Abstract
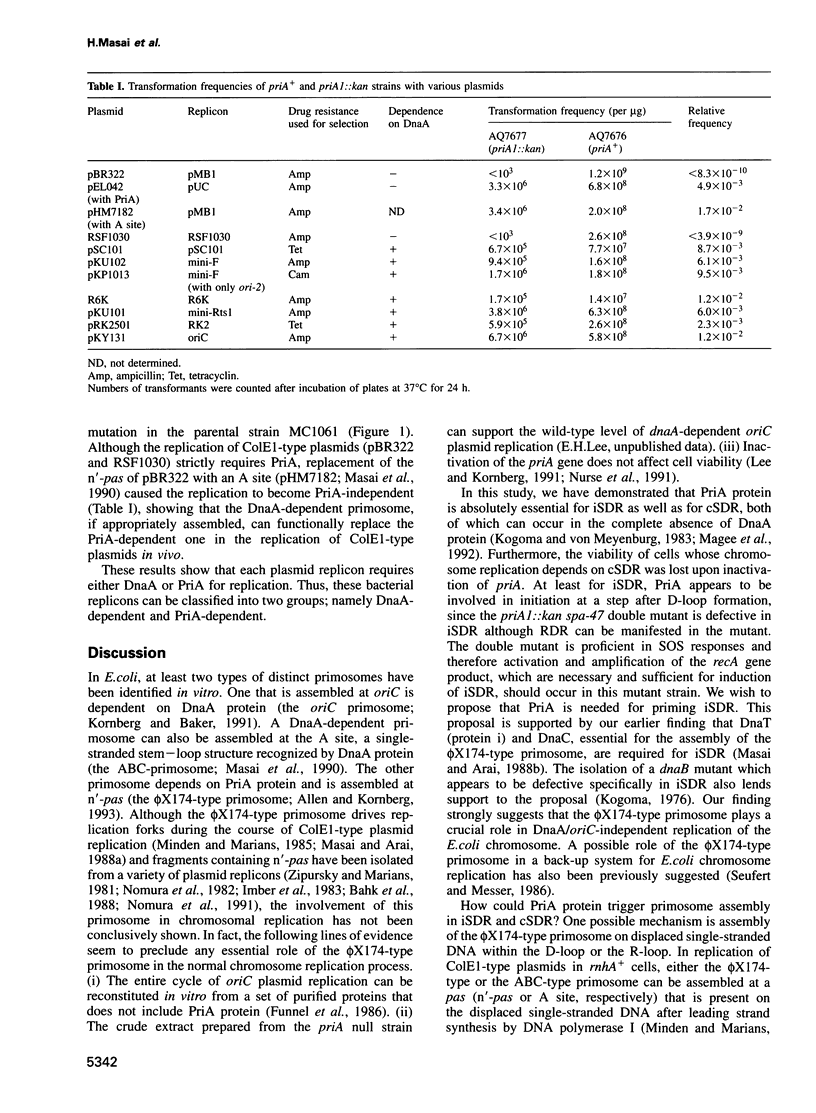
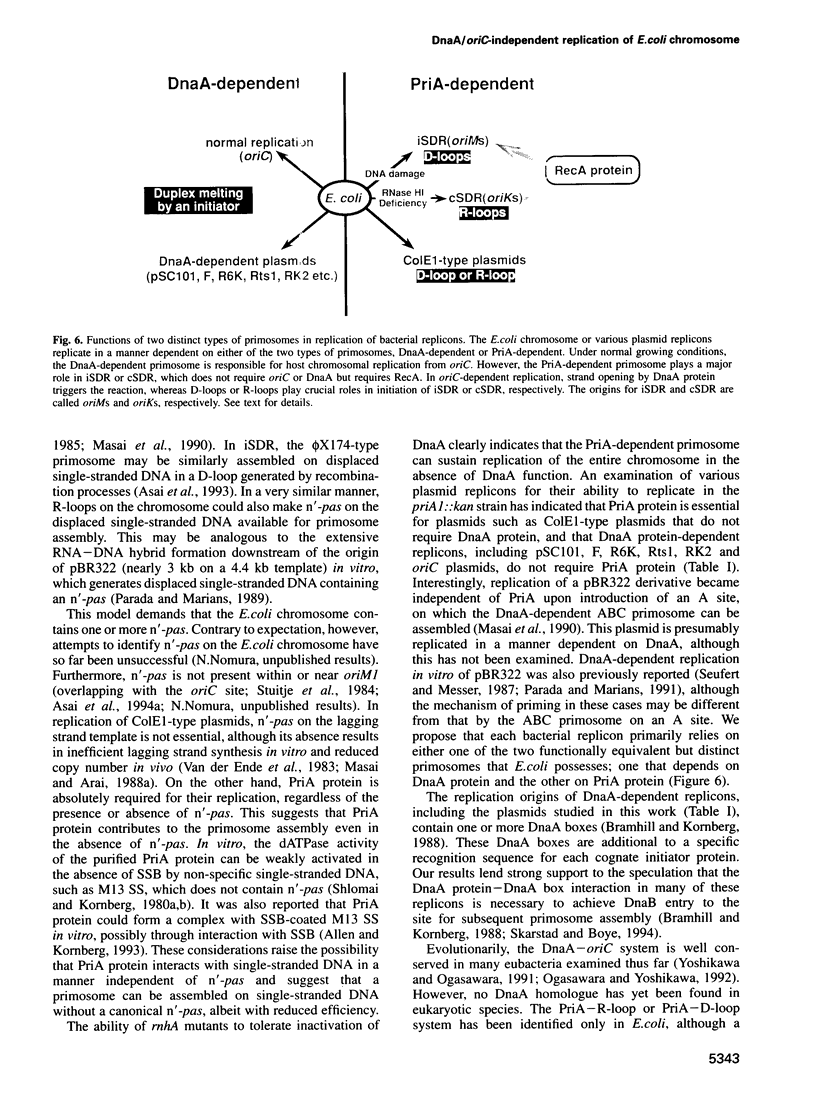
Under certain conditions, Escherichia coli cells exhibit either of two altered modes of chromosomal DNA replication. These are inducible stable DNA replication (iSDR), seen in SOS-induced cells, and constitutive stable DNA replication (cSDR), seen in rnhA mutants. Both iSDR and cSDR can continue to occur in the absence of protein synthesis. They are dependent on RecA protein, but do not require DnaA protein or the oriC site. Here we report the requirement for PriA, a protein essential for assembly of the phi X174-type primosome, for both iSDR and cSDR. In priA1(Null)::kan mutant cells, iSDR is not observed after induction by thymine starvation. Replication from one of the origins (oriM1) specific to iSDR is greatly reduced by the priA1::kan mutation. cSDR in rnhA224 mutant cells deficient in RNase HI is also completely abolished by the same priA mutation. In both cases, SDR is restored by introduction of a plasmid carrying a wild-type priA gene. Furthermore, the viability of an rnhA::cat dnaA46 strain is lost at 42 degrees C upon inactivation of the priA gene, indicating the lethal effect of priA inactivation on those cells whose viability depends on cSDR. These results demonstrate that a function of PriA protein is essential for iSDR and cSDR and suggest the involvement of the PriA-dependent phi X174-type primosome in these DnaA/oriC-independent pathways of chromosome replication. Whereas ColE1-type plasmids, known to be independent of DnaA, absolutely require PriA function for replication, DnaA-dependent plasmid replicons such as pSC101, F, R6K, Rts1 and RK2 are able to transform and to be maintained in the priA1::kan strain.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen G. C., Jr, Kornberg A. Assembly of the primosome of DNA replication in Escherichia coli. J Biol Chem. 1993 Sep 15;268(26):19204–19209. [PubMed] [Google Scholar]
- Asai T., Bates D. B., Kogoma T. DNA replication triggered by double-stranded breaks in E. coli: dependence on homologous recombination functions. Cell. 1994 Sep 23;78(6):1051–1061. doi: 10.1016/0092-8674(94)90279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Imai M., Kogoma T. DNA damage-inducible replication of the Escherichia coli chromosome is initiated at separable sites within the minimal oriC. J Mol Biol. 1994 Feb 4;235(5):1459–1469. doi: 10.1006/jmbi.1994.1101. [DOI] [PubMed] [Google Scholar]
- Asai T., Kogoma T. D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J Bacteriol. 1994 Apr;176(7):1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Sommer S., Bailone A., Kogoma T. Homologous recombination-dependent initiation of DNA replication from DNA damage-inducible origins in Escherichia coli. EMBO J. 1993 Aug;12(8):3287–3295. doi: 10.1002/j.1460-2075.1993.tb05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahk J. D., Sakai H., Komano T. Plasmid pACYC184 contains an ssi signal for initiation of single-strand phage DNA replication. Gene. 1988 May 15;65(1):93–99. doi: 10.1016/0378-1119(88)90420-9. [DOI] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986 Aug;167(2):594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Buhk H. J., Messer W. The replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene. 1983 Oct;24(2-3):265–279. doi: 10.1016/0378-1119(83)90087-2. