problem (Fig. 1A). Traditionally, solutions were sought ‘in-house’; however, through Crowdsourcing the seeker promulgates the problem through the W3. Thus, the problem is introduced to myriad potential solvers, with varying availability and ability (Fig. 1A). By harnessing the power of this diverse community, a robust resolution is often achieved [6, 7]. We suggest that this template is observable in the dynamic interplay of nuclear proteins and genome function. A particular nuclear activity, such as coordinate gene regulation, is both seeker and problem—a task that requires (solicits) factors for its action. In this vein, proximity (which does not exist for the W3, as we are all connected) is the availability of any given agent. On the other hand, ability is the potential that an agent can contribute to the initiated function. Thus, the striking multi-functionality of nuclear proteins can be considered a multi-agent system, in which problems/functions are addressed by a ‘crowd’ of proteins based upon their individual availability and ability (Fig. 1B). As physicists and mathematicians begin to analyze the nature of Crowdsourcing, the principles behind it may yield useful models for understanding the dynamic networks involved in protein multi-functionality. In any event, we offer the idea of Crowdsourcing as a guide for our review of the dynamic complexity and interconnected nature of genome organization.
Fig. 1.
Depiction of nuclear Crowdsourcing. (A) In traditional Crowdsourcing, company or agency (the seeker), advertises a given problem to the W3. The immense ‘crowd’ of individuals on the W3 then participate in creating a solution to the problem based upon their availability (they are aware of the challenge) and ability (they are capable of contributing to its solution). For example, red arrows represent solvers who are aware but unable to help, whereas green arrows indicate solvers who are both available and able. (B) Crowdsourcing of nuclear function would be similar, with the notable exception of the seeker and the problem being inextricable, as the problem is the seeking entity. In parallel with (A) the solvers of the function/’problem’ would also have varying availability and ability. In this case availability would be whether a given agent (protein) is proximal (or dynamic enough) to engage the problem, and ability would be its capacity to function in the given process or pathway. Circles, squares, and triangles represent different classes of protein agents, with red and green shapes indicating proteins either available and/or able to participate.
2. Genome organization
A central focus of the study of nuclear spatial organization involves the physical positioning of the genome within the nuclear space. The features of nuclear topology include the localization patterns of chromosomes, genes, and other DNA elements, as well as the interactions that occur between these components (Fig. 2). An important aspect of the spatial organization of the genome is that the observed patterns change during cell differentiation and regulation of gene expression. Here we discuss the recent findings associated with dynamic genome organization and the proteins involved in this process.
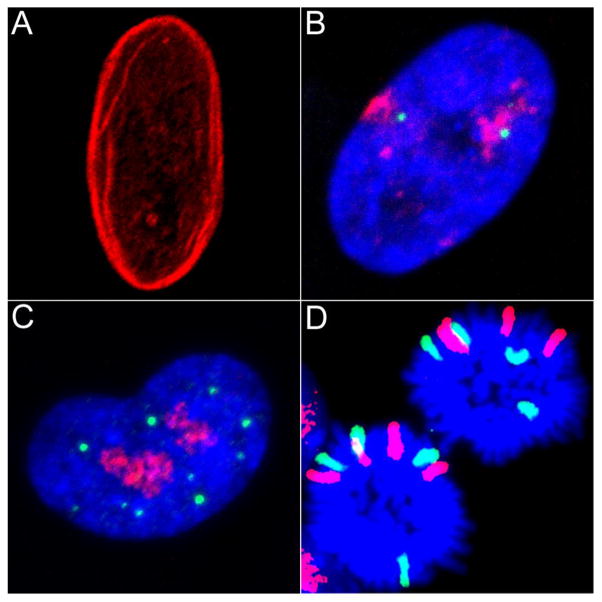
Fig. 2.
Various forms of nuclear organization. (A) Immunofluorescent staining of a human retinal pigmented epithelial cell (RPE1) nucleus with anti-lamin B1 (red) shows the localization of the nuclear lamina. (B) FISH staining of a human fetal lung fibroblast (IMR90) nucleus with a chromosome 1 paint (red) and the region around the MYOG gene on chromosome 1 (green) shows the radial positioning of CTs and individual loci as well as the relationship between these elements. Image courtesy of D. Neems. (C) Immunofluorescent staining of an IMR90 nucleus with anti-PML (green) and anti-nucleolin (red) shows the organization of PML nuclear bodies and nucleoli respectively. (D) FISH staining of dividing murine hematopoietic progenitor cells with 4n DNA content. Chromosome 11 (red) and chromosome 2 (green) are depicted. This image shows maintained organization of chromosome positioning in daughter cells during mitosis. In B–D, DAPI is used to visualize DNA.
2.1 Genome Organizers
When describing the spatial organization of the genome, there are a few key factors that repeatedly appear. These proteins can influence many different functions and are involved in numerous pathways, with the common thread being their ability to influence genome architecture. Here we describe three of the most prominent genome organizers: CTCF, cohesin, and nuclear lamins.
2.1.1 CTCF
CTCF is a ubiquitously expressed, zinc finger DNA-binding protein that was first described as a transcription factor based on its role in regulation of c-myc gene expression [8–10]. CTCF was later shown to block the communication between enhancers and promoters (enhancer blocking activity) and also to act as a boundary to the spread of chromatin domains (barrier function), and was therefore additionally defined as an insulator protein [11–13]. Genome-wide analysis revealed CTCF binding throughout the genome, and found that the majority of sites are invariant across various cell-types [14]. Many of these sites are involved in CTCF-mediated chromatin loops that correlate with chromatin domains, lamin-associated domains (LADs), and enhancer-promoter interactions [15]. In order to incorporate this array of functions, a more general view describes CTCF as a facilitator of chromatin looping and a global genome organizer [16]. In this role, CTCF participates in a variety of different pathways including regulation of transcription, imprinting, alternative splicing, X inactivation, telomere end protection, and somatic recombination (Fig. 3 and Table S1) [17–22]. This may be possible due to its vast number of interacting partners including transcription factors and chromatin regulatory proteins (YB1, Kaiso, and YY1), chromatin remodeling factors (CHD8), nucleolar components (nucleophosmin, UBF), and proteins involved in other nuclear functions (cohesin, PARP1, Suz12, RNAPII) [23, 24]. How interacting partners are determined at specific genomic locations is not understood, but it is likely influenced by the protein composition of the local nuclear environment. Based on its functional multiplicity, it is not surprising that CTCF is an essential protein and that CTCF deficient mice result in early embryonic lethality [25, 26].
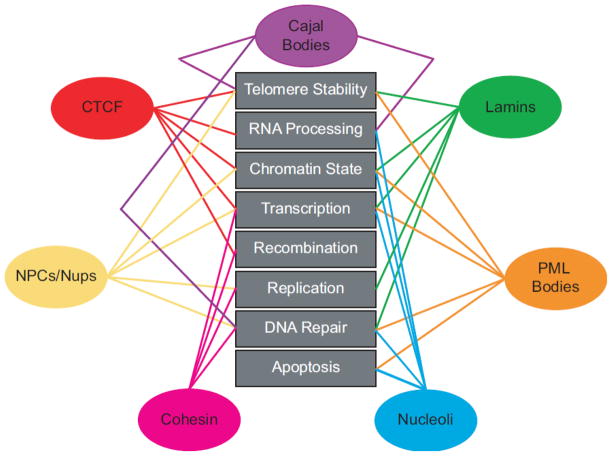
Fig. 3.
Schematic of the multi-functionality of nuclear proteins. Nuclear proteins are connected to the pathways and functions in which they are involved, depicting not only multiplicity of function, but also the vast amount of functional overlap between the various proteins involved in nuclear organization. A chart with references is included as Supplemental Table 1.
