Abstract
The sequence selectivity of 14 classical protein-tyrosine phosphatases (PTPs) (PTPRA, PTPRB, PTPRC, PTPRD, PTPRO, PTP1B, SHP-1, SHP-2, HePTP, PTP-PEST, TCPTP, PTPH1, PTPD1, and PTPD2) was systematically profiled by screening their catalytic domains against combinatorial peptide libraries. All of the PTPs exhibit similar preference for pY peptides rich in acidic amino acids and disfavor positively charged sequences, but differ vastly in their degrees of preference/disfavor. Some PTPs (PTP-PEST, SHP-1, and SHP-2) are highly selective for acidic over basic (or neutral) peptides (by >105-fold), whereas others (PTPRA and PTPRD) show no to little sequence selectivity. PTPs also have diverse intrinsic catalytic efficiencies (kcat/KM values against optimal substrates), which differ by >105-fold due to different kcat and/or KM values. Moreover, PTPs show little positional preference for the acidic residues relative to the pY residue. Mutation of Arg47 of PTP1B, which is located near the pY-1 and pY-2 residues of a bound substrate, decreased the enzymatic activity by 3–18-fold toward all pY substrates containing acidic residues anywhere within the pY-6 to pY+5 region. Similarly, mutation of Arg24, which is situated near the C-terminus of a bound substrate, adversely affected the kinetic activity of all acidic substrates. A co-crystal structure of PTP1B bound with a nephrin pY1193 peptide suggests that Arg24 engages in electrostatic interactions with acidic residues at the pY+1, pY+2, and likely other positions. These results suggest that long-range electrostatic interactions between positively charged residues near the PTP active site and acidic residues on pY substrates allow a PTP to bind acidic substrates with similar affinities and the varying levels of preference for acidic sequences by different PTPs are likely caused by the different electrostatic potentials near their active sites. The implications of the varying sequence selectivity and intrinsic catalytic activities with respect to PTP in vivo substrate specificity and biological functions are discussed.
Keywords: Combinatorial library, catalytic activity, kinetics, phosphotyrosine, PTP, substrate specificity
Protein-tyrosine phosphatases (PTPs) are a large family of enzymes that catalyze the hydrolysis of phosphotyrosine (pY) in proteins to tyrosine and inorganic phosphate. The human genome encodes 107 putative PTPs, with the class I cysteine-based PTPs constituting the largest group.1 This group is divided into 61 dual-specificity phosphatases (DUSPs) and 38 tyrosine-specific or ‘‘classical” PTPs. The latter can be further classified into the receptor and nonreceptor subgroups. Dysregulation of PTPs is associated with a multitude of diseases, and many members of the PTP family have been recognized as potential therapeutic targets.2,3 PTPs are actively involved in cell signaling, capable of either turning signaling pathways on or off.1–5 To date, over 15000 pY sites on >6000 mammalian proteins have been reported,6 which are presumably dephosphorylated by the 107 PTPs. The large number of PTPs and potential substrate proteins suggests that PTPs must have some level of substrate specificity in vivo in order to achieve fidelity for the signaling pathways in which they are involved. Currently, the prevailing view is that PTPs have “exquisite substrate specificity” in vivo. However, how PTPs achieve such specificity is not well understood. It has been proposed that the substrate specificity of PTPs is controlled combinatorially by the intrinsic sequence specificity of the catalytic domain and other regulatory mechanisms, including specific tissue distribution, restricted subcellular localization, posttranslational modification events (e.g., phosphorylation), and accessory or regulatory domains (e.g., KIM motifs or SH2 domains).7,8 For receptor PTPs, homodimerization (and possibly, heterodimerization) provides yet another potential mechanism to regulate the PTP activity.9
There have been many previous attempts to define the intrinsic sequence selectivity (or “active-site specificity”) of PTPs and its contribution to the overall PTP substrate specificity in vivo.10 However, this has been a challenging task because the PTP active site interacts with at least 3–5 residues on either side of pY; definition of PTP sequence selectivity requires the analysis of a large number of pY peptide sequences. We recently developed a combinatorial peptide library approach for systematically profiling the sequence selectivity of PTPs.10–12 In this study, we applied the peptide library method to profile the intrinsic sequence selectivity of 4 receptor [PTPRB (RPTPβ), PTPRC (CD45), PTPRD (RPTPδ), and PTPRO (GLEPP1)] and 6 nonreceptor PTPs [(HePTP (PTPN7), PTP-PEST (PTPN12), TCPTP (PTPN2), PTPH1 (PTPN3), PTPD1 (PTPN21), and PTPD2 (PTPN14)]. Surprisingly, these PTPs, as well as 3 out of the 4 PTPs we have studied previously (PTP1B, SHP-1, and SHP-2),11 all have a similar preference for pY peptides rich in acidic residues (e.g., Asp and Glu) and disfavor positively charged sequences, but do not exhibit simple consensus sequences. Kinetic analyses of the 14 PTPs against model pY-peptide substrates showed that the PTPs have vastly different catalytic properties. The molecular basis of this unusual sequence selectivity was further investigated by site-directed mutagenesis and X-ray crystallographic studies. Our results suggest that long-range electrostatic interactions between basic residues near the PTP active site and acidic residues flanking the pY residue on peptide substrates are responsible for the unusual sequence selectivity of PTPs. They also suggest that PTPs likely have varying levels of substrate specificity in vivo, and some PTPs might take advantage of their low intrinsic catalytic efficiencies to achieve selective dephosphorylation of target proteins.
MATERIALS AND METHODS
Materials
Reagents for peptide synthesis were purchased from Advanced ChemTech (Louisville, KY), AAPPTec (Louisville, KY), Chem-Impex (Wood Dale, IL), or Peptides International (Louisville, KY). N-(9-Fluorenylmethoxycarbonyloxy)succinimide was purchased from Advanced ChemTech. α-Cyano-4-hydroxycinnamic acid, phenyl isothiocyanate, and 3-methyl-2-benzothiazolinonehydrazone (MBTH) were obtained from Sigma-Aldrich. Nα-Fmoc-O-t-butyl-3,5-difluorotyrosine (F2Y) was synthesized as described previously.13 Tyrosinase from Streptomyces antibioticus was expressed in Escherichia coli and purified as described previously.10
Purification of PTPs
The catalytic domains of wild-type and mutant PTP1B (amino acids 1–321) and SHP-1 (aa 205–595) were expressed in Escherichia coli and purified as previously described.10,14 PTPD2 (aa 876–1187), PTPRA (aa 224–802), and PTPRC (aa 620–1236), all containing an N-terminal histidine tag, and SHP-2 (aa 199–593) and PTPD1 (aa 875–1174), each containing a C-terminal histidine tag, were expressed in E. coli and purified as described.15 TCPTP (aa 1–314), PTP-PEST (aa 5–300), PTPRB (aa 1686–1971), HEPTP (aa 22–335), PTPH1 (amino acids 628–909), PTPRD (aa 1299–1899), and PTPRO PTP domain were expressed in E. coli as fusion proteins with glutathione-S-transferase (GST) and purified as described previously.15,16 Protein concentrations were determined by the Bradford method, using bovine serum albumin (BSA) as the standard.
Library Screening
Peptide libraries I–V (Table 1) were synthesized on polyethylene glycol acrylamide resin (0.4 mmol/g; 150–300 µm diameter in water), as described previously.10 In a typical screening experiment, 15 mg of the appropriate peptide library (dry weight, ~45,000 beads) was placed in a plastic micro-BioSpin column (2 mL, Bio-Rad) and extensively washed with DMF and ddH2O. The resin was blocked for 1 h with blocking buffer (30 mM Hepes, pH 7.4, 150 mM NaCl, 0.01% Tween 20, and 0.1% gelatin). The library was then incubated with a given PTP (final concentration 0.5–800 nM) in blocking buffer containing 5 mM tris(carboxyethyl)phosphine (TCEP) (total volume 0.8 mL) at room temperature for 5–30 min with gentle mixing. The resin was drained, washed with 0.1 M KH2PO4 (pH 6.8), and resuspended in 1.6 mL of 0.1 M KH2PO4 (pH 6.8) containing 1.2 µM tyrosinase and 6 mM 3-methyl-2-benzothiazolinonehydrazone (MBTH). The resulting mixture was incubated at room temperature with gentle mixing and exposure to air. Intense pink/red color typically developed on positive beads after 20–60 min. Positive beads were retrieved from the library using a micropipette under a dissecting microscope, and sequenced by the partial Edman degradation-mass spectrometry (PED-MS) method.17 Control experiments without PTPs produced no red beads under otherwise identical conditions. Detailed conditions for individual screening experiments are provided in Table S1 in Supporting Information (SI).
Table 1.
Peptide Libraries Employed in This Work
| Library No. | Library Design | Amino Acid Composition at Random Positions (X) |
|---|---|---|
| I | ASXXXXXpYAABBRM | Ala, Arg, Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| II | Alloc-AXXXXXXXXpYAABBRM | Ala, Arg, Asn, Asp, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| III | Alloc-ASXXXXXpYAABBRM | Ala, Arg (10-fold reduced content), Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys (10-fold reduced content), Nle, Phe, Pro, Ser, Thr, Trp, Val |
| IV | Alloc-AApYXXXXXNNBBRM | Ala, Arg, Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| V | Alloc-AXXXXXpYXXXXXNNBBRM | Ala, Arg, Asn, Asp, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
Synthesis of Selected Peptides
Individual peptides were synthesized on 100 mg of CLEAR-amide resin using standard Fmoc/HBTU/HOBt chemistry. For coupling of pY, 2.0 equivalents of Fmoc-Tyr(PO3H2)-OH were employed, whereas 4.0 equivalents were used for all other amino acids. Resin-bound peptides were washed with dichloromethane, cleaved from the resin, and deprotected using modified reagent K [7.5% phenol, 5% water, 5% thioanisole, 2.5% ethanedithiol, and 1% anisole in trifluoroacetic acid (TFA)] at room temperature for 2 h. After evaporation of solvents, the mixture was triturated three times with 20 volumes of cold Et2O. Precipitates were collected and dried under vacuum, and crude peptides were purified by reversed-phase HPLC on a semi-preparative C18 column. The identity of each peptide was confirmed by MALDI-TOF mass spectrometric analysis.
PTP Assays
PTP activities were determined by two different methods. For method A, assay reactions were performed in a quartz microcuvette (total volume 120 µL) containing 100 mM HEPES, pH 7.4, 50 mM NaCl, 2 mM EDTA, 1 mM TCEP, 0.1 mg/mL BSA, and 0–1.4 mM pY peptide. The reaction was initiated by the addition of PTP (final concentration 0.5–20000 nM) and monitored continuously at 282 nm (Δε = 1102 M−1 cm−1 ) on a UV-VIS spectrophotometer. Initial rates were calculated from the early regions of the reaction progress curves (typically <60 s). Data fitting against the Michaelis-Menten equation V = Vmax · [S]/(KM + [S]) or the simplified equation V = kcat[E][S]/KM (when KM >> [S]) gave the kinetic constants kcat, KM, and/or kcat/KM. For method B, reactions were performed similarly, but reaction progress was monitored continuously at 282 nm until the substrate was depleted. The kinetic constants were obtained by directly fitting the reaction progress curves against equation:
where t is time, p is the product concentration at time t, [E] is the total enzyme concentration, and p∞ is the product concentration at infinity.18
Pre-Steady State Kinetics
Pre-steady-state kinetic experiments were carried out by rapidly mixing the PTP and p-nitrophenyl phosphate (pNPP) in a temperature-controlled Applied Photophysics SX20 stopped-flow spectrophotometer equipped with a detector of 5-mm path length, and monitoring reaction progression at 410 nm. The reactions contained 100 mM Tris, pH 7.4, 50 mM NaCl, 2 mM EDTA, 1 mM TCEP, and 20 mM pNPP (final concentration). Experiments with PTPD1 contained 10 µM enzyme (final concentration) and were carried out at 4 °C, whereas PTPD2 reactions were performed with 5 µM enzyme (final concentration) at 25 °C. The rate constants (kburst and kcat) were determined by fitting the experimental data (Abs values as a function of time) against the equation:
where A and B are the amplitudes of the bursts, kburst is the first-order rate constant of the initial burst, t is the time, k1’ is the first-order rate constant of the conformational change, C is the slope of the final linear phase of the curve, and D is the y-intercept of the linear phase of the curve. The kcat value was calculated from slope C according to the equation: kcat = C / (ε • [E]) where ε is the molar absorptivity of pNPP at 410 nm (10,000 M−1 cm−1) and [E] is the enzyme concentration. The kinetic constants k2 and k3 were determined using the equations kburst = k2 + k3 and kcat = k2 • k3/(k2 + k3).