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977 Nov;132(2):734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H., Luttropp L. K., Falkow S. Nature of R-factor replication in the presence of chloramphenicol. Proc Natl Acad Sci U S A. 1975 Feb;72(2):654–658. doi: 10.1073/pnas.72.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. Complete enzymatic replication of plasmids containing the origin of the Escherichia coli chromosome. J Biol Chem. 1986 Apr 25;261(12):5616–5624. [PubMed] [Google Scholar]
- Gaylo P. J., Turjman N., Bastia D. DnaA protein is required for replication of the minimal replicon of the broad-host-range plasmid RK2 in Escherichia coli. J Bacteriol. 1987 Oct;169(10):4703–4709. doi: 10.1128/jb.169.10.4703-4709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen E. B., Yarmolinsky M. B. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4423–4427. doi: 10.1073/pnas.83.12.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Imber R., Low R. L., Ray D. S. Identification of a primosome assembly site in the region of the ori 2 replication origin of the Escherichia coli mini-F plasmid. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7132–7136. doi: 10.1073/pnas.80.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamio Y., Tabuchi A., Itoh Y., Katagiri H., Terawaki Y. Complete nucleotide sequence of mini-Rts1 and its copy mutant. J Bacteriol. 1984 Apr;158(1):307–312. doi: 10.1128/jb.158.1.307-312.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline B. C. Aspects of plasmid F maintenance in Escherichia coli. Can J Microbiol. 1988 Apr;34(4):526–535. doi: 10.1139/m88-090. [DOI] [PubMed] [Google Scholar]
- Kline B. C., Kogoma T., Tam J. E., Shields M. S. Requirement of the Escherichia coli dnaA gene product for plasmid F maintenance. J Bacteriol. 1986 Oct;168(1):440–443. doi: 10.1128/jb.168.1.440-443.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. RNase H-defective mutants of Escherichia coli. J Bacteriol. 1986 May;166(2):361–363. doi: 10.1128/jb.166.2.361-363.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T. Two types of temperature sensitivity in DNA replication of an Escherichia coli dnaB mutant. J Mol Biol. 1976 May 5;103(1):191–197. doi: 10.1016/0022-2836(76)90059-0. [DOI] [PubMed] [Google Scholar]
- Kogoma T., von Meyenburg K. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 1983;2(3):463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lark K. G., Lark C. A. recA-dependent DNA replication in the absence of protein synthesis: characteristics of a dominant lethal replication mutation, dnaT, and requirement for recA+ function. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):537–549. doi: 10.1101/sqb.1979.043.01.059. [DOI] [PubMed] [Google Scholar]