2.1.2 Cohesin
Traditionally, the cohesin complex has been considered for its role in maintaining sister chromatid cohesion after DNA replication [27]. It is composed of four core subunits (Smc1, Smc3, Scc1/Rad21, Scc3/SA1/SA2), three of which (Smc1, Smc3, Scc1/Rad21) are thought to form a tripartite ring around sister chromatids after replication. Additionally, there are many accessory proteins that associate with the core cohesin subunits, mainly involved in loading and dissociation of the cohesin ring [28, 29]. Beyond its role in chromosome segregation, cohesin proteins also play a role in double stranded break repair, stabilization of transcription factor binding, somatic recombination, and the facilitation of short and long-range interactions involved in transcriptional regulation (Fig. 3 and Table S1) [27, 30–33]. The ability of cohesin to form a ring structure around DNA strands probably plays an integral part in its ability to form the chromatin loops involved in these processes. Many CTCF genomic binding sites are co-occupied by cohesin, and it is thought that CTCF is necessary to position cohesin on chromatin [34–37]. Whole-genome chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) in the developing mouse limb identified over 2,000 interactions between cohesin-associated loci and 65% of these involve CTCF [38]. This data set includes tissue-specific interactions as well as interactions that are found in multiple tissues with distinct expression profiles suggesting diverse functional outputs from cohesin-mediated interaction. In general, cohesin sites that co-localize with CTCF tend to be cell-type invariant while non-CTCF cohesin sites conversely demonstrate cell-type specific binding, co-localize with transcription factors, and facilitate transcription promoting enhancer-promoter interactions through co-localization with transcription factors, mediator, and the cohesin loading factor Nipbl [30–32].
2.1.3 Lamins
Lamins are nucleus-specific class V intermediate filament proteins that form the major component of the nuclear lamina, the proteinaceous layer found on the nucleoplasmic side of the nuclear envelope, as well as throughout the nucleoplasm (Fig. 2A). This distribution suggests a role for the lamina as a nucleoskeleton. There are three lamin genes in mammalian cells (LMNA, LMNB1, and LMNB2) that encode the A- and B- type lamins. At least one B-type lamin protein is thought to be expressed in all nucleated, metazoan cell types, but the A-type lamins, lamin A and C produced from alternative splicing of LMNA, are developmentally regulated [39, 40]. It was generally accepted that A-type lamins are not expressed in embryonic stem cells (ESCs) [41], but a recent study has detected low levels of expression at both the mRNA and protein level in murine ESCs [42], suggesting that there may be fundamental differences between mouse and human, and raising questions about the effect of expression level on lamin A/C function.
Lamins have been found to associate with large chromatin domains, and the functional outputs of these associations on genome organization are wide ranging. DamID analysis of lamin B1 classified roughly 40% of the genome as LADs in four different mouse cell types at different stages of neuronal differentiation [43]. A similar degree of lamin B1 association is observed in human cells [44]. LADs have been characterized to be gene poor, transcriptionally inactive, A/T rich regions of chromatin, and are highly conserved between murine and human studies. The majority of LADs were found to be constant across cell-types; however, a subset of regions shows changes in lamin association during differentiation [43, 45]. Recently, the occurrence of a GAGA motif bound by a cKrox/Lap2β/HDAC3 complex was shown to direct lamin association of specific, developmentally regulated loci in a transcription-dependent manner [46]. It is yet to be determined whether this is a general mechanism that directs lamin association, and how cKrox binding is regulated between cell-types. Despite the large amount of the genome associated with the nuclear lamina, mESCs lacking both LMNB1 and LMNB2 appear normal with only limited changes in gene expression within LADs [47], and triple knockout mESCs lacking lamin B1, lamin B2, and lamin A/C expression also show no defects in proliferation or differentiation [48]. Mice derived from the double knockout mice die at birth, suggesting that nuclear lamins are important for development and cell differentiation but not cell survival.
Lamin A/C association with chromatin has also been mapped and is highly correlated with the previously identified lamin B1 LADs [45, 49]. The agreement in lamin A/C and lamin B1 association is almost complete (98–99%) for constitutive LADs and less, but still substantial for cell-type variant LADs (83–86%) [45]. Various mutations in the LMNA gene lead to disruption in nuclear positioning, chromatin compaction, and chromatin state that correlate with changes in gene expression [49, 50]. Interestingly, murine rod cells do not express lamin A/C or lamin B receptor (LBR) causing an inverted genome organization in which the peripheral localization of heterochromatin is lost [51, 52]. It has recently been shown that a similar intranuclear heterochromatin repositioning can be induced in a variety of mouse cell types by loss of lamin A/C and LBR indicating that these proteins are essential to maintain proper heterochromatin organization in most, in not all, cell types [51].
Like CTCF and cohesin, lamins play a role in many different processes, including: transcriptional regulation, DNA replication elongation, maintenance of nuclear structure and shape, chromosome positioning and condensation, gene positioning, telomere positioning, and positioning of DNA damage foci (Fig. 3 and Table S1) [50, 53–56]. Due to the role of lamin proteins in so many different nuclear processes, disruption of these proteins leads to a phenotypically diverse set of disease states. Collectively, these are called laminopathies with most of the known disorders a result of mutations in LMNA, including cardiac and skeletal myopathies such as Emery-Dreifuss muscular dystrophy (EDMD), pre-mature aging disorders such as Hutchinson-Gilford progeria syndrome (HGPS) and atypical Werner syndrome, as well as peripheral nerve disorders and lipodystrophies [57]. Additionally, diseases associated with mutations in LMNB1 and LMNB2 have also been identified including adult-onset leukodystrophy and partial lipodystrophy [57]. Thus the importance and multi-functionality of nuclear lamins is manifested in the wide variety of disease states caused by mutations in lamin genes.
Clearly, these different genome organizers are involved in many different pathways. While likely candidates include interacting partners, the local chromatin environment, and/or covalent modifications, the mechanistic underpinnings of how the same proteins are able to function in varied pathways are unknown. Moreover, a persuasive model that seeks to address the basis for this multi-functionality remains to be fully developed.
2.2 Global Genome Organization
The majority of our understanding of genome organization comes from fluorescent in situ hybridization (FISH) analysis and chromosome conformation capture (3C) [58] based techniques. FISH has the advantage of single-cell analysis in the absence of a large degree of data manipulation and amplification of starting material while 3C-based techniques show increased resolution and, in combination with high-throughput analysis, are capable of monitoring many interactions over large regions simultaneously. Due to these different strengths, these techniques are best used in combination and it is important to be aware of the limitations of each technique when interpreting results.
2.2.1 Chromosome Territories
The organization of the genome into chromosomes plays a large role in dictating the three dimensional spatial organization of the nucleus. Chromosomes occupy discrete regions of the nucleus called chromosome territories (CTs) [59] (Fig. 2B). The radial positioning of CTs themselves is an important aspect of nuclear organization. Multiple factors are thought to influence radial positioning of CTs, including gene density and chromosome size. Gene poor chromosomes are found more peripherally and gene dense chromosomes are found more centrally in the nucleus [60–62]. Additionally, in some cases larger chromosomes are positioned at the periphery and smaller chromosomes more internally [61, 63, 64]. Altered gene expression can also influence radial positioning of CTs [65, 66]. For example, induction of cellular quiescence through serum starvation leads to the radial repositioning of many chromosomes. Interestingly, movement in response to serum starvation occurs rapidly and is dependent on actin and myosin, while relocalization of serum re-stimulated cells occurs more slowly and requires cell division, providing evidence that different mechanisms exist within the cell to regulate chromosome positioning [67, 68]. Furthermore, the relative positioning of chromosomes to one another is influenced by gene expression. This is observed in hematopoiesis during which proximity of chromosomes is related to the degree of coordinated gene regulation [4, 69]. Therefore, changes in expression profiles that occur during differentiation can result in cell-type specific CT organization.