Crystallization and Data Collection
The purified catalytically inactive C215S PTP1B (aa 1–321) was co-crystallized with the peptide Ac-AWGPLpYDEVQM-NH2 (where M is L-norleucine) at 4 °C by hanging drop vapor diffusion. The catalytic domain (10 mg/mL) in 0.1 M HEPES (pH 7.0) and 50 mM NaCl was mixed with 10-fold molar excess of peptide dissolved in water. The hanging drops were prepared by mixing 2 µL of the complex and 2 µL of the reservoir solution, which consisted of 14.4 % PEG 8000, 0.1 M HEPES (pH 7.5), and 0.2 M MgCl2. Thin needle crystals grew after one week. For X-ray data collection, crystals were transferred to a cryo-protectant solution containing the reservoir solution supplemented with 25% glycerol. A single crystal was mounted on a nylon loop and plunged in liquid nitrogen. X-ray diffraction data were collected at the LRL-CAT beamline 31-ID of the Advanced Photon Source, Argonne National Laboratory. Diffraction data were integrated and scaled using MOSFLM and SCALA of the CCP4 suite.19
Structure Determination
The structure was determined by molecular replacement with MOLREP of the CCP4 suite using the structure of the free PTP1B catalytic domain [Protein Data Bank (PDB) entry 1PTY] as the search model.19 After an initial round of refinement at 1.74 Å using REFMAC5 of the CCP4 suite,20 electron density maps revealed strong density for the pY residue of the nephrin peptide, which was built into the model using COOT.21 Several additional rounds of refinement and model building led to the final model consisting of residues 2–298 of the PTP1B catalytic domain (chain A), and residues pY–3 to pY+2 of the nephrin peptide (chain B). Data collection and refinement statistics are listed in Table 4. Structural figures were prepared using CCP4MG.22
Table 4.
SHP-1, SHP-2, and PTP1B Activity toward Acidic pY Peptides of Different Sequences
| Entry No. |
Enzyme | Substratea |
kcat (s−1) |
km (µM) |
kcat/KM x 106 (M−1 s−1) |
|---|---|---|---|---|---|
| 1 | SHP-1b | SASASpYSASA | 26 ± 4 | 1990 ± 420 | 0.013 |
| 2 | WAGDDpYAA | 57 ± 2 | 45 ± 5 | 1.3 | |
| 3 | FDIDIpYAA | 33 ± 1 | 13 ± 1 | 2.5 | |
| 4 | EIFDFpYAA | 32 ± 2 | 27 ± 4 | 1.2 | |
| 5 | FYDIDpYAA | 66 ± 2 | 28 ± 2 | 2.4 | |
| 6 | SHP-2b | SASASpYSASA | 42 ± 5 | 2300 ± 400 | 0.018 |
| 7 | WAGDDpYAA | 36 ± 3 | 68 ± 15 | 0.53 | |
| 8 | FDIDIpYAA | 30 ± 2 | 39 ± 5 | 0.77 | |
| 9 | EIFDFpYAA | 26 ± 1 | 54 ± 6 | 0.48 | |
| 10 | FYDIDpYAA | 50 ± 4 | 66 ± 13 | 0.75 | |
| 11 | PTP1B | SASASpYSASA | 28 ± 1 | 12 ± 1 | 2.3 |
| 12 | EEDNAWpYAA | 27 ± 1 | 1.3 ± 0.1 | 21 | |
| 13 | ASSDEpYAA | 31 ± 1 | 1.8 ± 0.1 | 17 | |
| 14 | AApYDLDEY | 34 ± 1 | 1.9 ± 0.3 | 18 | |
| 15 | AApYWAYDD | 34 ± 1 | 2.3 ± 0.4 | 15 | |
| 16 | AApYDDIDE | 37 ± 2 | 3.5 ± 0.1 | 11 | |
All substrates have an N-terminal acetyl group and a C-terminal amide.
Data from Ref. 10.
RESULTS
Peptide Library Design and Screening
Five different peptide libraries were employed in this work (Table 1). Each PTP was first screened against library I and/or II. Library I contains five random residues N-terminal to pY, ASXXXXXpYAABBRM-resin [where B is β-alanine and X is F2Y (used as a Tyr surrogate), norleucine (Nle or M, used as a replacement of Met), or any of the 19 proteinogenic amino acids except for Tyr, Met, or Cys]. Library II, which is similar to library I but contains eight random residues N-terminal to pY, was designed to test whether residues beyond pY-5 (relative to pY, which is defined as position 0) affect PTP activity and/or specificity. Library III has 10-fold reduced Arg and Lys content, but is otherwise identical to library I. When screened against library I or II, some PTPs (e.g., PTPRA10) selected predominantly Arg- and Lys-rich sequences, which had poor activity when tested against the PTPs in solution. This bias was likely caused by multi-dentate, electrostatic interactions between the positively charged sequences and negatively charged surface patches on the PTPs,23 resulting in recruitment of greater amounts of PTPs to the “false positive” beads. We previously showed that reduction of Arg and Lys content in the library by 10-fold effectively eliminated the Arg-and Lys-rich sequences, allowing the specificity profile of PTPRA to be determined.10 For most of the PTPs examined in this work (except for PTP-PEST), screening against library II or III produced sequence data of the highest quality, which are presented below. Library IV contains five random residues C-terminal to pY (Alloc-AApYXXXXXNNBBRM-resin), whereas library V contains five random residues on each side of pY (Alloc-AXXXXXpYXXXXXNNBBRM-resin). Library V was designed to test whether a PTP requires specific sequences on both N- and C-terminal sides of pY for activity and/or exhibits covariance between the N- and C-terminal sequences. Libraries I, III, and IV have a theoretical diversity of ~2.5 × 106, whereas libraries II and V have theoretical diversities of 1.7 × 1010 and 6.1 × 1012, respectively. All peptide libraries were synthesized in the one-bead-one-compound (OBOC) format on polyethylene glycol acrylamide (PEGA) resin (150–300 µm in water, ~3 × 106 beads/g of dry resin). Library screening involved treatment of a portion of the library beads (typically 15 mg of dry resin in each reaction) with a limited amount of PTP so that only beads that display the most efficient substrates underwent significant dephosphorylation. The optimal PTP concentration and reaction time were determined by trial and error and varied greatly depending on the catalytic efficiency of the PTP (Table S1). After termination of the PTP reaction (by washing the library beads with a buffer), the exposed Tyr side chain was oxidized into an orthoquinone by incubation with tyrosinase in the presence of O2.10,11 The orthoquinone was captured in situ by MBTH to form a reddish pigment, which remained covalently attached to the beads. Both tyrosinase and MBTH were used in excess to ensure that the coloration reaction went to completion. The tyrosinase exhibits no sequence selectivity.10 Positive (red) beads were isolated manually from the library with a micropipette and were sequenced individually by using the PED-MS method.17
PTPs Have Similar Specificity Profiles
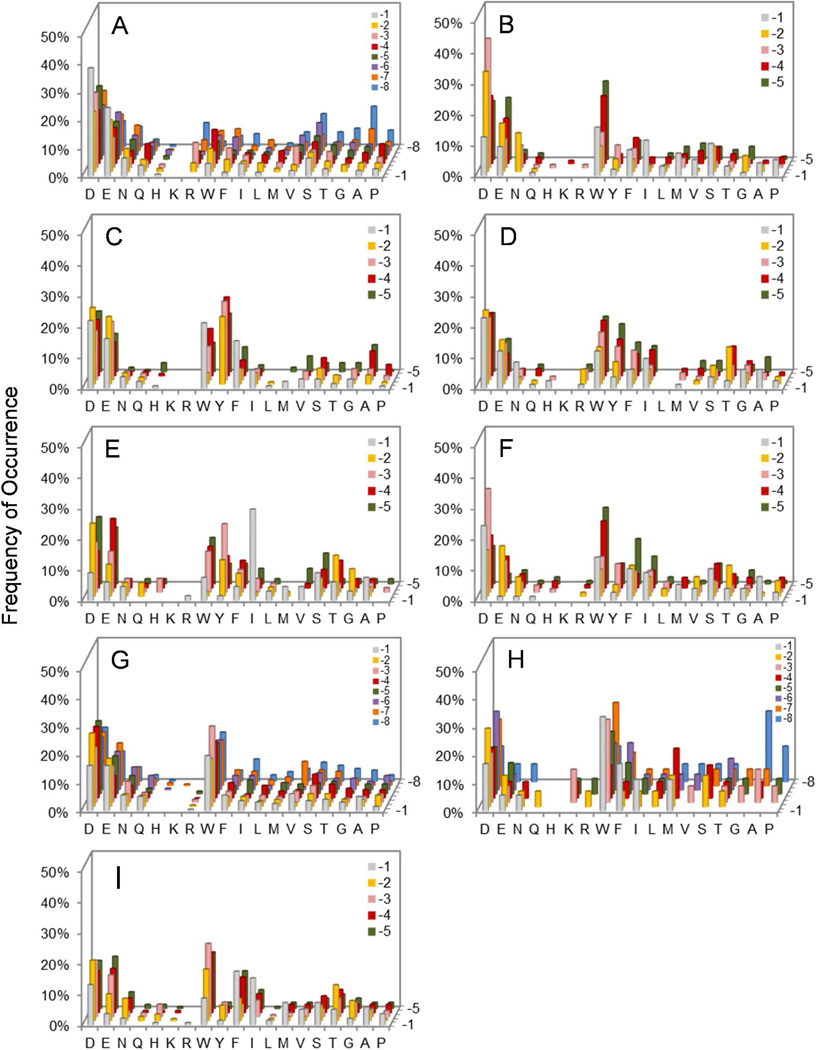
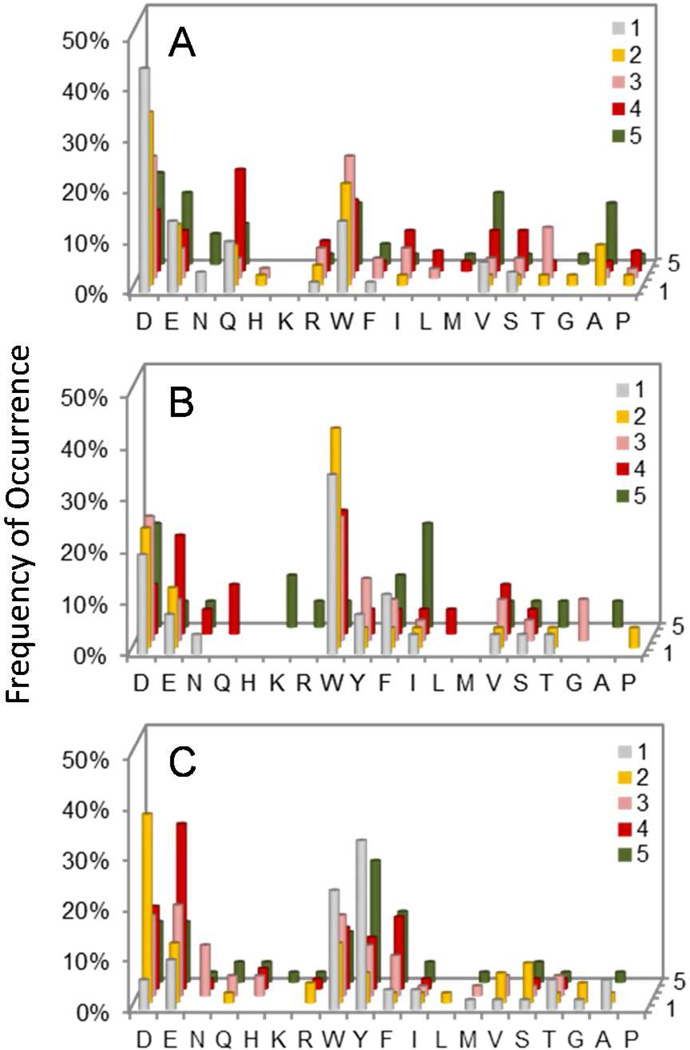
A total of 45 mg of library II was screened against 500 nM HePTP in three separate experiments. Sequencing of the intensely, medium, and lightly red beads gave 61, 106, and 99 complete (and some partial) sequences, respectively (Table S2). HePTP was also screened against library IV (30 mg resin in 2 experiments), producing 50 complete sequences (Table S3). HePTP strongly preferred a hydrophilic residue (especially Asp and Glu) at the pY-1 position (Figure 1a); 42 out of the 61 sequences (69%) derived from intensely colored beads, which represent the most efficient HePTP substrates, had Asp or Glu at this position, whereas most of the remaining sequences contained Asn, Gln, Thr, and Ser. The absence of large hydrophobic amino acids (e.g., Try and Phe) at the pY-1 position makes HePTP a notable exception among the classical PTPs studied so far. It also has strong preference for acidic residues at positions pY+1 and pY+2 and modest preference for acidic residues at all other positions (Figure 1a and Figure 2a). Note that the number of beads/sequences screened against HePTP represents only a small fraction of the entire sequence space (0.0008% and 3.6% for library II and IV, respectively). We have previously demonstrated that randomly sampling a small fraction of a combinatorial peptide library is sufficient to unambiguously determine the specificity profiles of protein binding domains,24 protein kinases,25 and protein phosphatases.10–12
Figure 1.
Histograms showing the sequence selectivity of HePTP (A), PTP-PEST (B), PTPH1 (C), TCPTP (D), PTPD2 (E), PTPRB (F), PTPRC (G), PTPRD/Class I (H), and PTPRO (I) on the N-terminal side of pY. The y axis represents the percentage of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY-8 to pY-1, on the z axis). Data shown were from libraries I and III for PTP-PEST, library II for HePTP, PTPRC, and PTPRD, and library III for PTPH1, TCPTP, PTPD2, PTPRB, PTPRO. M, Nle; Y, F2Y.
Figure 2.
Histograms showing the C-terminal selectivity of HePTP (A), TCPTP/Class I (B), and PTPD2/Class I (C) derived from Library IV. The y axis represents the percentage of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY+1 to pY+5, on the z axis). M, Nle; Y, F2Y.