- Lee E. H., Kornberg A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n' protein. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. H., Masai H., Allen G. C., Jr, Kornberg A. The priA gene encoding the primosomal replicative n' protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4620–4624. doi: 10.1073/pnas.87.12.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M. A., Sparks R. B., Helinski D. R. Bidirectional replication of plasmid R6K DNA in Escherichia coli; correspondence between origin of replication and position of single-strand break in relaxed complex. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2905–2909. doi: 10.1073/pnas.72.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee T. R., Asai T., Malka D., Kogoma T. DNA damage-inducible origins of DNA replication in Escherichia coli. EMBO J. 1992 Nov;11(11):4219–4225. doi: 10.1002/j.1460-2075.1992.tb05516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee T. R., Kogoma T. Requirement of RecBC enzyme and an elevated level of activated RecA for induced stable DNA replication in Escherichia coli. J Bacteriol. 1990 Apr;172(4):1834–1839. doi: 10.1128/jb.172.4.1834-1839.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S., Kuribayashi M., Miki T., Horiuchi T. An amber replication mutant of F plasmid mapped in the minimal replication region. Mol Gen Genet. 1983;191(2):231–237. doi: 10.1007/BF00334819. [DOI] [PubMed] [Google Scholar]
- Masai H., Arai K. Escherichia coli dnaT gene function is required for pBR322 plasmid replication but not for R1 plasmid replication. J Bacteriol. 1989 Jun;171(6):2975–2980. doi: 10.1128/jb.171.6.2975-2980.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Arai K. Initiation of lagging-strand synthesis for pBR322 plasmid DNA replication in vitro is dependent on primosomal protein i encoded by dnaT. J Biol Chem. 1988 Oct 15;263(29):15016–15023. [PubMed] [Google Scholar]
- Masai H., Arai K. Operon structure of dnaT and dnaC genes essential for normal and stable DNA replication of Escherichia coli chromosome. J Biol Chem. 1988 Oct 15;263(29):15083–15093. [PubMed] [Google Scholar]
- Masai H., Bond M. W., Arai K. Cloning of the Escherichia coli gene for primosomal protein i: the relationship to dnaT, essential for chromosomal DNA replication. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1256–1260. doi: 10.1073/pnas.83.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Nomura N., Arai K. The ABC-primosome. A novel priming system employing dnaA, dnaB, dnaC, and primase on a hairpin containing a dnaA box sequence. J Biol Chem. 1990 Sep 5;265(25):15134–15144. [PubMed] [Google Scholar]
- Meissner P. S., Sisk W. P., Berman M. L. Bacteriophage lambda cloning system for the construction of directional cDNA libraries. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4171–4175. doi: 10.1073/pnas.84.12.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985 Aug 5;260(16):9316–9325. [PubMed] [Google Scholar]
- Murialdo H. Lethal effect of lambda DNA terminase in recombination deficient Escherichia coli. Mol Gen Genet. 1988 Jul;213(1):42–49. doi: 10.1007/BF00333396. [DOI] [PubMed] [Google Scholar]