CT formation also influences genome organization by restricting broad genomic interactions occurring within a chromosome. Although there is some evidence for intermingling of CTs [70], this idea is controversial and more recent studies suggest that mixing of CTs occurs only at borders and is not extensive [71, 72]. Additionally, chromosome centromeres seem to form a barrier to interaction between chromosome arms [73–76]. This was clearly depicted by Tolhuis et al. 2011, through the use of a chromosome inversion across the centromere that resulted in altered interactions such that mainly intra-arm interactions are observed in both the normal and inverted state. This result suggests that a centromeric barrier, not cues from DNA sequence or chromatin state, dictates intra-chromosomal interactions. Though less common, inter-chromosomal interactions have also been observed, and regions with high inter-chromosomal interaction frequency have been suggested to loop out of CTs [75]. However, recent single-cell Hi-C analysis shows that regions displaying a high degree of inter-chromosomal interactions still maintain intra-chromosomal interactions indicating these regions do not loop dramatically away from the territory [77]. Furthermore, assaying these inter-chromosomal interactions by FISH shows that they are only observed in a very small percentage of cells [75]. Thus it is critical to keep in mind when interpreting population-based interaction profiles as maps of nuclear organization, and underscores the importance of single cell analysis in understanding nuclear organization.
2.2.2 Large-Scale Genomic Interactions
Beyond organization at the level of the chromosome, other properties of whole genome organization have also been described. Active and inactive chromatin demonstrate a high frequency of self-interaction. In this manner the genome can be separated into two compartments that represent active and inactive regions [75, 77–81]. This is influenced by nuclear lamins since a LMNA mutation associated with HGPS leads to disruption of this compartmentalization in late passages [49]. When exonic regions of a mouse chromosome were compared to CT signal, the exonic chromatin was found to occupy a larger, more internal nuclear space than the territory itself, supporting the idea of active chromatin being spatially distinct from inactive chromatin [82]. Furthermore, the inter-chromosomal interactions that have been identified tend to involve the active chromatin compartment, indicating that active chromatin is more mobile than inactive chromatin [75, 80].
Chromosome conformation capture carbon-copy (5C) and Hi-C analysis have revealed that chromatin interactions tend to cluster within local regions termed ‘topologically associating domains’ (TADs) [83] or ‘topological domains’ that are roughly 10–500 kb in Drosophila and 200 kb–1Mb in mammalian cells [74, 80, 83, 84]. TADs were found to be relatively stable across different cell types, and most cell-type specific interactions are found to occur within a TAD. Recently, single-cell Hi-C analysis was performed revealing that TADs are also fairly consistent within a population of cells, while inter-domain interactions show a higher degree of cell-to-cell variability [77]. Additionally, TADs tend to correlate with the compartmentalization of the genome into active versus inactive regions, which also can be coincident with LADs [83, 84]. The components necessary to establish a TAD are not completely understood; however, some common elements have been reported at TAD borders. Drosophila TAD borders tend to be gene dense, contain DNase hypersensitive sites, and are enriched for RNA Polymerase II (RNAPII), insulator proteins, and the mitotic spindle protein Chromator [74, 80]. Mammalian TAD borders have similar components in that they tend to be enriched for CTCF, cohesin, promoters of housekeeping genes, tRNA genes, and SINE family repeat elements [83, 84]. Interestingly, while CTCF is enriched at these regions, only 15% of the total CTCF sites are found at TAD borders suggesting that a CTCF site alone is not sufficient to dictate a TAD border [84]. A critical experiment by Nora et al. 2012 found that deletion of roughly 60 kb at a TAD border within the Xist/Tsix locus resulted in increased association between TADs and was accompanied by misregulation of gene expression providing evidence that a border region is essential to create independent interacting domains [83]. It is still uncertain what elements within this region actually maintain border activity, and a greater focus on genome disruption analysis is necessary to understand the formation and maintenance of TADs and the effect they have on chromatin structure.
A recent high-resolution 5C analysis of mESCs during neuroectoderm differentiation verified the existence of constitutive TADs, but was also able to identify sub-TADs that show more cell-type specific variability [85]. Interactions were found to be anchored by CTCF, cohesin, and mediator, and the presence of these factors was shown to be functional, such that knockdown of their expression led to a loss of interaction. Specific combinations of these proteins were correlated with the distance between interacting loci as well as the cell type variability of an interaction. In general, CTCF with or without cohesin was found to facilitate large, constitutive interactions, while mediator with or without cohesin was found to facilitate small and intermediate, developmentally regulated interactions. Although this statement oversimplifies the complex interplay of proteins present at interaction sites, it suggests that it is actually the combination of factors at a specific region that dictates the spatial organization. Additionally, since the same protein can be involved in different kinds of interactions, this strongly supports the pervasive multi-functionality of nuclear proteins.
2.3 Genome Organization of Specific Loci
2.3.1 Locus Organization
Chromatin looping within a locus has also been studied in depth at a few specific loci, and chromatin interactions within these model loci have been detected that lead to a variety of functions [17]. The imprinted H19/Igf2 locus is one of the best-understood regions in terms of regulation of chromatin looping. Within this region CTCF binding at the imprinting control region is inhibited by DNA methylation of the paternal allele [18, 86, 87]. This allele-specific CTCF binding leads to different chromatin interactions within the locus that dictate enhancer-promoter interactions and gene expression [88–90]. Regulation of genomic imprinting is further discussed in this issue [Weaver and Bartolomei].
Chromatin looping has also been described at lymphocyte antigen receptor loci, and in this context chromatin interactions facilitate somatic recombination. CTCF and cohesin have been implicated at these loci and deletion of these proteins or their binding sites leads to impaired recombination. Current data show a role for CTCF and cohesin in regulating transcription within these loci by facilitating long-range interactions, and it is unclear if they are also directly involved in bringing distal gene segments together for recombination [91].
Another well-studied locus is the murine HoxD cluster, which shows linear regulation of gene expression due to chromatin looping during development [92]. This region represents an interesting pattern of chromatin organization as the HoxD cluster is located within the boundary of two TADs [93]. During limb development HoxD genes participate in interactions with regulatory elements in the surrounding TADs, and a switch in some HoxD genes from interaction with the telomeric to the centromeric TAD correlates with changes in chromatin state and gene expression [93, 94].
In interpreting these data for locus organization, it is important to note that observed interactions indicate contacts that occur at a given point in time, not necessarily stable chromatin structures. Therefore, individual regulatory elements and gene promoters may undergo many different interactions and it remains to be determined how the identified interactions co-exist within a population. Use of the recently developed single-cell Hi-C analysis technique will provide important insight into this topic [77]. Nonetheless, it is clear that although the same proteins often facilitate chromatin interactions that dictate locus organization, they can be regulated in different manners and can result in different functional outputs.
2.3.2 Gene Positioning
Patterns of spatial organization of the genome have also been identified at the level of individual genes. Clustering of co-regulated genes through self-organization has been proposed as a thermodynamically favorable form of nuclear organization [95–97]. This type of clustering has been observed and in some cases occurs at RNAPII enriched transcription factories [98–104]. Additionally, co-regulated gene clustering in a transcription factor dependent manner was observed in erythroid cells with genes regulated by the transcription factor Klf1 co-localizing with Klf1 as well as RNAPII [105]. Importantly, not all co-regulated genes are observed to cluster [106]. This bias may be related to the effects of the linear chromatin environment, including such features as gene density, transcriptional activity of neighboring genes, and position on the chromosome affecting gene loci mobility and localization [102, 103].
Gene positioning in relation to the nuclear periphery is another important aspect of nuclear organization. Peripheral gene localization is associated with gene silencing for some loci [107–109] and a subset of gene loci demonstrates cell-type specific lamin association [43, 110]. Furthermore, artificially tethering a locus to the nuclear periphery can lead to gene silencing, although this phenomenon is gene specific as peripheralization is not sufficient to regulate transcription of all genes [111]. The complexity of radial gene positioning is further exemplified in mouse olfactory neurons where clustering of transcriptionally silent olfactory receptor (OR) genes in heterochromatin foci is observed at the nuclear interior [112]. This clustering is dependent on the loss of LBR and is necessary for proper monogenic expression of OR genes.