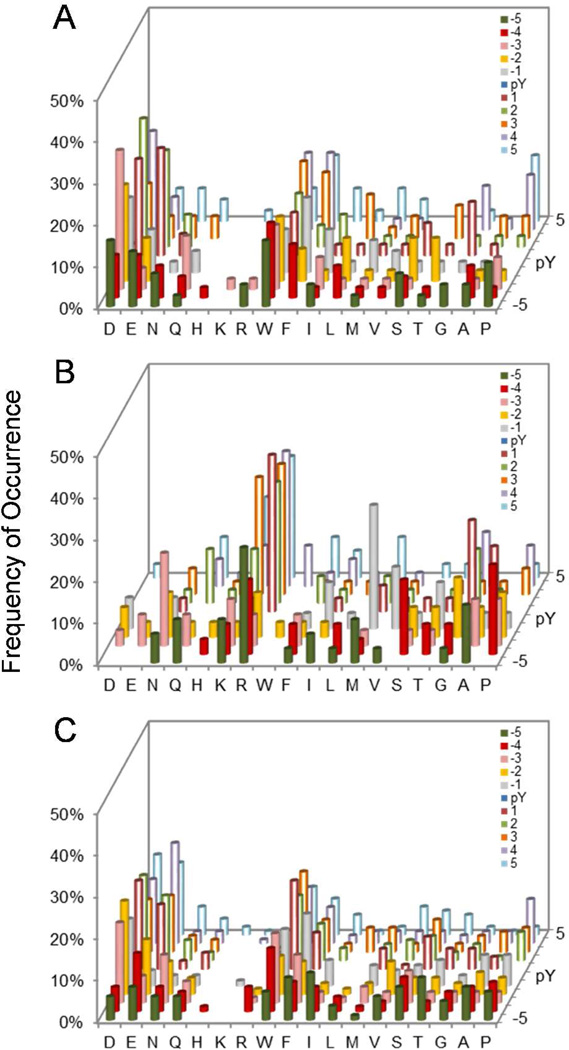
Screening of the PTP-PEST catalytic domain against libraries I and III produced 96 sequences (Table S4 in SI). PTP-PEST strongly prefers acidic residues, Asn, or Ser at the pY-2 and pY-3 positions (Figure 1b). Unlike HePTP, it prefers a hydrophobic residue (e.g., Trp, Ile, Phe) at the pY-1 position and either acidic or hydrophobic residues at the pY-4 and pY-5 positions. To determine the C-terminal specificity, PTP-PEST was also screened against library V, which features five random residues on each side of pY. Again, PTP-PEST exhibited overwhelming preference for acidic sequences; each of the 39 selected sequences contained two or more acidic residues, which occurred most frequently at the pY+2, pY+1, pY-3, and pY-2 positions (Figure 3a and Table S5). Only 7/135 sequences selected from the two libraries contained an Arg, Lys, and/or His residue, and these residues usually occupied less critical positions (e.g., pY-5 and pY+5).
Figure 3.
Histograms showing the sequence selectivity of PTP-PEST (A), PTPRB (B), and PTPRO/Class I (C) derived from library V. The y axis represents the percentage of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY-5 to pY+5, on the z axis). M, Nle.
Initial screening of PTPH1 against library II generated exclusively Arg- and Lys-rich sequences, which had poor activity when tested against PTPH1 in solution (Table S6 and vide infra). To alleviate this screening bias, we screened PTPH1 against library III, which has a reduced Arg and Lys content (by 10-fold). Screening of library III against PTPH1 produced 138 preferred sequences, which were rich in aromatic hydrophobic (e.g., Trp, Tyr, and Phe) and, to a lesser extent, acidic residues (Table S7 and Figure 1c). Remarkably, no Arg or Lys was found in any of the 138 sequences, indicating that PTPH1 strongly disfavors basic pY peptides as substrate. The X-ray structure of PTPH1 shows that it has a highly positively charged surface surrounding the active site, but highly negatively charged surfaces elsewhere.15
TCPTP behaved similarly to PTPH1 in that screening against library I produced many Arg- and Lys-rich sequences (Table S8). When screened against library III, it selected peptides rich in acidic and aromatic hydrophobic residues. However, despite the 10-fold reduced Arg and Lys content, the selected peptides contained a fair number of Arg and His residues, suggesting that TCPTP is more tolerant to basic amino acids than PTPH1 (Table S9 and Figure 1d). Screening of the C-terminal library (library IV) gave 26 acidic sequences and 45 basic sequences, consistent with the notion that TCPTP has very broad sequence specificity and is relatively tolerant to basic sequences (Table S10 and Figure 2b).
PTPD2 was screened against libraries III and IV (Table S11 and Table S12, respectively). Compared to other PTPs, substantially higher PTPD2 concentration (800 nM) and longer reaction time (60 min) were necessary to generate positive beads (Table S1). It strongly prefers Ile or other hydrophobic residues at the pY-1 position and has moderate preference for acidic and aromatic hydrophobic residues at the pY-2 to pY-5 positions (Figure 1e). Like PTPH1, PTPD2 appears to strongly disfavor basic sequences on the N-terminal side of pY. On the C-terminal side, it favors Tyr or Trp at the pY+1 position and acidic residues at the pY+2 position (Figure 2c). Library screening against PTPD1 failed to produce positive beads under the conditions employed in this work (500 nM PTPD1 for 60 min), suggesting that it has low catalytic activity toward all peptide substrates.
The four receptor PTPs showed very similar, broad specificity profiles (Table S13–18, Figure 1f–1i, and Figure 3b–3c). They all have moderate to weak preference for acidic and aromatic hydrophobic residues. PTPRB is unique in that it prefers an Asp (but not Glu) residue at the pY-1 position and tolerates small hydrophilic (e.g., Ser, Thr, Asn) but not basic (Arg, Lys, and His) residues at this position. It has moderate preference for acidic residues at pY-2 and pY-3 and aromatic hydrophobic residues at the pY-4 and pY-5 positions. PTPRD appears to be somewhat more tolerant to basic residues than the other 3 receptor PTPs, as indicated by the larger number of Arg and Lys residues selected.
Overall, the 10 PTPs examined in this study and three out of the four PTPs previously investigated (SHP-1, SHP-2, and PTP1B) show similar specificity profiles: they all prefer acidic and large hydrophobic amino acids and disfavor basic residues. The only exception is PTPRA, which exhibits little sequence selectivity.10 Surprisingly, none of the 14 PTPs exhibit any consensus sequence(s); the preferred acidic and hydrophobic residues were found at essentially any of the positions proximal to the pY residue (pY-8 to pY+5).
PTPs Exhibit Diverse Kinetic Properties and Different Degrees of Preference for Acidic Sequences
Since most of the PTPs (except for PTPRA) prefer acidic/hydrophobic sequences and disfavor basic ones, we assayed the 14 PTPs against a generic set of peptide substrates, Ac-YDEDFpYDYEF-NH2 (acidic), Ac-SASASpYSASA-NH2 (neutral), and Ac-YRKRFpYRYKF-NH2 (basic) to quantitatively assess their preference for acidic sequences and tolerance for basic ones. Remarkably, and in contrast to their similar specificity profiles, these PTPs displayed vastly different kinetic properties, with their intrinsic catalytic efficiencies differing by up to 2 × 105-fold (Table 2). The most active enzyme, PTP-PEST, had a kcat/KM value of 2.2 × 108 M−1 s−1 toward the acidic peptide, whereas the least active enzyme (PTPD1) had a value of only 1.2 × 103 M−1 s−1. The different catalytic efficiencies were caused both by differences in kcat, which ranged from 470 s−1 (PTP-PEST) to 0.032 s−1 (PTPD1) and/or by different KM values, which varied from 0.28 to 280 µM (or 1000-fold). Overall, PTP-PEST, SHP-1, SHP-2, PTP1B, TCPTP, PTPH1, PTPRB, PTPRC, and PTPRO are highly efficient enzymes (kcat/KM values ≥6.8 × 106 M−1 s−1), whereas HePTP, PTPD1, PTPRA, and PTPRD have much lower intrinsic activity (kcat/KM values ≤7.5 × 104 M−1 s−1) (Table 2). Interestingly, PTPD2, which was previously reported to lack significant activity in vitro,15 is actually a rather efficient catalyst, having an intrinsic activity (kcat/KM) of 1.1 × 105 M−1 s−1. However, like PTPD1, it has an unusually low kcat value (0.073 s−1), which probably made previous kinetic measurements (performed with less efficient substrates) difficult. Second, while all of the PTPs (with the exception of PTPRA) preferred acidic over basic sequences (Figure 1), their degree of preference/disfavor varied greatly. On one end of the spectrum, PTP-PEST, SHP-1, and SHP-2 were highly selective for acidic substrates, having kcat/KM values for Ac-YDEDFpYDYEF-NH2 ~105-fold higher than that of Ac-YRKRFpYRYKF-NH2 (Table 2 and 3). Their intermediate activities toward the neutral substrate (Ac-SASASpYSASA-NH2) indicate that these PTPs engage in attractive and repulsive interactions with acidic and basic sequences, respectively, as expected from the highly positive electrostatic potential surrounding their active sites.15 By contrast, PTPRA was almost completely insensitive to the substrate sequence, having essentially the same activity toward acidic, neutral, and basic peptides. Other PTPs showed intermediate selectivity for acidic sequences (21–2600-fold higher kcat/KM values for acidic than basic substrate). For some of the PTPs (e.g., PTPD1 and PTPRD), the selectivity is presumably the result of attractive interactions with acidic substrates, because they have similar activities toward neutral and basic substrates (Table 3). Other PTPs (e.g., PTPH1 and PTPD2) show relatively little discrimination between acidic and neutral substrates but strongly disfavor basic sequences. As a result of these specificity differences, the 14 PTPs have markedly different abilities for dephosphorylating various types of substrates. For example, PTP-PEST is 2930-fold more active than PTPRA toward peptide Ac-YDEDFpYDYEF-NH2 but is 30-fold less active toward peptide Ac-YRKRFpYRYKF-NH2.
Table 2.
Kinetic Constants of PTPs toward Acidic, Neutral, and Basic Peptide Substrates
| YDEDFpYDYEFa | SASASpYSASAa | YRKRFpYRYKFa | |||||||
|---|---|---|---|---|---|---|---|---|---|
| PTP | kcat (s −1) |
KM (µM) |
kcat/KM x106 (M−1 s−1 ) |
kcat (s−1) |
KM (µM) |
kcat/KM x106 (M−1 s−1 ) |
kcat (s−1) |
KM (µM) |
kcat/KM x106 (M−1 s−1 ) |
| PEST | 470 ± 10 | 2.2 ± 0.5 | 220 | 160 ± 2 | 800 ± 10 | 0.20 | − | − | 0.0015 |
| PTP1B | 21 ± 0.5 | 0.38 ± 0.02 | 55 | 28 ± 0.7 | 12 ± 0.3 | 2.3 | − | − | 0.037 |
| PTPRC | 300 ± 20 | 7.0 ± 0.9 | 43 | 340 ± 50 | 170 ± 20 | 2.0 | − | − | 0.049 |
| PTPRO | 200 ± 10 | 5.7 ± 0.8 | 35 | 240 ± 8 | 250 ± 10 | 0.94 | − | − | 0.053 |
| PTPRB | 55 ± 2 | 1.8 ± 0.4 | 31 | 56 ± 2 | 26 ± 1 | 2.2 | − | − | 0.028 |
| TCPTPb | 9.0 ± 0.4 | 0.31 ± 0.01 | 29 | 17 ± 2 | 10 ± 1 | 1.7 | − | − | 0.032 |
| SHP-1b | 75 ± 2c | 3.2 ± 0.3c | 23c | 26 ± 4c | 2000 ± 400c | 0.013 | − | − | 0.00023 |
| PTPH1 | 2.4 ± 0.06 | 0.28 ± 0.02 | 8.8 | 2.5 ± 0.2 | 5.8 ± 0.7 | 0.44 | − | − | 0.0033 |
| SHP-2b | 43 ± 1c | 6.3 ± 0.4c | 6.8c | 42 ± 5c | 2300 ± 400c | 0.018c | − | − | 0.00015 |
| PTPD2 | 0.073 ± 0.003 | 0.70 ± 0.09 | 0.11 | 0.12 ± 0.006 | 9.4 ± 0.4 | 0.013 | − | − | 0.000082 |
| PTPRA | 19 ± 0.6 | 260 ± 30 | 0.075 | − | − | 0.023 | − | − | 0.045 |
| HePTP | 7.0 ± 0.3 | 280 ± 25 | 0.025 | − | − | 0.00060 | − | − | 0.000010 |
| PTPRD | >0.86 | >60 | 0.014 | − | − | 0.00068 | − | − | 0.00070 |
| PTPD1 | 0.032 ± 0.005 | 26 ± 5 | 0.0012 | − | − | 0.0000038 | − | − | 0.0000057 |
All peptides have an N-terminal acetyl group and a C-terminal amide.
The peptide WDEDFpYDWEF was used in lieu of YDEDFpYDYEF
From ref (10)
Table 3.
Differential Preference of PTPs for Acidic over Neutral and Basic Sequencesa
| enzyme | Acidic/Basic | Acidic/Neutral | Neutral/Basic |
|---|---|---|---|
| PEST | 150000 | 1100 | 140 |
| SHP1 | 100000 | 1800 | 57 |
| SHP2 | 46000 | 370 | 120 |
| PTPH1 | 2600 | 20 | 130 |
| HePTP | 2500 | 42 | 59 |
| PTP1B | 1500 | 24 | 61 |
| PTPD2 | 1300 | 8 | 160 |
| PTPRB | 1100 | 14 | 77 |
| TCPTP | 910 | 17 | 53 |
| PTPRC | 880 | 22 | 41 |
| PTPRO | 660 | 37 | 18 |
| PTPD1 | 220 | 330 | 0.7 |
| PTPRD | 21 | 21 | 1 |
| PTPRA | 2 | 3 | 0.5 |
Ratio of kcat/KM values for peptides Ac-YDEDFpYDYEF-NH2 (acidic), Ac-SASASpYSASA-NH2 (neutral), and Ac-YRKRFpYRYKF-NH2 (basic).
PTPs are Insensitive to the Position of Acidic Residues
To confirm the observation from library screening that PTPs prefer acidic and hydrophobic residues but are insensitive to their positions relative to pY, we examined the kinetic activities of SHP-1, SHP-2, and PTP1B toward a panel of peptide substrates that contain a similar number of acidic and hydrophobic residues at different positions. As shown in Table 4, SHP-1 had similar activity toward peptides WAGDDpY, FDIDIpY, EIFDFpY, and FYDIDpY (≤2-fold difference in kcat/KM values), which each contain two acidic residues and 1–3 hydrophobic residues at different positions N-terminal to pY. The importance of the acidic residues in substrate binding and catalysis is underscored by its 100–200-fold lower activity toward the neutral peptide SASASpYSASA. SHP-2 showed the same trend (Table 4). PTP1B, which has moderate preference for acidic sequences, also had similar kcat/KM values toward a panel of acidic pY peptides.10 We chose PTP1B for further analysis, because its intermediate level of preference for acidic sequences is more representative of the classical PTP subfamily and various PTP1B mutants were already available.14,26 To this end, we synthesized and tested five additional pY peptides that contained acidic residues at very different positions, from the extreme N-terminus (EEDNAWpYAA) to the C-terminus (AApYWAYDD), against PTP1B (Table 4, entries 12–16). Again, all five acidic peptides were highly active toward PTP1B and had very similar kinetic activities (with kcat/KM values of 1–2 × 107 M−1 s−1), which were 5–10-fold higher than that of the neutral peptide SASASpYSASA. On the basis of these kinetic and specificity profiling data,10,27 we propose that the general preference for acidic sequences that lack a specific consensus motif may be a common feature of all PTPs.
Arg47 and Arg24 of PTP1B Engage in Long-Range Interactions with Acidic Residues in pY Substrates
A potential explanation for the unique sequence selectivity pattern of PTPs may be that the acidic residues in pY substrates engage in long-range electrostatic interactions with basic residues (Arg and Lys) surrounding the PTP active site. Because charge-charge interaction, which decreases linearly with the distance between opposite charges, is relatively distance insensitive, an acidic residue on a bound pY peptide may interact with all of the Arg and Lys residues near the PTP active site and generate similar overall binding energies irrespective of its precise location in the substrate. Conversely, a positively charged residue near the PTP active site may interact with all acidic residues that are proximal to the pY. Thus, the degree of preference/disfavor for acidic/basic sequences by a PTP would be dictated by the overall electrostatic potential surrounding its active site. To test this notion, we assayed a panel of pY peptides containing acidic amino acids at varying positions against WT, R47E, and R24M PTP1B variants. Arg47 is located ~8 Å from the pY-binding pocket (i.e., the distance between Cα atoms of Arg47 and pY of a bound peptide) and interacts electrostatically with acidic residues at the pY-1 and pY-2 positions.28–30 Arg24 is located near the C-terminal region of a bound pY peptide and is ~15 Å away from the pY residue (Cα-Cα). When a bound substrate contains a tandem pY motif (pYpY), Arg24 has been shown to interact with the side chain of the second pY residue.30 However, whether Arg47 and Arg24 interact with other substrate residues was previously unknown.
Mutation of Arg47 or Arg24 had little effect on the PTP1B catalytic activity toward pNPP or the neutral peptide SASASpYSASA (Tables 5 and 6), indicating that the R47E and R24M mutations do not affect the overall stability or the active-site structure of PTP1B. By contrast, the R47E mutation decreased the catalytic activity of PTP1B by 3–18-fold toward all acidic substrates, including those containing acidic residues at the extreme N- (e.g., EEDNAWpYAA) or C-terminus (e.g., AApYAAADD) (Table 5). The largest decrease occurred for peptide ASSDEpYAA (18-fold), which contains acidic residues at the pY-1 and pY-2 positions. This is consistent with the previous observation that Arg-47 is located near the pY-1 and pY-2 residues of a bound substrate.28–30 Note that both WT and R47E mutant PTP1B variants had, within the experimental error, essentially the same kcat value (~35 s−1) toward all peptide substrates, and that the mutation primarily affected the KM values. These results suggest that Arg47 engages in favorable electrostatic interactions with acidic residues located anywhere within the pY-6 to pY+5 region to enhance the substrate binding affinity. Consistent with this notion, the R47E mutation resulted in increased PTP1B activity toward the positively charged peptide ARKRIpYAA (by 5-fold), suggesting that either Arg47 engages in repulsive interactions with the positively charged residues in the pY peptide and/or Glu47 in the mutant enzyme favorably interacts with the Arg/Lys residues of the substrate peptide. Previous studies with R47A and R47E mutants demonstrated that both repulsive and attractive interactions contribute to the difference in activity toward peptide DADEpYLIPQQG.14
Table 5.
Kinetic Properties of WT and R47E PTP1B against pY Peptides
| Entry No. |
Sequencea | WT | R47E | WT/R47E | ||||
|---|---|---|---|---|---|---|---|---|
| kcat | KM | kcat/KM | kcat | KM | kcat/KM | |||
| (s−1) | (µM) | x 107 (M−1 s−1 ) | (s−1) | (µM) | x 107 (M−1 s−1 ) | |||
| pNPP | 18 ± 1 | 2600 ± 200 | 0.00069 | 16 ± 1 | 2800 ± 300 | 0.00057 | 1.2 | |
| 6 | SASASpYSASA | 28 ± 1 | 12 ± 1 | 0.23 | 35 ± 1 | 12 ± 1 | 0.29 | 0.79 |
| 7 | EEDNAWpYAA | 27 ± 1 | 1.3 ± 0.1 | 2.1 | 32 ± 1 | 9.5 ± 0.4 | 0.33 | 6.4 |
| 8 | ASSDEpYAA | 31 ± 1 | 1.8 ± 0.1 | 1.7 | 31 ± 1 | 32 ± 1 | 0.097 | 18 |
| 9 | AApYDDIDE | 37 ± 2 | 3.5 ± 0.1 | 1.1 | 43 ± 1 | 56 ± 2 | 0.076 | 14 |
| 10 | ARKRIpYAA | 41 ± 1 | 166 ± 6 | 0.025 | 37 ± 2 | 31 ± 4 | 0.12 | 0.21 |
| 11 | AApYIRKRA | 37 ± 1 | 89 ± 2 | 0.042 | 34 ± 2 | 89 ± 3 | 0.038 | 1.1 |
| 12 | AApYDDAAA | 28 ± 1 | 6.5 ± 0.1 | 0.43 | 33 ± 2 | 34 ± 2 | 0.097 | 4.4 |
| 13 | AApYADDAA | 30 ± 1 | 2.2 ± 0.1 | 1.4 | 34 ± 1 | 12 ± 1 | 0.29 | 4.8 |
| 14 | AApYAADDA | 29 ± 1 | 2.6 ± 0.1 | 1.1 | 36 ± 2 | 11 ± 2 | 0.32 | 3.4 |
| 15 | AApYAAADD | 29 ± 1 | 3.0 ± 0.1 | 0.97 | 35 ± 1 | 10 ± 1 | 0.35 | 2.8 |
| 16 | AWGPLpYDEVQMb | 31 ± 1 | 1.8 ± 0.1 | 1.7 | 34 ± 2 | 5.6 ± 0.4 | 0.61 | 2.8 |
| 17 | DPRGIpYDQVAG | 27 ± 1 | 7.1 ± 0.4 | 0.38 | 35 ± 1 | 23 ± 1 | 0.15 | 2.5 |
All sequences have an N-terminal acetyl group and a C-terminal amide.
The amino acid norleucine was used in lieu of methionine.
Table 6.
Kinetic Properties of WT and R24M PTP1B against pY Peptides
| Entry No. |
Sequencea | WT | R24M | WT/R24M | ||||
|---|---|---|---|---|---|---|---|---|
| kcat | KM | kcat/KM | kcat | KM | kcat/KM | |||
| (s−1) | (µM) | x 107 (M−1 s−1) | (s−1) | (µM) | x 107 (M−1 s−1) | |||
| pNPP | 18 ± 1 | 2600 ± 200 | 0.00069 | 17 ± 1 | 3.0 ± 0.1 | 0.00057 | 1.2 | |
| 6 | SASASpYSASA | 28 ± 1 | 12 ± 1 | 0.23 | 42 ± 2 | 14 ± 0.6 | 0.30 | 0.77 |
| 7 | EEDNAWpYAA | 27 ± 1 | 1.3 ± 0.1 | 2.1 | 35 ± 1 | 1.9 ± 0.1 | 1.8 | 1.2 |
| 8 | ASSDEpYAA | 31 ± 1 | 1.9 ± 0.1 | 1.7 | 32 ± 1 | 3.8 ± 0.2 | 0.84 | 2.0 |
| 9 | AApYDDIDE | 37 ± 2 | 3.5 ± 0.1 | 1.1 | 42 ± 1 | 37 ± 1 | 0.11 | 10 |
| 11 | AApYIRKRA | 37 ± 1 | 89 ± 2 | 0.042 | 34 ± 1 | 52 ± 3 | 0.065 | 0.62 |
| 12 | AApYDDAAA | 28 ± 1 | 6.5 ± 0.1 | 0.43 | - | - | 0.027 | 16 |
| 13 | AApYADDAA | 30 ± 1 | 2.2 ± 0.1 | 1.4 | 38 ± 1 | 6.4 ± 0.2 | 0.59 | 2.4 |
| 14 | AApYAADDA | 29 ± 1 | 2.6 ± 0.1 | 1.1 | 38 ± 1 | 6.9 ± 0.1 | 0.55 | 2.0 |
| 15 | AApYAAADD | 29 ± 1 | 3.0 ± 0.1 | 0.97 | 37 ± 1 | 8.7 ± 0.4 | 0.43 | 2.3 |
| 16 | AWGPLpYDEVQMb | 31 ± 1 | 1.8 ± 0.1 | 1.7 | 36 ± 1 | 5.0 ± 0.5 | 0.72 | 2.3 |
| 17 | DPRGIpYDQVAG | 27 ± 1 | 7.1 ± 0.4 | 0.38 | 36 ± 1 | 71 ± 1 | 0.051 | 7.6 |
All sequences have an N-terminal acetyl group and a C-terminal amide.
The amino acid norleucine was used in lieu of methionine.
Similarly, the R24M mutation decreased the PTP1B activity toward all acidic substrates, but not toward the neutral peptide SASASpYSASA (Table 6). In general, the mutation had a greater effect on substrates containing acidic residues on the C-terminal side of the pY residue, especially those with acidic residues at the pY+1 and pY+2 positions, consistent with its physical proximity to these two residues in the crystal structure.28–30 As expected, the R24M mutation slightly increased the PTP1B activity toward peptide AApYIRKRA (by 1.6-fold), indicating that Arg24 engages in repulsive interaction with the positively charged residues C-terminal to pY. Note that peptides containing an acidic amino acid at the pY+1 position generally had poorer activity toward PTP1B, especially against the R24M mutant (e.g., peptides AApYDDIDE and AApYDDAAA). This is in agreement with a previous report that PTP1B disfavors acidic residues at the pY+1 position,27 but in apparent contradiction with our observation that Arg24 interacts most strongly with an acidic residue at the pY+1 or pY+2 position.