An additional complication in understanding the functional consequences of peripheral localization is that nuclear pores are dispersed throughout the nuclear membrane creating a lack of homogeneity at the nuclear periphery. Beyond their role in nuclear transport, nuclear pore complexes (NPCs), composed of nucleoporins (Nups), are also involved in the organization of gene loci [113, 114]. NPCs and Nups interact with specific chromosome regions, and unlike other peripheralized chromatin, these regions are usually enriched for active chromatin. In yeast, association with NPCs is often specified by DNA sequences termed ‘DNA zip codes’, and genes with similar zip codes cluster together [115–117]. This localization is necessary for robust gene activation, transcriptional memory that allows efficient re-activation, and potentially even rapid gene silencing after activation [118–120]. In Drosophila, Nups can also interact with gene loci contributing to gene activation; however, this interaction can occur in the nucleoplasm away from the nuclear periphery [121–123]. Recently, similar functional interactions were found to be conserved in human cells [124]. In addition to this role in genome organization, NPCs and Nups play a role in DNA replication, telomere stability, and DNA repair (Fig. 3 and Table S1) [125]. Together, this suggests that Nups are another class of nuclear protein that exhibits a multitude of functions and has a critical role in the spatial organization of the genome.
2.4 Dynamics of genome organization
Our understanding of the dynamics of genome organization is growing, yet the relationship between nuclear activities and changes in genome organization are not fully characterized.
2.4.1. Dynamic genome organization and transcription
A key aspect of the dynamics of genome organization is its relationship with transcription, and an active area of research is determining whether genome organization plays a causal role in regulating transcription, vice versa, or most likely, both. As mentioned in Section 2.3.2, tethering of some loci to the nuclear periphery can result in altered gene expression indicating a causal influence of genome organization on transcription [111]. However, these experiments are extremely artificial and may not accurately represent the normal temporal association of genome reorganization and changes in transcription.
The β-globin locus exhibits dynamic, developmentally regulated chromatin looping and gene positioning, and includes examples of changes in genome organization that proceed changes in gene expression as well as the reverse. Within this locus stage specific regulated interactions have been identified between a locus control region (LCR) and β-globin genes that are linearly arranged on the chromosome in the order of their activation [126, 127]. These LCR-promoter interactions are mediated by various transcription factors that regulate β-globin gene expression [128–130]. Additional CTCF mediated interactions are also present surrounding the locus and are detected prior to LCR-promoter interaction and activation of gene expression [131, 132]. Inhibition of transcription does not have large effects on the interactions made by the active β-globin gene locus, suggesting that transcription is not necessary to maintain these interactions [133, 134]. These studies do not rule out a role for transcription in initiating interactions, and since these experiments were performed over relatively short time scales (0.5–5 hours), they do not test the role of transcription in maintenance of interactions through cell division. Interestingly, artificially induced chromatin looping in the absence of the transcription factor GATA-1 was found to be sufficient to induce chromatin interactions and resulted in activation of β-globin gene expression [135]. Therefore, at least in this situation, genome organization plays a causal role in regulation of gene expression. However, analysis of the β-globin locus at the level of gene positioning reveals a less direct relationship with transcription. Movement of the β-globin locus away from the nuclear interior begins after initiation of transcription indicating genome reorganization is not necessary to alter gene expression [108]. Together these data for β-globin genome organization exemplify the complex interplay that exists between long-range interactions, gene positioning, and transcription.
Changes in genome organization can also be associated with transcriptional silencing. For example, in the Drosophila embryo the hunchback gene is expressed early in neuronal differentiation and is then silenced. Although this transcriptional repression is accompanied by nuclear peripheralization, lamin-dependent gene movement is not observed for three cell divisions after transcriptional repression [136]. This movement was shown to coincide with permanent transcriptional silencing, but not the original onset of gene repression. Therefore, like the relationship with transcriptional activation, the relationship between transcriptional silencing and genome organization is not straightforward.
2.4.2 Dynamic genome organization and other nuclear functions
In addition to the connection that dynamic genome organization exhibits with transcription, there are also notable links with other nuclear functions including replication timing and DNA repair.
DNA replication occurs asynchronously throughout the genome such that euchromatic chromatin located in the nuclear interior replicates during early S phase, and peripheralized heterochromatin replicates during late S phase [137]. Additionally, genome-wide maps of replication timing correlate extremely well with genomic interaction profiles and suggest the spatial compartmentalization of replication domains [138]. Importantly, replication timing of some genomic regions is developmentally regulated, and changes in replication timing correlate with changes in spatial organization, indicating that dynamic genome organization in relation to replication timing may play a role in cell differentiation [138–141]. In support of this idea, cell populations that fail to reprogram during the formation of induced pluripotent stem cells (iPSCs) lack reprograming of replication timing, suggesting that this aspect of dynamic genome organization may be necessary to establish pluripotency [140].
The role of dynamic genome organization with DNA repair is unique in that chromatin mobility can theoretically both facilitate and hinder genomic stability after DNA damage [142]. Increased mobility of damaged DNA can increase the rate of finding templates for homologous recombination (HR) and other broken ends for non-homologous end joining (NHEJ) [143–147]. At the same time, It has been demonstrated that the spatial proximity of genes correlates with their translocation frequencies [148, 149] and increased mobility can lead to translocations [150]. For this reason, nuclear factors such as lamin A/C act to reduce mobility of DNA damage foci, probably in order to prevent deleterious events [55, 151]. Despite this disparity in the effect of DNA damage on locus mobility, it was recently demonstrated that at least some gene-rich CTs reveal nuclear repositioning in response to DNA damage, indicative of global effects on genome organization [152].
2.4.3 Dynamic genome organization and the cell cycle
Cell division introduces an additional layer of complexity to our understanding of the dynamics of genome organization (Fig. 2D). For example, patterns of nuclear organization can vary throughout the cell cycle. This behavior is observed in telomere positioning with telomeres localizing to the nuclear periphery during nuclear reassembly after mitosis, but not at other stages of the cell cycle [153].
There is also some evidence that cell division may be necessary to allow major rearrangements in genome organization. A recent study developed a new “molecular contact memory” approach using an extension of DamID technology to monitor LADs over time in live-cells [154]. This technique revealed relatively stable association of chromatin with the nuclear lamina within a single cell cycle, but a large reorganization after mitosis. Additionally, this study identified a large degree of cell-to-cell variability in LADs, and found that this variability often correlates with the transcriptional status of genes in individual cells. These results again highlight the importance of single-cell analysis in understanding the spatial organization of the nucleus and suggest that reorganization of LADs occurs during mitosis. Therefore, it is possible that after changes in the transcriptional status of a gene occur, the cell must divide before large changes in spatial organization are observed. This brings into question how much regulation of spatial organization occurs in terminally differentiated cells that do not undergo cell division.
3. Nuclear bodies
A critical form of the spatial organization of the nucleus is found in the non-membranous structures referred to as nuclear bodies (NBs). Considered broadly, NBs are thought to manifest a potential for consolidated activity: the constituent proteins, DNA, and RNA occupy a discrete location to facilitate a collective function [155]. Although NBs appear to be stable structures, their components are in fact highly dynamic, freely exchanging with a nucleoplasmic pool of protein. Nuclear bodies often associate with chromatin, and the extent of nuclear body movement is determined by the accessibility and dynamics of the surrounding chromatin [156].
Some NBs are found in a wide variety of cell types, while others are present in only a limited number of cell types and/or are formed only under certain conditions [157]. In general, assigning a specific function to a particular NB has been difficult due to the large variety of constituent proteins and their involvement in many different pathways, as well as the fact that many NBs share protein interacting partners. This problem will probably only become more challenging since mathematical modeling of known and predicted nuclear protein localization revealed that various nuclear compartments and NBs are probably even more functionally diverse than currently known [158]. In the following sections, we will discuss recent findings of the multi-functionality of NBs and discuss how this allows for efficient interactions in a confined nuclear space.
3.1 Promeyelocytic leukemia bodies
Promyelocytic leukemia (PML) nuclear bodies (also known as ND 10s or Kremer bodies) (Fig. 2C) are functionally promiscuous and dynamic structures that have been implicated in such processes as proliferation, senescence, apoptosis, genomic stability, telomere maintenance, and the DNA damage response (Fig. 3 and Table S1) [159]. PML bodies were first observed by electron microscopy in the 1960s, and later as nuclear dots by immunofluorescence [160, 161]. These nuclear dots became referred to as PML oncogenic domains (PODs), or simply PML bodies, after the discovery of the localization of the promyelocytic leukemia protein (PML), so named as it was first characterized through its fusion with the retinoic acid receptor alpha (RARA) in acute promyelocytic leukemia (APL) patients. Over 100 proteins have been shown to localize to PML bodies [162]. While it was initially unclear whether they simply functioned as a depository of proteins, recent work strongly indicates the role of PML bodies as functional structures whose activity is dictated by the combination of factors present at a given time. Here we will discuss some of the functions of PML bodies by describing various interacting partners.
PML bodies are thought to be involved in apoptosis and cell survival through death-associated protein 6 (DAXX), a transcriptional co-repressor and histone chaperone [163]. In support of the importance of a functional role for the interaction of DAXX with PML bodies, DAXX-induced apoptosis is abrogated in PML−/− cells while acquiring a diffuse spatial localization [164]. Phosphorylation of DAXX promotes SUMO1 binding, and this in turn enhances its interaction with PML as well as its physical recruitment to anti-apoptotic genes [165]. Although DAXX is mostly found at PML bodies, it also forms small foci at centromeric and pericentromeric (CEN/periCEN) heterochromatin in some cells where it is thought to play a role in gene expression through deposition of H3.3. Under heat shock conditions, the balance of DAXX localization shifts from PML bodies to CEN, exemplifying the dynamics of PML body composition [166]. DAXX recruits H3.3 as well as soluble H3.3–H4 dimers to PML bodies, and it is believed that this association allows pairing with other histone chaperones for deposition onto chromatin [167, 168].
The association of PML with p53 contributes to its function in apoptosis and transcriptional activity [169–172]; however, the role that PML bodies play in p53 function is not fully understood. A recent study revealed that after DNA damage, monocytic leukemia zinc finger protein (MOZ), a histone acetyltransferase (HAT), relocalizes to PML bodies. This interaction greatly increases MOZ-mediated acetylation of p53 and, thus, expression of p21, G1 arrest, and cellular senescence [173]. The interaction of MOZ and PML bodies is inhibited by the direct phosphorylation of MOZ at its PML binding domain, which in turn negatively regulates p53 acetylation [173]. This result underscores the importance of PML body association to mediate function, and suggests that PML bodies play an active role in regulating p53 activity.
PML bodies have been shown to associate with genetic loci, such as the major histocompatibility complex (MHC) [174]. Recently, it was demonstrated that upon IFN-gamma treatment, the spatial proximity between PML bodies and the MHC II gene cluster is increased [175, 176]. IFN-gamma treatment also caused PML bodies to associate with and stabilize CIITA, an MHC class II transactivator, and this association promotes the transcription of MHC II genes [176]. PML bodies also play a role in transcriptional memory within the MHC II gene cluster through interaction with a methyltransferase, which can prime genes for faster transcriptional reactivation after IFN-gamma treatment through the establishment of a permissive epigenetic mark [175]. Using a novel immuno-TRAP technique to probe for nuclear body-chromatin interactions, new PML body-locus associations were recently identified including the PML locus itself [177]; however, the function of PML bodies at these sites is unclear. Although PML bodies associate with genomic regions of high transcriptional activity, they themselves are not sites of transcription [178]. Instead, RNA associates with the periphery of PML bodies, and highly acetylated chromatin typically surrounds PML bodies [179].
The heterogeneity of PML bodies, in both form and function, illustrates the difficulty in assigning a clear activity to this type of NB. Instead of describing PML bodies as a specific nuclear compartment it may be more appropriate to think of them as a class of NBs, which can be further defined based on their individual components and the surrounding nuclear environment. Nonetheless, the vast array of activities involving PML bodies perfectly exemplifies the functional multiplicity of NBs.
3.2 Cajal bodies
Due to their high concentration of factors such as small nuclear ribonucleic particles (snRNPs) and other RNA processing factors, Cajal bodies are thought to predominantly function in post-translational modification of spliceosomal components. First referred to as coiled bodies given their fibrillar appearance under EM, they were renamed Cajal bodies after Santiago Rámon y Cajal who first described them in the early 1900s. Over 60 proteins and over 25 RNA species, including small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and small Cajal body-specific RNAs (scaRNAs), have been found to associate with Cajal bodies [180]. Cajal bodies form by stochastic self-organization. Tethering individual Cajal body components to chromatin results in the de novo formation of a mature Cajal body [3]. Recent evidence indicates that Cajal bodies are more functionally diverse than previously considered (Fig. 3 and Table S1).
For example, Cajal bodies have been implicated in the maturation of snRNPs and snRNAs. The integrator complex, which is involved in 3′ end processing of snRNA, is essential for Cajal body integrity, as defects in its activity cause dispersal of Cajal body components [181]. Recent evidence indicates that the nuclear matrix associated heterogeneous ribonucleoprotein U (hnRNP U) regulates Cajal body morphology and U2 snRNP maturation [182]. Cajal bodies, like PML bodies, have been shown to transiently associate with gene loci. Cajal bodies associate with the U1, U2, and U4 snRNP gene clusters and the U11 and U12 snRNA loci [183]. Evidence for a functional association has been shown in that interaction of Cajal bodies with an inducible U2 gene array is dependent on transcription [184]. As mentioned previously, it is hypothesized that Cajal bodies act to recruit factors necessary for the biogenesis and maturation of snRNPs and snRNAs suggesting that the association with these gene loci aids in the efficient processing of nascent transcripts.
Cajal bodies have also been shown to be important in telomere maintenance. Telomerase contains a WD40-repeat protein that allows it to bind to Cajal bodies, and this association appears to be necessary for telomere synthesis [185]. Recently, identification of an hnRNP A2 splice variant, hnRNP A2*, was found to promote telomere extension by telomere G-quadruplex unfolding activity [186]. hnRNP A2* localized with Cajal bodies at telomeres, and it is suggested hnRNP A2* associates with the telomerase holoenzyme at Cajal bodies before localization to telomeres [186]. Dyskeratosis congentia patients with a mutation in the telomerase protein TCAB1 are defective in localizing telomerase to Cajal bodies, and instead it localizes to nucleoli which prevents telomere maintanence [187].
3.3 Nucleoli
In mammalian cells, nucleoli form around tandem repeats of rDNA genes, known as nucleolar organizing regions (NORs), that are spread across five acrocentric chromosomes: HSA13, HSA14, HSA15, HSA21, and HSA22 (Fig. 2C). Although the nucleolus was formally described in the 1800s, its primary function in ribosome biosynthesis was not discovered until the 1960s [188]. The position of the nucleolus is determined by the presence of transcribing NORs, as well as the process of ribosome biogenesis itself [189]. Proteomic analysis has identified over 300 proteins associated with the nucleolus, and the majority of these proteins have no known role in ribosome biogenesis [190–192]. Furthermore, the nucleolar proteome is dynamic and changes in response to cellular growth conditions [193]. As evidenced below, beyond the well-defined role in ribosome biogenesis, the multi-functionality of the nucleolus is becoming increasingly clear (Fig. 3 and Table S1) [194].
The nucleolus has roles in protein sequestration and stress sensing. The tumor suppressor p53 has a short life span, partly maintained by its association with the E3 ubiquitin protein ligase Mdm2, while the stabilization of p53 is necessary for its activation. A wide variety of ribosomal proteins have been identified that, upon stress, are relocalized from the nucleolus and interact with Mdm2 to stabilize p53 [195]. Non-coding RNA (ncRNA) transcribed from a large intergenic spacer (IGS) region that separates rRNA tandem repeat genes, are able to directly target proteins with a nucleolar detention sequence (NoDS) to nucleoli for sequestration [196]. Different stimuli, such as acidosis, heat shock, or transcriptional stress are able to cause the expression of IGS ncRNA from various IGS loci, and target proteins such as Mdm2 to the nucleolus for immobilization [196].