Structural Basis for Long-Range Interactions between Arg47/Arg24 and Acidic Residues in pY Substrates
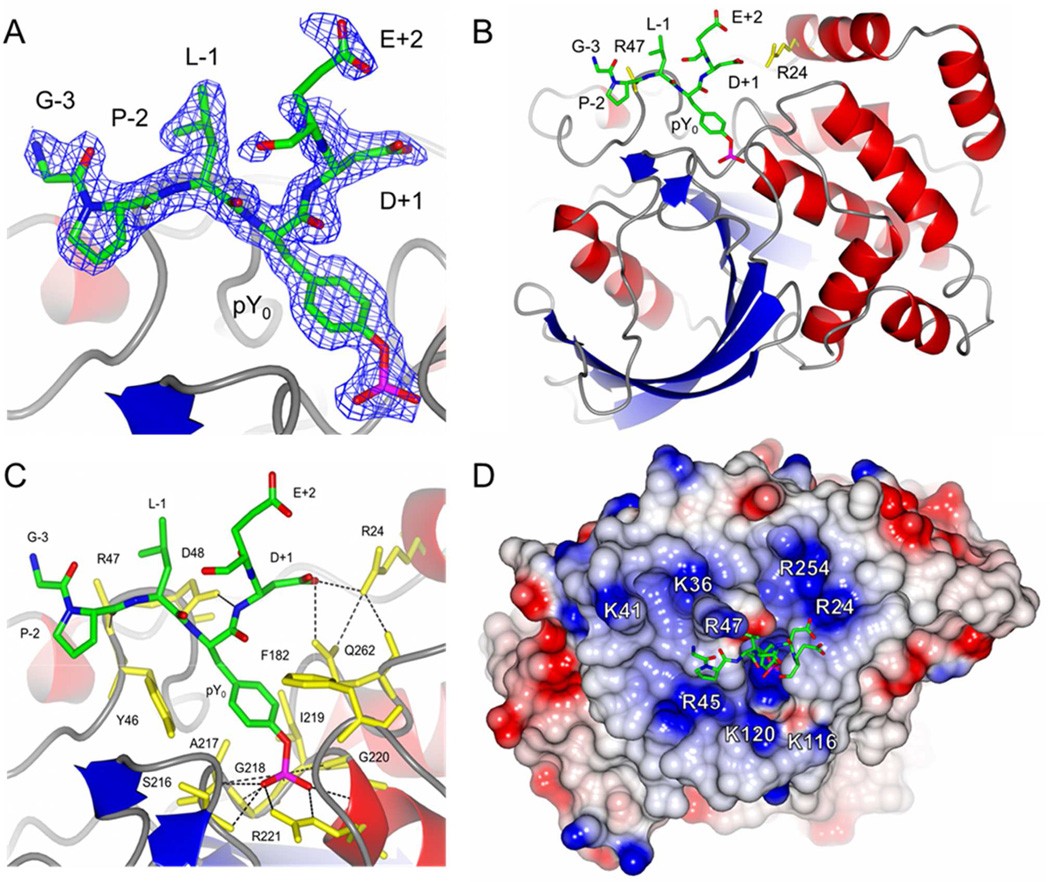
Many PTP-pY peptide co-crystal structures have been reported, most of which involved substrates containing acidic residues N-terminal to pY.28–34 To gain a structural basis for the observed interactions between the Arg24/Arg47 residues of PTP1B and acidic residues C-terminal to pY, and to reconcile the apparent contradiction mentioned above, we determined the X-ray crystal structure of PTP1B in complex with a peptide containing acidic residues on the C-terminal side of pY. A recently discovered PTP1B substrate, nephrin, contains two pY sites with acidic residues immediately C-terminal to pY, AWGPLpY1193DEVQM and DPRGIpY1217DQVAG.35 Both peptides were efficient substrates of PTP1B and the R24M mutation resulted in substantial reduction of their activities toward PTP1B (2.3- and 7.6-fold, respectively) (Table 6). The nephrin pY1193 peptide was co-crystallized with catalytically inactive C215S mutant PTP1B and the structure of the complex was solved at 1.7 Å resolution to an R factor of 0.170 (Table 7). In the structure, PTP1B residues 2–298 and nephrin peptide residues pY-3 to pY+1 were clearly resolved and these five substrate residues were fit unambiguously to the electron density (Figure 4A). A region of electron density was also observed next to the pY+1 residue and tentatively fit with the sidechain carboxyl group of pY+2 Glu. As observed in previous PTP1B-pY peptide structures,28–30 the nephrin peptide bound to PTP1B in the canonical orientation with the peptide backbone in an extended β-strand conformation (Figure 4B). The pY residue was bound to the pY-binding loop in the usual manner with the phosphate forming close interactions with the guanidino group of Arg-221, several backbone amides of the pY-binding loop (residues 217–221), and the N-terminal end of the α-helix that follows (Figure 4C and Figure S1 in SI). The pY peptide backbone forms three hydrogen bonds with PTP1B, two from the carboxylate group of Asp-48 to the NH groups of the pY and the pY+1 residues, and a third from the backbone carbonyl of the pY-2 residue to the backbone amide of Arg-47 (Figure 4C and S1). Based on the electron density maps, the Arg24 side chain was modeled in two alternative conformations (Figure S2 in SI). In the first conformation, the guanidinium group of Arg24 forms a hydrogen bond with the carboxylate group of the pY+1 Asp, which is also hydrogen bonded to the side chain amine of Gln262 (Figure 4C and S1). When in the alternative conformation, the Arg24 side chain points toward and presumably interacts electrostatically with the pY+2 Glu side chain (Figure S2). The side chain of Arg-47, which is positioned near the pY-1 residue, was not well resolved in the structure, and was therefore truncated to alanine in the final refined model. However, the side chain of Leu at the pY-1 position adopts a similar conformation as previously reported by Sarmiento et al.,29 which is favorable for hydrophobic interactions with the aliphatic portion of the Arg-47 side chain. The disorder of the Arg-47 side chain in this structure potentially reflects the fact that the residues at the pY-1 and −2 positions were Leu and Pro, respectively, instead of acidic residues. As observed in previous structures,28–30 Arg-47 can make specific interactions with acidic residues at these positions.
Table 7.
Crystallographic and Refinement Statistics
| X-ray Diffraction Data | |
| Space group | P21 |
| Cell dimensions | |
| a (Å) | 43.8 |
| b (Å) | 88.7 |
| c (Å) | 50.0 |
| β (°) | 97.3 |
| Resolution (Å) | 19.95–1.74 (1.83–1.74) |
| No. observations | 147,218 |
| No. unique | 38,860 |
| Completeness (%) | 99.7 (99.7) |
| I/σI | 6.2 (1.9) |
| Rmerge | 0.120 (0.744) |
| Refinement Statistics | |
| Resolution (Å) | 19.95–1.74 |
| No. reflections | 36891 |
| Rwork/Rfree | 0.170 / 0.207 |
| No. non- hydrogen atoms | 2740 |
| Mean B factor (Å2) | 24.7 |
| Root-mean-square deviation for bond lengths (Å) | 0.023 |
| Root-mean-square deviation for bond angles (deg) | 2.15 |
Figure 4.
Structure of C215S PTP1B in complex with nephrin peptide AWGPLpY1193 DEVQM. A) The nephrin peptide was contoured with the Fobs – Fcalc omit map at 1.0 σ. B) Ribbon diagram of the PTP1B–nephrin peptide complex. The Arg24 and Arg47 residues of PTP1B (yellow) are shown in stick representation. C) Interactions between the nephrin peptide (green) and PTP1B (yellow). Hydrogen bonds are shown as black dashed lines. D) The electrostatic surface potential of PTP1B. Basic residues near the active site are shown in blue and labeled. In (B) and (C), the side chain of Arg47 was truncated (to Ala) due to the absence of clear electron density for the side chain.
To assess the potential contribution of long-range electrostatic interactions between Arg47/24 and acidic residues on pY substrates to catalysis, we replaced each residue of the nephrin pY1193 peptide in the co-crystal structure with an Asp and measured the minimal and maximal possible distances between the Asp and Arg side chains, assuming that both Arg and Asp side chains can rotate freely into different energetically allowable conformations. The same modeling exercise was also carried out with a previously reported co-crystal structure of PTP1B bound with a consensus peptide Ac-ELEFpYMDYE-NH2, in which substrate residues from pY-3 to pY+4 were well resolved.29 The distances derived from the two structures were rather similar and consistent with the kinetic results (Table 8). Arg47 had the shortest distance from Asp at the pY-3 and pY-1 positions (~1 Å), but was also within 3.5–10 Å from Asp residues at the pY-2 to pY+4 positions. Given that the backbone of the pY peptides binds to the surface of PTP1B in essentially an identical manner in all structures so far determined, it is unlikely that Arg47 could make direct hydrogen bonding interactions with acidic residues at the pY+1 to pY+5 positions. We conclude that Arg47 engages in long-range electrostatic interactions with the acidic residues C-terminal to pY, enhancing the kinetic activity of the peptide substrate. Similarly, Arg24 can potentially form a hydrogen bond with an Asp residue at the pY+1 position, but is too far for direct hydrogen bonding with acidic residues at other positions. Again, the distances between Arg24 and these other acidic residues (6–15 Å) are well within the range for significant electrostatic interactions. The PTP1B-nephrin peptide structure shows that several other basic residues are within 17 Å distance from the active site, including Arg45, Arg254, Lys36, Lys41, Lys116, and Lys120 (Figure 4D). These residues likely also engage in favorable electrostatic interactions with acidic residues of pY peptides, although the amount of binding energy they provide might be smaller, given their overall longer distances from a bound substrate.
Table 8.
Side Chain Distances (Å) between Arg (in PTP1B) and Asp Residues (in pY Peptide)a
| Asp position | Arg47 | Arg24 |
|---|---|---|
| pY-3 | 0.88–7.0 (1.1–7.3) | 14–24 (15–23) |
| pY-2 | 4.2–12 (3.7–11) | 14–23 (15–24) |
| pY-1 | 0.94–11 (1.2–11) | 8.2–17 (8.9–17) |
| pY+1 | 5.4–16 (6.0–15) | 3.8–11 (4.2–11) |
| pY+2 | 3.5–14 (9.6–18) | 7.2–14 (6.3–14) |
| pY+3 | — (4.6–15) | — (6.2–15) |
| pY+4 | — (8.8–20) | — (11–17) |
Distances were derived from the structure of PTP1B bound with Ac-AWGPLpYDEVQM-NH2. Values in parenthesis were derived from the structure of PTP1B bound with Ac-ELEFpYMDYE-NH2 (PDB file: 1EEO).
Finally, we examined two previously reported PTP-pY peptide structures that contained acidic residues C-terminal to pY (Figure S3 in SI). We found that in the structure of lymphoid-specific PTP (LYP) bound with peptide YGEEpYDDLY (pdb: 3OLR),34 both Asp residues of the bound peptide point toward the side chain of Lys32 (Figure S3 in SI). Similarly, in the structure of PTPRC (CD45) in complex with peptide REEpYDV (pdb: 1YGR),31 the side chains of pY-1 Glu and pY+1 Asp both point toward the side chain of Arg637 (Figure S3). We propose that long-range electrostatic interactions are involved in the E•S complex formation for these PTPs as well.
C-Terminal Sequence Is the Primary Specificity Determinant for PTP-PEST
Screening of PTP-PEST against library V resulted in a greater number of acidic residues on the C-terminal side of pY than the N-terminal side (15, 13, 19, and 21 Asp/Glu residues at positions pY-3, pY-2, pY+1, and pY+2, respectively), suggesting that the C-terminal sequence is the primary determinant of PTP-PEST substrate specificity (Table S5). This notion was confirmed by assaying PTP-PEST against peptides containing acidic residues on either the N- or C-terminal side of pY: Ac-ASSDEpYAA-NH2 and Ac-AApYDLDEY-NH2). These two substrates exhibited kcat/KM values of 7.2 × 106 and 1.3 × 108 M−1 s−1, respectively; notably, the latter value approaches that of the most active PTP-PEST substrate (2.2 × 108 M−1 s−1 for Ac-YDEDFpYDYEF-NH2). Therefore, PTP-PEST is unique among the classical PTPs, which usually have their major specificity determinant on the N-terminal side of pY (e.g., PTP1B, SHP-1, and SHP-2).10,36 The only other PTP superfamily member known to have its major specificity determinant located on the C-terminal side of pY is the dual specificity phosphatase VH1.37
PTPD1 and PTPD2 Are Highly Active Under Single Turnover Conditions
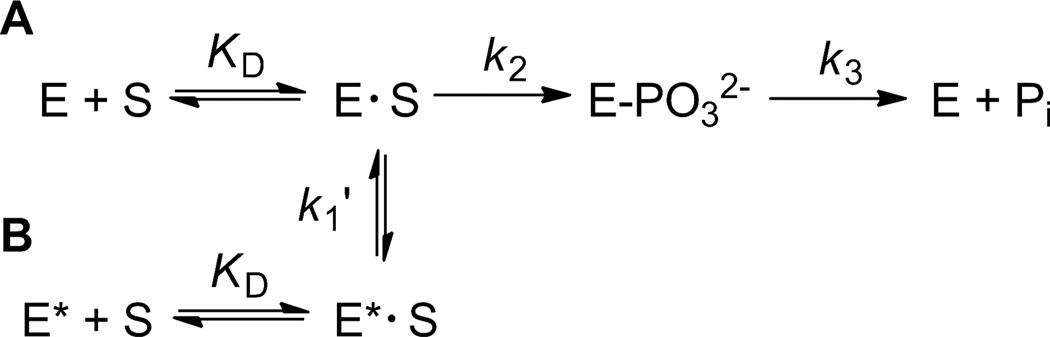
Compared with most of the other PTPs, PTPD1 and PTPD2 have very low intrinsic catalytic efficiencies in vitro due to unusually low kcat values. Nevertheless, their catalytic activities are both necessary and sufficient for their in vivo functions.38,39 In an attempt to reconcile this apparent contradiction, we conducted pre-steady-state kinetic analysis to determine the reason for their low kcat values. PTP catalysis involves the formation of a covalent phosphocysteinyl-enzyme intermediate (E-PO32-), and the overall reaction rate (kcat) is determined by both its formation (k2) and decay (k3) (Scheme 1A).40,41
Scheme 1.
PTP Kinetic Mechanisms
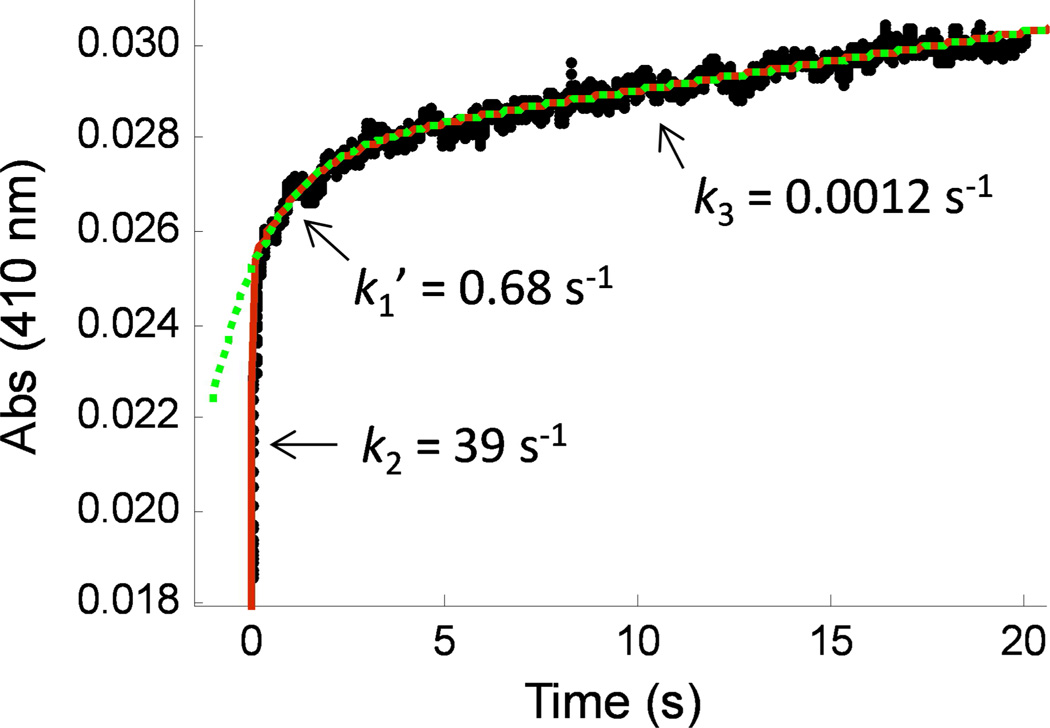
Rapid mixing of PTPD1 (10 µM) and a saturating concentration of pNPP (20 mM) in a stopped-flow apparatus at 4 °C produced a reaction progress curve that consisted of a rapid initial phase (0–100 ms) and a slow final phase (>3 s) (Figure 5). Experiments at 25 °C failed to resolve the initial phase, which was too fast for the instrument used in this work (k >100 s−1 ). Our initial attempt to fit the experimental data to the kinetic model in Scheme 1A was unsuccessful (due to inability to fit data in the 0–0.2 s region). The data, however, could be fit to a model that assumes a mixture of two different populations of PTPD1, with the first having the kinetic mechanism illustrated in Scheme 1A and the other having the mechanism in Scheme 1B, which predicts an additional conformational change converting either the free enzyme (E*) or the initial E*•S complex into the catalytically competent form (E or E•S). Curve fitting yielded rate constants for formation of the covalent intermediate (kburst = 39 s−1), conformational change (k1’ = 0.68 s−1 ), and decay of the phosphoenzyme intermediate (kcat = 0.0012 s−1). The microscopic rate constants k2 and k3 of PTPD1 were determined from the kburst and kcat values, and are 39 s−1 and 0.0012 s−1 , respectively. The amplitudes of the two burst phases (A = 0.027 and B = 0.0025 absorbance units) suggest an ~10:1 ratio of the two enzyme populations. The putative conformational change might involve the cis-trans isomerization of a peptidyl-prolyl bond (e.g., that of the WPD/E motif that provides a critical general acid/base during catalysis4). There are many examples of proteins existing as a mixture of peptidyl-prolyl cis and trans isomers.42 Additional studies will be necessary to reveal the structural basis of the putative conformational change, which is not the focus of this study. Experiments with PTPD2 showed similar burst kinetics and a very fast initial phase (k2 ≥110 s−1). Thus, for both PTPD1 and PTPD2, breakdown of the covalent intermediate (k3, or a step after that) is rate-limiting and responsible for their unusually low kcat values (kcat ≈ k3). However, both enzymes are highly active under single turnover conditions (high k2 values). In fact, their k2 values are similar to those of other highly proficient PTPs (e.g., a k2 value of 193 s−1 at 3.5 °C for Yersinia PTP, which is one of the most active PTPs known to date43). Note that the k2 values of PTPD1 and PTPD2 are expected to be still higher for preferred peptide and protein substrates. The fact that PTPD1 and PTPD2 have similar kcat values of toward Ac-YDEDFpYDYEF-NH2, Ac-SASASpYSASA-NH2, and pNPP, which have vastly different kcat/KM values (Table 2 and Table S19 in SI), is consistent with the above conclusion.
Figure 5.
Reaction progress curve showing the burst kinetics of PTPD1-catalyzed hydrolysis of pNPP (20 mM) at 4 °C. Data were fitted to a double exponential equation (red, solid line) or to a single exponential equation (green, dotted line). Fitting of the entire progress curve to the mechanism in Scheme 1 using the double exponential equation: P = A • exp(-kburst • t) + B • exp(-k1’ • t) + C • t + D, where kburst = k2 + k3, kcat = C/(ε • [E]) = k2 • k3/(k2 + k3). The data shown is the average of three independent experiments performed under the same conditions.
DISCUSSION
In this and a previous study,10 we have systematically profiled the sequence selectivity for 14 out of the 38 classical human PTPs by screening combinatorial peptide libraries. Surprisingly, 13 of the PTPs exhibit similar specificity profiles in that they all prefer acidic and large hydrophobic amino acids and disfavor basic residues, although they do have subtle differences at certain positions (e.g., HePTP prefers an acidic residue, whereas PTP-PEST, PTPD2, PTPRD, and PTPRO prefer a hydrophobic amino acid at the pY-1 position). On the other hand, these PTPs differ drastically in their intrinsic catalytic efficiency and degree of preference/disfavor for acidic/basic sequences. Furthermore, unlike proteases and protein kinases, the PTPs do not exhibit any consensus sequence(s), even for those that are highly selective for acidic sequences (e.g., PTP-PEST, SHP-1, and SHP-2). Apparently, acidic residues located anywhere between the pY-8 and pY+5 positions (the region tested in this study) contribute favorably to substrate binding and catalysis.
What is the molecular basis of this unusual sequence selectivity? We found that removal of the side chain of Arg47 or Arg24 reduced the catalytic activity of PTP1B toward pY peptides containing acidic residues anywhere between the pY-6 and pY+5 positions. The co-crystal structures of PTP1B bound with two different peptides revealed that acidic residues located at most of the above positions are too distant to form direct hydrogen bonds with the arginine residues. Therefore, we conclude that Arg47 and Arg24 engage in long-range electrostatic interactions with acidic residues on the pY peptides and the R47E and R24M mutations eliminate the favorable electrostatic interactions and, in the case of the former, likely generate repulsive interactions with the acidic substrates. Although not examined in this study, Arg45, Arg254, Lys36, Lys41, Lys116 and Lys120 are also located near the bound pY substrates (Figure 4D) and expected to engage in similar electrostatic interactions with acidic residues in the substrates. Unlike other weak interactions (e.g., van der Waals forces), charge-charge interaction is relatively insensitive to the distance (1/r) and remains significant even when a charged pair is separated by 10–20 Å. We propose that the presence of several Arg/Lys residues “strategically” positioned around the substrate-binding site is responsible for PTP1B’s lack of significant positional preference for acidic residues. An acidic residue located anywhere on a bound substrate (pY-6 to pY+5) is expected to interact with all of the positively charged residues near the PTP active site, but with different strengths. For example, an acidic residue near the N-terminus (e.g., at position pY-5) would interact more strongly with Lys41, Arg45, and Arg47 due to their physical proximity to the pY-5 residue, and weakly with the other, more distal Arg/Lys residues. On the other hand, an acidic residue at the C-terminus of the peptide (e.g., at position pY+2) may gain most of its binding energy through interactions with Arg24, Arg47, and Arg254, with less contribution from Lys41 and Arg45 (Figure 4D). Thus, an Asp residue at position pY-5 or pY+2 may enhance the overall binding affinity and the kcat/KM value of a substrate by a similar amount, even though in each case the acidic residue interacts with a different set of PTP1B residues. This lack of positional preference for acidic residues, together with the absence of any well-defined substrate-binding pockets/grooves on the PTP surface (other than the pY-binding pocket),15,28–34 likely endows PTPs with the ability to accommodate a broad range of peptide sequences (or inability to recognize specific sequence motifs) and therefore similar specificity profiles. The differential degrees of preference/disfavor for acidic/basic sequences are likely caused by the different electrostatic potentials at their substrate-binding sites. Indeed, PTPs that strongly prefer acidic over neutral and basic substrates (e.g., PTP-PEST, SHP-1, SHP-2) all have highly positively charged surfaces near their active sites, whereas PTPRA, PTPRD, and PTPD1, which show only weak preference for acidic substrates, have a mixture of basic and acidic amino acids and overall neutral electrostatic potential at their substrate-binding sites.15
There have been conflicting reports with regard to PTP1B’s specificity at the pY+1 position. While several studies showed that a second pY residue at the pY+1 position substantially enhances PTP1B activity,30,44 inverse alanine scanning suggested that PTP1B prefers a hydrophobic residue (e.g., Met) and disfavors acidic residues (Asp and Glu) at the pY+1 position.27 The kinetic data in this work confirmed PTP1B’s slight disfavor for Asp and Glu at the pY+1 position (Table 5, compare entries 12 and 13). Yet, the R24M mutation caused the largest activity reduction toward substrates containing an acidic residue at the pY+1 position, suggesting that Arg24 contributes significantly to substrate binding and catalysis. The X-ray structure of PTP1B bound to the nephrin pY1193 peptide provides a potential explanation for this apparent contradiction. PTP1B possesses a shallow amphipathic pocket next to the pY-binding pocket, formed by the side chains of Asp48, Val49, Ile219, and Met258 on the pocket floor and the side chains of Gln262 and Arg24 on one side (Figure 4C and S4). A hydrophobic side chain such as that of methionine is well accommodated by the pocket and enhances substrate binding and catalysis through hydrophobic effects (Figure S4B).29 A pY residue also fits snugly into the pocket, because its phenyl ring interacts with the hydrophobic surface and the negatively charged phosphate forms salt bridges with Arg24 (Figure S4A).30 Asp and Glu are not preferred because their side chains do not interact effectively with the hydrophobic surface and unlike pY, their side chains are too short to interact favorably with Arg24 (Figure S4C). In order to hydrogen bond with the pY+1 Asp and neutralize its negative charge (and therefore accommodate it in a hydrophobic pocket), the side chain of Arg24 moves toward the Asp residue by ~2.3 Å into a presumably less stable conformation (Figure 4C and S2). The R24M mutation eliminates the charge neutralization effect, leaving a negatively charged Asp side chain in a largely hydrophobic pocket. Indeed, the R24M mutant enzyme has unusually low activity toward peptides containing an acidic pY+1 residue (Table 6).
Despite PTPs’ broad, similar specificities toward peptide substrates in vitro, numerous genetic “knock-in” and knockout experiments have demonstrated that at least some PTPs exhibit substantial substrate specificity in vivo. What factors, then, may enhance the PTPs’ substrate specificity in vivo? We propose that PTPs may utilize their diverse catalytic properties as yet another strategy, along with sequence selectivity, subcellular localization, spatial and temporal expression, posttranslational modification, and protein-protein interaction to achieve in vivo substrate specificity. On the basis of their intrinsic catalytic efficiency, we divide the 14 PTPs into five different groups and briefly speculate how the catalytic efficiency may influence their in vivo substrate specificity.
Group I (PTP-PEST, SHP-1, and SHP-2)
These three enzymes are highly active and selective for acidic sequences, while having poor activity toward neutral sequences and extremely poor activity for basic sequences (Table 2 and Table 3). The intrinsic sequence selectivity of their PTP domains likely plays a key role in controlling their in vivo substrate specificity. For highly negatively charged pY sites, which are numerous in the human proteome, substrate recruitment may not be necessary to achieve efficient dephosphorylation; rather, it will be important to keep their highly proficient PTP domains in check to avoid unintended reactions. Both SHP-1 and SHP-2 exist in an auto-inhibited state, in which their N-terminal SH2 domain binds intramolecularly to their PTP active site, sterically blocking substrate access to the catalytic site.45,46 Binding of their N-SH2 domain to a high-affinity pY ligand activates the PTP domain by disrupting the auto-inhibitory mechanism and recruits the PTP domain to the vicinity of the pY ligand.47,48 This mechanism allows the selective dephosphorylation of acidic pY motifs on the pY-ligand protein and/or other pY proteins in the same signaling complex. For less acidic or neutral pY motifs (which could contain either all neutral residues or a combination of positively and negatively charged residues), substrate recruitment mediated by the SH2 domains (SHP-1 and SHP-2) or proline-rich sequences (PTP-PEST) should further increase their activity toward specific substrates. However, we envision that it is unlikely for substrate recruitment to overcome their extremely poor activity toward highly basic pY sites. Indeed, all of the in vivo SHP-1 and SHP-2 substrates reported to date contain highly negatively charged pY motifs.10 The reported PTP-PEST substrates, SRC (IEDNEpY424TARQ),49 FAK (SEIDDpY397AEIID),50 PYK2 (IESDIpY402AEIPD and IEDEDpY579pY580KASVT),51 WASP (TSKLIpY291DFIED),52 and PSTPIP1 (RNELVpY344ASIEV),53 also all contain negatively charged residues surrounding the pY sites.