Identification of nucleolus-associated domains (NADs) suggests that nucleoli play a role in genome organization [197, 198]. In addition to rRNA genes, nucleoli predominantly interact with tRNA genes, centromeres, specific satellite repeats, and repressive heterochromatin. Interestingly, there is significant overlap between NADs and LADs, and single cell analysis has revealed exchange between nucleolar and nuclear lamina localization after mitosis suggesting functional similarities between them [154, 197].
It is apparent that nuclear bodies are a form of nuclear organization that facilitate a wide range of nuclear activities. It is important to note that there are other types of nuclear bodies from those mentioned here. Nuclear speckles are NBs enriched in RNA splicing factors and are believed to be storage sites for these factors [199]. Paraspeckles are structurally formed by the ncRNA NEAT1 and are suggested to predominantly play a role in gene expression [200]. Polycomb group bodies are formed by repressive complexes that act by post-translationally modifying histones for gene repression [201]. These examples further reinforce the idea that NBs are multi-functional structures that are implicated in myriad of nuclear processes.
4. Nuclear Structure
When discussing nuclear organization, in addition to describing the arrangement of elements within the nucleus, we must also consider the nucleus as it relates to the rest of the cell. This includes nuclear size and shape, and interactions between the nucleus and components in the cytoplasm.
Although nuclear size correlates to some extent with genome length, cytoplasmic components seem to be the predominant determinants [202, 203]. Furthermore, in Xenopus it was found that nuclear import plays a major role dictating nuclear size, which may be at least in part to due to import of a B-type lamins [204]. This finding suggests that nuclear size may be regulated by the availability of structural proteins. Indeed, knockdown of lamin proteins in C. elegans leads to reduced nuclear size [205]. Interestingly, there is a correlation between nuclear size and mitotic chromosome length, and during Xenopus development nuclear diameter and chromosome length both decrease. In this system, in vitro manipulation of nuclear size revealed that progression through a cell cycle is necessary to induce a corresponding change in chromosome length, suggesting that cell division is important for dynamic genome organization [206]. Although these studies are beginning to shed light on factors that dictate nuclear and chromosome size, the functional implications of this regulation in terms of nuclear organization, gene expression, and genomic stability are still not clear. Interestingly, alterations in nuclear size are often observed in cancer cells and this feature is used to stage various cancers, highlighting the importance of this area of research [207].
The linker of the nucleus to the cytoskeleton (LINC) complex directly connects the cytoplasm to the nucleoplasm and provides a platform for communication between nuclear processes and the rest of the cell. The LINC complex spans the nuclear membrane and consists of nesprins that transverse the outer nuclear membrane (ONM) and bind to cytoplasmic actin, intermediate filaments, and microtubules; SUN domain proteins span the inner nuclear membrane (INM) and bind to the nuclear lamina and nuclear membrane-associated proteins [208]. The LINC complex mediates mechanotransduction forces to the nucleus that are, for the most part, mediated by actin. A perinuclear actin cap, for instance, regulates nuclear shape in response to the shape of the cell, and this regulation by actin is dependent on the LINC complex and lamin A/C [209]. Furthermore, actin also mediates lateral compressive forces in regulating nuclear shape [210]. Interestingly, nuclear volume loss caused by nuclear shape change is marked by chromatin condensation and a decreased cell proliferation rate, indicating that nuclear form and function are directly coupled to extra-nuclear forces [210]. Given the role the nuclear lamina plays in establishing genome organization and its direct association with the LINC complex, the extent of nuclear regulation by mechanical forces is becoming increasingly clear. Mechanical force application to the plasma membrane, for instance, alters actin force transmission to the nucleus that causes a LINC complex-dependent chromatin decondensation [211]. In addition, force transmission through the integrin-actin-LINC-lamin A/C interface has been shown to dissociate Cajal bodies, indicating that nuclear processes, beyond just genome organization patterns, can be coupled to external forces [212]. It will be interesting to see how structural cues regulate such processes such as transcription and splicing.
5. Concluding remarks
While we are unable to review all of its various components, we have focused on the dynamic complexity of nuclear organization and function. A recurrent theme encountered in the study of genome organization is that it is rather difficult to assign a given factor, such as CTCF, or a particular NB, such as the PML body, a single function. Rather, they often are associated with varied and disparate functions; thus, understanding what they really ‘do’ is hard to pin down. How, then, can we begin to understand this pervasive multi-functionality as we consider nuclear organization from a systems biology perspective? As suggested in the introduction, we believe that the phenomenon of Crowdsourcing, an outcome of the Web 2.0, provides an intriguing corollary to the multitude of functions observed for many nuclear factors. The basic paradigm of Crowdsourcing is simple (Fig. 1A). Given the enormous interconnectivity provided by the W3, a task or problem can be presented to a large audience. While not everyone online will be aware of this problem (posted on a particular website), which we consider to be availability, a large enough crowd will exist with enough individuals who have different abilities to act. As a community, the problem or task can then be solved in a decentralized fashion. Indeed, there are now websites dedicated to Crowdsourcing various tasks, enriching for talents directly relevant to the problem at hand. Additionally, there is also the important component of a reward, be it financial or simply self-satisfaction for contributing to a greater good [6].
Let us take the PML body as an example of how Crowdsourcing might pertain to its many functions (Fig. 1B). The PML protein is the canonical member of the body, and it has seven characterized isoforms. In a sense, this core constituent may represent the means by which the need for a function/problem is presented. Based upon which isoform may predominate in a PML body, proteins that are both available (i.e. in proximity of this call to function) and have the ability (i.e. can act on the function presented, such as p53 regulation) would then associate. The proteins that respond to the particular function may not always be the same, but would necessarily have the requisite capacity to act in the given pathway. Thus, a decentralized assembly of proteins performs a function, but another assembly might perform a different function.
So, how useful is our comparison of Crowdsourcing to protein multi-functionality? As it is still in its infancy, the concept of Crowdsourcing is only now attracting the attention of physicists and mathematicians who will use their skill sets to probe its behavior and model its features [213]. Thus, it is beyond the purview of this review for us to speculate on how this will unfold. As an example, however, we believe that evolutionary game theory models might provide useful tools. In particular, the ‘Tragedy of the Commons’ encompasses the salient features of Crowdsourcing. In its simplest form, the model describes a heterogeneous population of agents, whom have a choice to contribute to a public good. If enough choose to participate, then even those who did not will benefit [214]. With this basic platform, modulating various parts of the premise can create different models. For example, the agents involved can be defined, and each actor can behave according to its own rules; these rules might represent the localization or functional capacity of a given agent (i.e. protein). Indeed, analysis of Crowdsourcing will allow us to formalize these rules, as we begin to understand its dynamic and complex behavior. Finally, in an intriguing twist, the idea of Crowdsourcing has now yielded crowdfunding, in which a heterogeneous group of individuals donate to an identified cause or activity. There are now such sites that crowdfund biological studies. Perhaps in the future we might crowdfund our analysis of Crowdsourcing and nuclear protein multi-functionality.
Supplementary Material
Highlights.