Group II (HePTP, PTPRA, and PTPRD)
PTPs in this group are two to four orders of magnitude less active than most of the other PTPs (e.g., group I PTPs) due to high KM values, indicating that their active sites bind poorly to pY substrates. Their “normal” kcat values (i.e., similar to those of most other PTPs) suggest that once being recruited to a specific substrate via protein-protein interaction (and thus a reduced KM value), they have the potential to act as highly efficient enzymes (high kcat/KM values). The lack of sequence selectivity of the PTPRA catalytic domain predicts that (1) the in vivo substrate specificity of PTPRA is dictated by substrate recruitment and (2) PTPRA may be capable of hydrolyzing any pY motif recruited to its vicinity, including highly positively charged sequences. HePTP has moderate sequence selectivity (e.g., it prefers an acidic amino acid at the pY-1 and pY+1 positions), and its in vivo substrate specificity is likely determined by both substrate recruitment and the specific peptide sequence surrounding the pY residue. Therefore, due to their low intrinsic catalytic activity and inability to hydrolyze pY proteins in the absence of substrate recruitment, HePTP and PTPRA are expected to function as highly specific enzymes in vivo. This model is in excellent agreement with the results of previous studies. For example, PTPRA selectively dephosphorylates SRC at pY527.54 It was proposed that the SH2 domain of SRC binds to the pY789 of PTPRA and recruits it to the active site of PTPRA. Other PTPRA substrates, such as LYN and FYN kinases, contain SH2 domains and are likely recruited via the same mechanism.55,56 Similarly, HePTP has a kcat/KM value of 4.8 × 103 M−1 s−1 toward the ERK2 peptide DHTGFLpTEpY187VATRW but dephosphorylates the phospho-ERK2 protein at the same site with a rate of 2.6 × 106 M−1 s−1, because the N-terminal kinase interaction motif (KIM domain) of HePTP interacts with and recruits ERK2.57 PTPRD also showed low catalytic activity; unfortunately, we were not able to accurately determine its kcat or KM value. We tentatively assign it to Group II on the basis of its relatively high kcat (>0.86 s−1) and KM values (>60 µM) (Table 2).
Group III (PTPD1 and PTPD2)
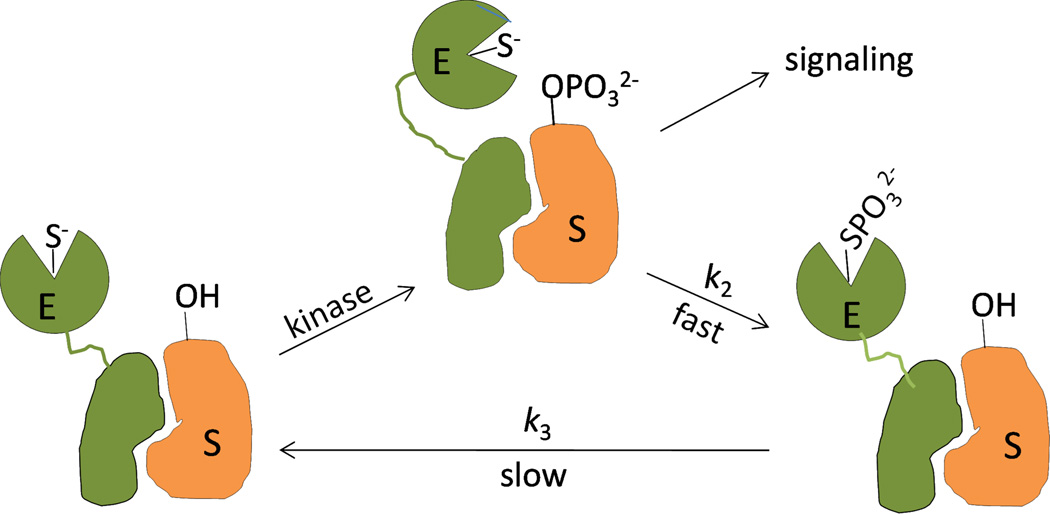
PTPD1 and PTPD2 are also orders of magnitude less active than most of the other PTPs, because of their unusually low kcat values (Table 2). Steady-state and pre-steady-state kinetic data indicate that slow breakdown of the covalent intermediate (k3, or a step after that) is responsible for the low kcat values. The slow k3 step is likely due to the presence of atypical residues in their active sites.15 The highly conserved aspartate residue of the WPD loop, which acts as a critical general base during the k3 step, is replaced by a glutamate in PTPD1. PTPD2 lacks a highly conserved tyrosine residue in the pY recognition loop. Because the k3 step is common for all substrates, they will all have the same kcat value (except for very poor substrates that have k2 as the rate-limiting step), no matter how tightly they bind to the PTP active site. Thus, substrate recruitment per se is not expected to increase the maximal reaction rate (Vmax) of PTPD1 or PTPD2 under steady-state conditions, although we cannot rule out the possibility that substrate binding, interaction with regulatory proteins, or posttranslational modification may change the conformation of the PTP domain and increase the k3 value. We propose that PTPD1 and PTPD2 function as single turnover enzymes in vivo. Because they can only efficiently turnover a substrate once, we predict that effective dephosphorylation of a specific substrate would require the formation of a stable, stoichiometric complex with the substrate protein through their non-catalytic elements (e.g., the FERM domain), and dephosphorylation would be limited to a single pY residue on the substrate (Figure 6). Since the reaction product presumably remains associated with the PTP, there is no need (or evolutional driving force) for the phosphoenzyme intermediate to undergo rapid hydrolysis. In fact, slow regeneration of the active enzyme enhances substrate specificity because the catalytically inactive phosphoenzyme cannot dephosphorylate other pY residues on the substrate protein or other pY proteins in the same signaling complex. Again, previous publications support our proposal. PTPD1 constitutively associates with SRC and activates the kinase by selective dephosphorylation of pY527, enhancing growth factor-induced signal transduction.58,59 Presumably, formation of the SRC/PTPD1 complex juxtaposes the pY527 residue (but not pY416, which is required for full activation of SRC60,61) with the PTPD1 active site. PTPD2 lacking the PTP domain stably associates with β-catenin and overexpression of this dominant negative mutant induces tyrosine phosphorylation of β-catenin and cell migration.39 PTPD2 also selectively dephosphorylates pY128 of p130Cas in vitro and in vivo.62
Figure 6.
Proposed model showing regulation of cell signaling by PTPD1 (or PTPD2) under single-turnover conditions. E, PTPD1 or PTPD2; S, pY substrate protein.
Group IV (PTP1B, TCPTP, and PTPH1)
PTPs in this group are highly active and have limited sequence selectivity. They can efficiently dephosphorylate acidic and neutral pY sequences and have respectable activity even toward the least reactive, highly basic peptides (kcat/KM values of 103–104 M−1 s−1). Although these enzymes are restricted to specific subcellular locations (PTP1B to the endoplasmic reticulum,63 TCPTP to the endoplasmic reticulum and nucleus,64,65 and PTPH1 to the cytoskeleton66), their catalytic domains are still accessible to a wide variety of pY proteins in the cytoplasm and/or nucleus. With kcat/KM values of >106 M−1 s−1 for acidic and neutral pY sequences, they can dephosphorylate 50% of a cellular protein containing acidic/neutral pY motifs in <5 s (assuming a PTP concentration of 100 nM, which is the average cellular concentration of signaling proteins67). We propose that PTPs in this group dephosphorylate a wide variety of pY proteins in their immediate environments to maintain an overall low level of tyrosyl phosphorylation of cellular proteins. During active cell signaling, cells are known to produce reactive oxygen species (e.g., H2O2), which may transiently inactivate these PTPs to allow signal transduction to take place. Notably, a large number of PTP1B and TCPTP protein substrates have been reported and feature diverse sequences surrounding the dephosphorylated pY sites.10, 68
Group V (PTPRB, PTPRC, and PTPRO)
The catalytic domains of these receptor PTPs are similar to the group IV enzymes, in that they are highly efficient PTPs with low sequence selectivity. Obviously, their membrane localization should facilitate their dephosphorylation of membrane-associated proteins or proteins transiently recruited to the membrane during cell signaling. How their potent PTP activity is kept in check is less clear at the present time. Some of them contain a second, catalytically inactive PTP domain, which can affect the physiological function of the active PTP domain.69,70 Dimerization provides another mechanism for regulating the activity of receptor PTPs.9,71 Receptor PTPs may also be regulated by redox signals.72 Finally, their large extracellular domains may respond to environmental signals and regulate the activity of the PTP domain.
In conclusion, systematic profiling of 14 classical PTPs allowed us to conclude that most of the classical PTPs prefer acidic over basic pY sequences as substrates, but differ vastly in their degree of sequence preference and intrinsic catalytic efficiency. Long-range electrostatic interactions between positively charged residues near the PTP active site and acidic residues of a peptide substrate render PTPs insensitive to the positions of acidic residues in substrates and therefore all having a similar preference for acidic substrates. Their different levels of preference for acidic substrates are likely caused by the different overall electrostatic potentials of their substrate-binding sites. Finally, our results suggest that PTPs may have different levels of substrate specificity in vivo, which are brought about by a combination of several mechanisms including substrate recruitment and utilization of low intrinsic catalytic efficiency.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Z. Suo and B. Maxwell for providing equipment and technical assistance for the pre-steady-state experiments and Dr. S. Jacob for providing the PTPRO expression vector. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357. Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Company, which operates the facility.
Funding
This work was supported by National Institutes of Health (CA69202, CA132855, and GM062820). R.L. was supported by a predoctoral fellowship from the Minister of Science and Technology, Thailand. B.G.N. is a Canada Research Chair, Tier 1, and is partially supported by the Ontario Ministry of Health and Long Term Care and the Princess Margaret Cancer Foundation. S.K. is grateful for support from the SGC, a registered charity (number 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation, the Canadian Institutes for Health Research, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust [092809/Z/10/Z].
ABBREVIATIONS
2-aminobutyrate;
- F2Y
3,5-difluorotyrosine
- Fmoc-OSu
N-(9-fluorenylmethoxycarbonyloxy) succinimide
- HBTU
O-benzotriazole-N,N,N’,N’-tetramethyluronium hexafluorophosphate
- HOBt
1-hydroxybenzotriazole hydrate
- MBTH
3-methyl-2-benzothiazolinonehydrazone
- Nle (M)
norleucine
- OBOC
one-bead-one-compound
- PED-MS
partial Edman degradation-mass spectrometry
- PEGA
polyethyleneglycol acrylamide
- PITC
phenyl isothiocyanate
- PTP
protein tyrosine phosphatase
- pY
phosphotyrosine
- TFA
trifluoroacetic acid
Footnotes
The authors declare no competing financial interest.
Supporting Information Available: Tables containing the peptides sequences selected against the three PTPs and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Tonks NK. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendriks WJ, Elson A, Harroch S, Pulido R, Stoker A, den Hertog J. Protein tyrosine phosphatases in health and disease. FEBS J. 2013;280:708–730. doi: 10.1111/febs.12000. [DOI] [PubMed] [Google Scholar]
- 4.Zhang ZY. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu. Rev. Pharmacol. Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 5.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem. J. 2007;402:1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucl. Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr. Opin. Cell Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 8.Soulsby M, Bennett AM. Physiological signaling specificity by protein tyrosine phosphatases. Physiology. 2009;24:281–289. doi: 10.1152/physiol.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Wijk T, Blanchetot C, den Hertog J. Regulation of receptor protein-tyrosine phosphatase dimerization. Methods. 2005;35:73–79. doi: 10.1016/j.ymeth.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Ren L, Chen X, Luechapanichkul R, Selner NG, Meyer TM, Wavreille AS, Chan R, Iorio C, Zhou X, Neel BG, Pei D. Substrate specificity of protein tyrosine phosphatases 1B, RPTPalpha, SHP-1, and SHP-2. Biochemistry. 2011;50:2339–2356. doi: 10.1021/bi1014453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garaud M, Pei D. Substrate profiling of protein tyrosine phosphatase PTP1B by screening a combinatorial peptide library. J. Am. Chem. Soc. 2007;129:5366–5367. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, Ren L, Kim S, Carpino N, Daniel JL, Kunapuli SP, Tsygankov AY, Pei D. Determination of the substrate specificity of protein-tyrosine phosphatase TULA-2 and identification of Syk as a TULA-2 substrate. J. Biol. Chem. 2010;285:31268–31276. doi: 10.1074/jbc.M110.114181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gopishetty B, Ren L, Waller TM, Wavreille AS, Lopez M, Thakkar A, Zhu J, Pei D. Synthesis of 3,5-difluorotyrosine-containing peptides: application in substrate profiling of protein tyrosine phosphatases. Org. Lett. 2008;10:4605–4608. doi: 10.1021/ol801868a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarmiento M, Zhao Y, Gordon SJ, Zhang ZY. Molecular basis for substrate specificity of protein-tyrosine phosphatase 1B. J. Biol. Chem. 1998;273:26368–26374. doi: 10.1074/jbc.273.41.26368. [DOI] [PubMed] [Google Scholar]
- 15.Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motiwala T, Datta J, Kutay H, Roy S, Jacob ST. Lyn kinase and ZAP70 are substrates of PTPROt in B-cells: Lyn inactivation by PTPROt sensitizes leukemia cells to VEGF-R inhibitor pazopanib. J. Cell. Biochem. 2010;110:846–856. doi: 10.1002/jcb.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thakkar A, Wavreille AS, Pei D. Traceless capping agent for peptide sequencing by partial edman degradation and mass spectrometry. Anal. Chem. 2006;78:5935–5939. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZY, Maclean D, Thieme-Sefler AM, Roeske RW, Dixon JE. A continuous spectrophotometric and fluorimetric assay for protein tyrosine phosphatase using phosphotyrosine-containing peptides. Anal. Biochem. 1993;211:7–15. doi: 10.1006/abio.1993.1224. [DOI] [PubMed] [Google Scholar]
- 19.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystal. Section D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystal. Section D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystal. Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystal. D. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Tan PH, Zhang Y, Pei D. On-bead screening of combinatorial libraries: reduction of nonspecific binding by decreasing surface ligand density. J. Comb. Chem. 2009;11:604–611. doi: 10.1021/cc9000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sweeney MC, Wavreille A-S, Park J, Butchar J, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: Sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 25.Trinh TB, Xiao Q, Pei D. Profiling the Substrate Specificity of Protein Kinases by On-Bead Screening of Peptide Libraries. Biochemistry. 2013;52:5645–5655. doi: 10.1021/bi4008947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo XL, Shen K, Wang F, Lawrence DS, Zhang ZY. Probing the molecular basis for potent and selective protein-tyrosine phosphatase 1B inhibition. J. Biol. Chem. 2002;277:41014–41022. doi: 10.1074/jbc.M207347200. [DOI] [PubMed] [Google Scholar]
- 27.Vetter SW, Keng YF, Lawrence DS, Zhang ZY. Assessment of protein-tyrosine phosphatase 1B substrate specificity using "inverse alanine scanning”. J. Biol. Chem. 2000;275:2265–2268. doi: 10.1074/jbc.275.4.2265. [DOI] [PubMed] [Google Scholar]
- 28.Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 29.Sarmiento M, Puius YA, Vetter SW, Keng YF, Wu L, Zhao Y, Lawrence DS, Almo SC, Zhang ZY. Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry. 2000;39:8171–8179. doi: 10.1021/bi000319w. [DOI] [PubMed] [Google Scholar]
- 30.Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol. Cell. 2000;6:1401–1412. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 31.Nam HJ, Poy F, Saito H, Frederick CA. Structural basis for the function and regulation of the receptor protein tyrosine phosphatase CD45. J. Exp. Med. 2005;201:441–452. doi: 10.1084/jem.20041890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Cheng Z, Niu T, Liang X, Zhao ZJ, Zhou GW. Structural basis for substrate specificity of protein-tyrosine phosphatase SHP-1. J. Biol. Chem. 2000;275:4066–4071. doi: 10.1074/jbc.275.6.4066. [DOI] [PubMed] [Google Scholar]
- 33.Critton DA, Tortajada A, Stetson G, Peti W, Page R. Structural basis of substrate recognition by hematopoietic tyrosine phosphatase. Biochemistry. 2008;47:13336–13345. doi: 10.1021/bi801724n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Chen M, Zhang S, Yu ZH, Sun JP, Wang L, Liu S, Imasaki T, Takagi Y, Zhang ZY. Substrate specificity of lymphoid-specific tyrosine phosphatase (Lyp) and identification of Src kinase-associated protein of 55 kDa homolog (SKAP-HOM) as a Lyp substrate. J. Biol. Chem. 2011;286:30526–30534. doi: 10.1074/jbc.M111.254722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoudjit L, Jiang R, Lee TH, New LA, Jones N, Takano T. Podocyte Protein, Nephrin, Is a Substrate of Protein Tyrosine Phosphatase 1B. J. Signal Transduct. 2011;2011:376543. doi: 10.1155/2011/376543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z-Y, Maclean D, McNamara DJ, Sawyer TK, Dixon JE. Protein tyrosine phosphatase substrate specificity: Size and phosphotyrosine positioning requirements in peptide substrates. Biochemistry. 1994;33:2285–2290. doi: 10.1021/bi00174a040. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Q, Luechapanichkul R, Zhai Y, Pei D. Specificity Profiling of Protein Phosphatases toward Phosphoseryl and Phosphothreonyl Peptides. J. Am. Chem. Soc. 2013;135:9760–9767. doi: 10.1021/ja401692t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlucci A, Gedressi C, Lignitto L, Nezi L, Villa-Moruzzi E, Avvedimento EV, Gottesman M, Garbi C, Feliciello A. Protein-tyrosine phosphatase PTPD1 regulates focal adhesion kinase autophosphorylation and cell migration. J. Biol. Chem. 2008;283:10919–10929. doi: 10.1074/jbc.M707248200. [DOI] [PubMed] [Google Scholar]
- 39.Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol. Biol. Cell. 2003;14:2520–2529. doi: 10.1091/mbc.E02-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang ZY, Wang Y, Dixon JE. Dissecting the catalytic mechanism of protein-tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1624–1627. doi: 10.1073/pnas.91.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denu JM, Lohse DL, Vijayalakshmi J, Saper MA, Dixon JE. Visualization of intermediate and transition-state structures in protein-tyrosine phosphatase catalysis. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2493–2498. doi: 10.1073/pnas.93.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 43.Zhang ZY, Palfey BA, Wu L, Zhao Y. Catalytic function of the conserved hydroxyl group in the protein tyrosine phosphatase signature motif. Biochemistry. 1995;34:16389–16396. doi: 10.1021/bi00050a020. [DOI] [PubMed] [Google Scholar]
- 44.Puius YA, Zhao Y, Sullivan M, Lawrence DS, Almo SC, Zhang Z-Y. Identification of second aryl phosphate-binding site in protein-tyrosine phosphates 1B: A paradigm for inhibitor design. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13420–13425. doi: 10.1073/pnas.94.25.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Liu L, He D, Song X, Liang X, Zhao ZJ, Zhou GW. Crystal structure of human protein-tyrosine phosphatase SHP-1. J. Biol. Chem. 2003;278:6516–6520. doi: 10.1074/jbc.M210430200. [DOI] [PubMed] [Google Scholar]
- 47.Pei D, Lorenz U, Klingmuller U, Neel BG, Walsh CT. Intramolecular regulation of protein tyrosine phosphatase SH-PTP1: a new function for Src homology 2 domains. Biochemistry. 1994;33:15483–15493. doi: 10.1021/bi00255a030. [DOI] [PubMed] [Google Scholar]
- 48.Pluskey S, Wandless TJ, Walsh CT, Shoelson SE. Potent stimulation of SH-PTP2 phosphatase activity by simultaneous occupancy of both SH2 domains. J. Biol. Chem. 1995;270:2897–2900. doi: 10.1074/jbc.270.7.2897. [DOI] [PubMed] [Google Scholar]
- 49.Mathew S, George SP, Wang YH, Siddiqui MR, Srinivasan K, Tan LZ, Khurana S. Potential molecular mechanism for C-Src kinase-mediated regulation of intestinal cell migration. J. Biol. Chem. 2008;283:22709–22722. doi: 10.1074/jbc.M801319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng Y, Xia Y, Hawke D, Halle M, Tremblay ML, Gao X, Zhou XZ, Aldape K, Cobb MH, Xie K, He J, Lu Z. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Mol. Cell. 2009;35:11–25. doi: 10.1016/j.molcel.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lyons PD, Dunty JM, Schaefer EM, Schaller MD. Inhibition of the catalytic activity of cell adhesion kinase beta by protein-tyrosine phosphatase-PEST-mediated dephosphorylation. J. Biol. Chem. 2001;276:24422–24431. doi: 10.1074/jbc.M011080200. [DOI] [PubMed] [Google Scholar]
- 52.Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA. Fyn and PTP-PEST-mediated regulation of Wiskott-Aldrich syndrome protein (WASp) tyrosine phosphorylation is required for coupling T cell antigen receptor engagement to WASp effector function and T cell activation. J. Exp. Med. 2004;199:99–112. doi: 10.1084/jem.20030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Côté JF, Chung PL, Théberge JF, Hallé M, Spencer S, Lasky LA, Tremblay ML. PSTPIP is a substrate of PTP-PEST and serves as a scaffold guiding PTP-PEST toward a specific dephosphorylation of WASP. J. Biol. Chem. 2002;277:2973–2986. doi: 10.1074/jbc.M106428200. [DOI] [PubMed] [Google Scholar]
- 54.Zheng XM, Resnick RJ, Shalloway D. A phosphotyrosine displacement mechanism for activation of Src by PTPalpha. EMBO J. 2000;19:964–978. doi: 10.1093/emboj/19.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young RM, Zheng X, Holowka D, Baird B. Reconstitution of regulated phosphorylation of FcepsilonRI by a lipid raft-excluded protein-tyrosine phosphatase. J. Biol. Chem. 2005;280:1230–1235. doi: 10.1074/jbc.M408339200. [DOI] [PubMed] [Google Scholar]
- 56.Bhandari V, Lim KL, Pallen CJ. Physical and functional interactions between receptor-like protein-tyrosine phosphatase alpha and p59fyn. J. Biol. Chem. 1998;273:8691–8698. doi: 10.1074/jbc.273.15.8691. [DOI] [PubMed] [Google Scholar]
- 57.Huang Z, Zhou B, Zhang Z-Y. Molecular Determinants of Substrate Recognition in Hematopoietic Protein Tyrosine Phosphatase. J. Biol. Chem. 2004;279:52150–52159. doi: 10.1074/jbc.M407820200. [DOI] [PubMed] [Google Scholar]
- 58.Møller NP, Møller KB, Lammers R, Kharitonenkov A, Sures I, Ullrich A. Src kinase associates with a member of a distinct subfamily of protein-tyrosine phosphatases containing an ezrin-like domain. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7477–7481. doi: 10.1073/pnas.91.16.7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cardone L, Carlucci A, Affaitati A, Livigni A, DeCristofaro T, Garbi C, Varrone S, Ullrich A, Gottesman ME, Avvedimento EV, Feliciello A. Mitochondrial AKAP121 binds and targets protein tyrosine phosphatase D1, a novel positive regulator of src signaling. Mol. Cell. Biol. 2004;24:4613–4626. doi: 10.1128/MCB.24.11.4613-4626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piwnica-Worms H, Saunders KB, Roberts TM, Smith AE, Cheng CH. Tyrosine phosphorylation regulates the biochemical and biological properties of pp60c–src. Cell. 1987;49:75–82. doi: 10.1016/0092-8674(87)90757-4. [DOI] [PubMed] [Google Scholar]
- 61.Kmiecik TE, Shalloway D. Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell. 1987;49:65–73. doi: 10.1016/0092-8674(87)90756-2. [DOI] [PubMed] [Google Scholar]
- 62.Zhang P, Guo A, Possemato A, Wang C, Beard L, Carlin C, Markowitz SD, Polakiewicz RD, Wang Z. Identification and functional characterization of p130Cas as a substrate of protein tyrosine phosphatase nonreceptor 14. Oncogene. 2013;32:2087–2095. doi: 10.1038/onc.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG. The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 64.Lorenzen JA, Dadabay CY, Fischer EH. COOH-terminal sequence motifs target the T cell protein tyrosine phosphatase to the ER and nucleus. J. Cell Biol. 1995;131:631–643. doi: 10.1083/jcb.131.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tiganis T, Flint AJ, Adam SA, Tonks NK. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J. Biol. Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- 66.Han S, Williams S, Mustelin T. Cytoskeletal protein tyrosine phosphatase PTPH1 reduces T cell antigen receptor signaling. Eur. J. Immunol. 2000;30:1318–1325. doi: 10.1002/(SICI)1521-4141(200005)30:5<1318::AID-IMMU1318>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 67.Moran U, Phillips R, Milo R. SnapShot: key numbers in biology. Cell. 2010;141:1262–1262. doi: 10.1016/j.cell.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 68.Bourdeau A, Dube N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr. Opin. Cell Biol. 2005;17:203–209. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 69.Wallace MJ, Fladd C, Batt J, Rotin D. The second catalytic domain of protein tyrosine phosphatase delta (PTP delta) binds to and inhibits the first catalytic domain of PTP sigma. Mol. Cell. Biol. 1998;18:2608–2616. doi: 10.1128/mcb.18.5.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanchetot C, Tertoolen LG, Overvoorde J, den Hertog J. Intra- and intermolecular interactions between intracellular domains of receptor protein-tyrosine phosphatases. J. Biol. Chem. 2002;277:47263–47269. doi: 10.1074/jbc.M205810200. [DOI] [PubMed] [Google Scholar]
- 71.Majeti R, Xu Z, Parslow TG, Olson JL, Daikh DI, Killeen N, Weiss A. An inactivating point mutation in the inhibitory wedge of CD45 causes lymho proliferation and autoimmunity. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 72.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxidants & Redox Signaling. 2005;7:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.