The spatial organization of the nucleus is fundamental in ensuring its proper function
Nuclear proteins and nuclear bodies are multifunctional
The organization of the genome is inherently tied to the multi-functionality of nuclear proteins
Aberrant nuclear organization can give rise to disruptions in genome stability
We suggest that crowdsourcing is a useful model to analyze the dynamic nature of nuclear protein interactions
Acknowledgments
We thank Daniel S. Neems for sharing images, and Ellen Rice for thoughtful discussions and ideas. This work is funded by a Career Award in the Biomedical Sciences from the Burroughs Welcome Fund, an Ellison Medical Foundation New Scholar Award, and an NIH New Innovator Award. A.W. is supported by a postdoctoral fellowship from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Misteli T. Self-organization in the genome. Proc Natl Acad Sci U S A. 2009;106:6885–6886. doi: 10.1073/pnas.0902010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 4.Rajapakse I, Perlman MD, Scalzo D, Kooperberg C, Groudine M, Kosak ST. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc Natl Acad Sci U S A. 2009;106:6679–6684. doi: 10.1073/pnas.0900986106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimi T, Butin-Israeli V, Adam SA, Goldman RD. Nuclear lamins in cell regulation and disease. Cold Spring Harb Symp Quant Biol. 2010;75:525–531. doi: 10.1101/sqb.2010.75.045. [DOI] [PubMed] [Google Scholar]
- 6.Brabham DC. Crowdsourcing as a Model for Problem Solving: An Introduction and Cases. Convergence. 2008;14:75–90. [Google Scholar]
- 7.Beucheler TLR, Ruchslin RM, Pfeifer R. Modeling and simulating Crowdsourcing as a complex biological system: Human crowds manifesting collective intelligence on the internet. In: Lenaerts TGM, Bersini H, Bourgine P, Dorigo M, Doursat R, editors. The Eleventh European Conference on the Sythesis and SImulation of Living Systems. Vol. 11. MIT Press; Boston: 2011. pp. 109–116. [Google Scholar]
- 8.Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- 11.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 12.Felsenfeld G, Burgess-Beusse B, Farrell C, Gaszner M, Ghirlando R, Huang S, Jin C, Litt M, Magdinier F, Mutskov V, Nakatani Y, Tagami H, West A, Yusufzai T. Chromatin boundaries and chromatin domains. Cold Spring Harb Symp Quant Biol. 2004;69:245–250. doi: 10.1101/sqb.2004.69.245. [DOI] [PubMed] [Google Scholar]
- 13.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holwerda S, de Laat W. Chromatin loops, gene positioning, and gene expression. Front Genet. 2012;3:217. doi: 10.3389/fgene.2012.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 19.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer RJ, del Rosario BC, Pinter SF, Lessing D, Sadreyev RI, Lee JT. A boundary element between Tsix and Xist binds the chromatin insulator Ctcf and contributes to initiation of X-chromosome inactivation. Genetics. 2011;189:441–454. doi: 10.1534/genetics.111.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih HY, Krangel MS. Chromatin architecture, CCCTC-binding factor, and V(D)J recombination: managing long-distance relationships at antigen receptor loci. J Immunol. 2013;190:4915–4921. doi: 10.4049/jimmunol.1300218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, Riethman H, Lieberman PM. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012;31:4165–4178. doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolaev LG, Akopov SB, Didych DA, Sverdlov ED. Vertebrate Protein CTCF and its Multiple Roles in a Large-Scale Regulation of Genome Activity. Curr Genomics. 2009;10:294–302. doi: 10.2174/138920209788921038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 25.Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling K-W, Gribnau J, Renkawitz R, Grosveld F, Hendriks RW, Galjart N. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 27.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 28.Mehta GD, Rizvi SM, Ghosh SK. Cohesin: a guardian of genome integrity. Biochim Biophys Acta. 2012;1823:1324–1342. doi: 10.1016/j.bbamcr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 29.Mehta GD, Kumar R, Srivastava S, Ghosh SK. Cohesin: Functions beyond sister chromatid cohesion. FEBS Lett. 2013;587:2299–2312. doi: 10.1016/j.febslet.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 30.Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 2012;22:2163–2175. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 38.Demare LE, Leng J, Cotney J, Reilly SK, Yin J, Sarro R, Noonan JP. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 2013;23:1224–1234. doi: 10.1101/gr.156570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res. 2007;313:2157–2166. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 41.Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- 42.Eckersley-Maslin MA, Bergmann JH, Lazar Z, Spector DL. Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus. 2013;4:53–60. doi: 10.4161/nucl.23384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Gräf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M, Wessels L, van Steensel B. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MM, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 45.Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim Y, Zheng X, Zheng Y. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell research. 2013;23:1420–1423. doi: 10.1038/cr.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23:260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, Dellefave L, Pytel P, Selig S, Labno CM, Reddy K, Singh H, McNally E. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One. 2010;5:e14342. doi: 10.1371/journal.pone.0014342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, Herrmann H, Blum H, Engelkamp D, Stewart CL, Leonhardt H, Joffe B. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Suarez I, Redwood AB, Perkins SM, Vermolen B, Lichtensztejin D, Grotsky DA, Morgado-Palacin L, Gapud EJ, Sleckman BP, Sullivan T, Sage J, Stewart CL, Mai S, Gonzalo S. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009;28:2414–2427. doi: 10.1038/emboj.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masny PS, Bengtsson U, Chung SA, Martin JH, van Engelen B, van der Maarel SM, Winokur ST. Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum Mol Genet. 2004;13:1857–1871. doi: 10.1093/hmg/ddh205. [DOI] [PubMed] [Google Scholar]
- 55.Mahen R, Hattori H, Lee M, Sharma P, Jeyasekharan AD, Venkitaraman AR. A-type lamins maintain the positional stability of DNA damage repair foci in Mammalian nuclei. PLoS One. 2013;8:e61893. doi: 10.1371/journal.pone.0061893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol. 2000;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 59.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 61.Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- 62.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cremer M, Küpper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Branco MR, Branco T, Ramirez F, Pombo A. Changes in chromosome organization during PHA-activation of resting human lymphocytes measured by cryo-FISH. Chromosome Res. 2008;16:413–426. doi: 10.1007/s10577-008-1230-x. [DOI] [PubMed] [Google Scholar]
- 67.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:R5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol. 2000;10:149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- 69.Kosak ST, Scalzo D, Alworth SV, Li F, Palmer S, Enver T, Lee JS, Groudine M. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Markaki Y, Smeets D, Fiedler S, Schmid VJ, Schermelleh L, Cremer T, Cremer M. The potential of 3D-FISH and super-resolution structured illumination microscopy for studies of 3D nuclear architecture: 3D structured illumination microscopy of defined chromosomal structures visualized by 3D (immuno)-FISH opens new perspectives for studies of nuclear architecture. Bioessays. 2012;34:412–426. doi: 10.1002/bies.201100176. [DOI] [PubMed] [Google Scholar]
- 72.Olivares-Chauvet P, Fennessy D, Jackson DA, Maya-Mendoza A. Innate structure of DNA foci restricts the mixing of DNA from different chromosome territories. PLoS One. 2011;6:e27527. doi: 10.1371/journal.pone.0027527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dietzel S, Jauch A, Kienle D, Qu G, Holtgreve-Grez H, Eils R, Münkel C, Bittner M, Meltzer PS, Trent JM, Cremer T. Separate and variably shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res. 1998;6:25–33. doi: 10.1023/a:1009262223693. [DOI] [PubMed] [Google Scholar]
- 77.Nagano T, Lubling Y, Stevens T, Schoenfelder S, Yaffe E, Dean W, Laue E, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 80.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, Cremer C, Bult CJ, O’Brien TP. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol. 2006;174:27–38. doi: 10.1083/jcb.200603083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011;19:901–909. doi: 10.1007/s10577-011-9245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Blüthgen N, Dekker J, Heard E. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, Bland MJ, Wagstaff W, Dalton S, McDevitt TC, Sen R, Dekker J, Taylor J, Corces VG. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 87.Szabó P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 88.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 89.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoon Y, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seitan VC, Krangel MS, Merkenschlager M. Cohesin, CTCF and lymphocyte antigen receptor locus rearrangement. Trends Immunol. 2012;33:153–159. doi: 10.1016/j.it.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- 93.Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 94.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 95.Laster K, Kosak ST. Genomic Pangea: coordinate gene regulation and cell-specific chromosomal topologies. Curr Opin Cell Biol. 2010;22:314–319. doi: 10.1016/j.ceb.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 96.Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Junier I, Martin O, Képès F. Spatial and topological organization of DNA chains induced by gene co-localization. PLoS Comput Biol. 2010;6:e1000678. doi: 10.1371/journal.pcbi.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 100.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 102.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kocanova S, Kerr EA, Rafique S, Boyle S, Katz E, Caze-Subra S, Bickmore WA, Bystricky K. Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet. 2010;6:e1000922. doi: 10.1371/journal.pgen.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 108.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 111.Deniaud E, Bickmore WA. Transcription and the nuclear periphery: edge of darkness? Curr Opin Genet Dev. 2009;19:187–191. doi: 10.1016/j.gde.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 112.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Capelson M, Doucet C, Hetzer MW. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb Symp Quant Biol. 2010;75:585–597. doi: 10.1101/sqb.2010.75.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 115.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brickner DG, Brickner JH. Interchromosomal clustering of active genes at the nuclear pore complex. Nucleus. 2012;3:487–492. doi: 10.4161/nucl.22663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Green EM, Jiang Y, Joyner R, Weis K. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell. 2012;23:1367–1375. doi: 10.1091/mbc.E11-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 123.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bukata L, Parker SL, D’Angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Curr Opin Cell Biol. 2013;25:378–386. doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 126.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 127.Carter D, Chakalova L, Osborne CS, Dai Y-f, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 128.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 130.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 132.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, Rathjen PD, Gilbert DM. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, Fisher AG. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 142.Dion V, Gasser SM. Chromatin movement in the maintenance of genome stability. Cell. 2013;152:1355–1364. doi: 10.1016/j.cell.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 143.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14:502–509. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 144.Neumann FR, Dion V, Gehlen LR, Tsai-Pflugfelder M, Schmid R, Taddei A, Gasser SM. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 2012;26:369–383. doi: 10.1101/gad.176156.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol. 2013;15:694–699. doi: 10.1038/ncb2745. [DOI] [PubMed] [Google Scholar]
- 146.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krawczyk PM, Borovski T, Stap J, Cijsouw T, ten Cate R, Medema JP, Kanaar R, Franken NA, Aten JA. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci. 2012;125:2127–2133. doi: 10.1242/jcs.089847. [DOI] [PubMed] [Google Scholar]
- 148.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dion V, Kalck V, Seeber A, Schleker T, Gasser SM. Cohesin and the nucleolus constrain the mobility of spontaneous repair foci. EMBO reports. 2013;14:984–991. doi: 10.1038/embor.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mehta IS, Kulashreshtha M, Chakraborty S, Kolthur-Seetharam U, Rao BJ. Chromosome territories reposition during DNA damage-repair response. Genome Biol. 2013;14:R135. doi: 10.1186/gb-2013-14-12-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Crabbe L, Cesare AJ, Kasuboski JM, Fitzpatrick JA, Karlseder J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell reports. 2012;2:1521–1529. doi: 10.1016/j.celrep.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 155.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Görisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc Natl Acad Sci U S A. 2004;101:13221–13226. doi: 10.1073/pnas.0402958101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carmo-Fonseca M, Berciano MT, Lafarga M. Orphan nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000703. doi: 10.1101/cshperspect.a000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bauer DC, Willadsen K, Buske FA, Lê Cao KA, Bailey TL, Dellaire G, Bodén M. Sorting the nuclear proteome. Bioinformatics. 2011;27:i7–14. doi: 10.1093/bioinformatics/btr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 160.Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lindsay CR, Morozov VM, Ishov AM. PML NBs (ND10) and Daxx: from nuclear structure to protein function. Frontiers in bioscience: a journal and virtual library. 2008;13:7132–7142. doi: 10.2741/3216. [DOI] [PubMed] [Google Scholar]
- 164.Zhong S, Salomoni P, Ronchetti S, Guo A, Ruggero D, Pandolfi PP. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. The Journal of experimental medicine. 2000;191:631–640. doi: 10.1084/jem.191.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chang CC, Naik MT, Huang YS, Jeng JC, Liao PH, Kuo HY, Ho CC, Hsieh YL, Lin CH, Huang NJ, Naik NM, Kung CC, Lin SY, Chen RH, Chang KS, Huang TH, Shih HM. Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell. 2011;42:62–74. doi: 10.1016/j.molcel.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 166.Morozov VM, Gavrilova EV, Ogryzko VV, Ishov AM. Dualistic function of Daxx at centromeric and pericentromeric heterochromatin in normal and stress conditions. Nucleus. 2012;3:276–285. doi: 10.4161/nucl.20180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Delbarre E, Ivanauskiene K, Kuntziger T, Collas P. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013;23:440–451. doi: 10.1101/gr.142703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Corpet A, Olbrich T, Gwerder M, Fink D, Stucki M. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell cycle (Georgetown, Tex) 2013;13 doi: 10.4161/cc.26988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 170.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 172.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 173.Rokudai S, Laptenko O, Arnal SM, Taya Y, Kitabayashi I, Prives C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci U S A. 2013;110:3895–3900. doi: 10.1073/pnas.1300490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 175.Gialitakis M, Arampatzi P, Makatounakis T, Papamatheakis J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol Cell Biol. 2010;30:2046–2056. doi: 10.1128/MCB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ulbricht T, Alzrigat M, Horch A, Reuter N, von Mikecz A, Steimle V, Schmitt E, Krämer OH, Stamminger T, Hemmerich P. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J Cell Biol. 2012;199:49–63. doi: 10.1083/jcb.201112015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ching RW, Ahmed K, Boutros PC, Penn LZ, Bazett-Jones DP. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J Cell Biol. 2013;201:325–335. doi: 10.1083/jcb.201211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley interdisciplinary reviews. RNA. 2013;4:17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 181.Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci. 2012;125:166–175. doi: 10.1242/jcs.090837. [DOI] [PubMed] [Google Scholar]
- 182.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor K, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang F, Tang ML, Zeng ZX, Wu RY, Xue Y, Hao YH, Pang DW, Zhao Y, Tan Z. Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc Natl Acad Sci U S A. 2012;109:20413–20418. doi: 10.1073/pnas.1200232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Leung AK, Lamond AI. The dynamics of the nucleolus. Critical reviews in eukaryotic gene expression. 2003;13:39–54. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.40. [DOI] [PubMed] [Google Scholar]
- 190.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 191.Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Leung AK, Andersen JS, Mann M, Lamond AI. Bioinformatic analysis of the nucleolus. The Biochemical journal. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 194.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 195.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 197.van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Langst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Levy DL, Heald R. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol. 2012;28:113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- 203.Edens LJ, White KH, Jevtic P, Li X, Levy DL. Nuclear size regulation: from single cells to development and disease. Trends Cell Biol. 2013;23:151–159. doi: 10.1016/j.tcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 204.Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Meyerzon M, Gao Z, Liu J, Wu JC, Malone C, Starr D. Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Developmental biology. 2009;327:433–446. doi: 10.1016/j.ydbio.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kieserman EK, Heald R. Mitotic chromosome size scaling in Xenopus. Cell Cycle. 2011;10:3863–3870. doi: 10.4161/cc.10.22.17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shimi T, Butin-Israeli V, Goldman RD. The functions of the nuclear envelope in mediating the molecular crosstalk between the nucleus and the cytoplasm. Curr Opin Cell Biol. 2012;24:71–78. doi: 10.1016/j.ceb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Versaevel M, Grevesse T, Gabriele S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nature communications. 2012;3:671. doi: 10.1038/ncomms1668. [DOI] [PubMed] [Google Scholar]
- 211.Iyer KV, Pulford S, Mogilner A, Shivashankar GV. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophysical journal. 2012;103:1416–1428. doi: 10.1016/j.bpj.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Poh YC, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N. Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nature communications. 2012;3:866. doi: 10.1038/ncomms1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yuxiang Z, Qinghua Z. Information Systems Frontiers. 2012. Evaluation on crowdsourcing research: Current status and future direction. [Google Scholar]
- 214.Moreira JA, Pacheco JM, Santos FC. Evolution of collective action in adaptive social structures. Sci Rep. 2013;3:1521. doi: 10.1038/srep01521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.