- Nomura N., Low R. L., Ray D. S. Identification of ColE1 DNA sequences that direct single strand-to-double strand conversion by a phi X174 type mechanism. Proc Natl Acad Sci U S A. 1982 May;79(10):3153–3157. doi: 10.1073/pnas.79.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N., Masai H., Inuzuka M., Miyazaki C., Ohtsubo E., Itoh T., Sasamoto S., Matsui M., Ishizaki R., Arai K. Identification of eleven single-strand initiation sequences (ssi) for priming of DNA replication in the F, R6K, R100 and ColE2 plasmids. Gene. 1991 Dec 1;108(1):15–22. doi: 10.1016/0378-1119(91)90482-q. [DOI] [PubMed] [Google Scholar]
- Nurse P., DiGate R. J., Zavitz K. H., Marians K. J. Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4615–4619. doi: 10.1073/pnas.87.12.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., Zavitz K. H., Marians K. J. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J Bacteriol. 1991 Nov;173(21):6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Yoshikawa H. Genes and their organization in the replication origin region of the bacterial chromosome. Mol Microbiol. 1992 Mar;6(5):629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pickett G. G., Kogoma T., Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C. A., Marians K. J. Mechanism of DNA A protein-dependent pBR322 DNA replication. DNA A protein-mediated trans-strand loading of the DNA B protein at the origin of pBR322 DNA. J Biol Chem. 1991 Oct 5;266(28):18895–18906. [PubMed] [Google Scholar]
- Parada C. A., Marians K. J. Transcriptional activation of pBR322 DNA can lead to duplex DNA unwinding catalyzed by the Escherichia coli preprimosome. J Biol Chem. 1989 Sep 5;264(25):15120–15129. [PubMed] [Google Scholar]
- Seufert W., Messer W. DnaA protein binding to the plasmid origin region can substitute for primosome assembly during replication of pBR322 in vitro. Cell. 1987 Jan 16;48(1):73–78. doi: 10.1016/0092-8674(87)90357-6. [DOI] [PubMed] [Google Scholar]
- Seufert W., Messer W. Initiation of Escherichia coli minichromosome replication at oriC and at protein n' recognition sites. Two modes for initiating DNA synthesis in vitro. EMBO J. 1986 Dec 1;5(12):3401–3406. doi: 10.1002/j.1460-2075.1986.tb04656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. A prepriming DNA replication enzyme of Escherichia coli. II. Actions of protein n': a sequence-specific, DNA-dependent ATPase. J Biol Chem. 1980 Jul 25;255(14):6794–6798. [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K., Boye E. The initiator protein DnaA: evolution, properties and function. Biochim Biophys Acta. 1994 Mar 1;1217(2):111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Stuitje A. R., Weisbeek P. J., Meijer M. Initiation signals for complementary strand DNA synthesis in the region of the replication origin of the Escherichia coli chromosome. Nucleic Acids Res. 1984 Apr 11;12(7):3321–3332. doi: 10.1093/nar/12.7.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey T. A., Kogoma T. Genetic analysis of constitutive stable DNA replication in rnh mutants of Escherichia coli K12. Mol Gen Genet. 1987 Jul;208(3):420–427. doi: 10.1007/BF00328133. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Linskens M. H., Kowalski D., Huberman J. A. New beginnings in studies of eukaryotic DNA replication origins. Biochim Biophys Acta. 1989 Jan 23;1007(1):1–14. doi: 10.1016/0167-4781(89)90123-1. [DOI] [PubMed] [Google Scholar]
- Van der Ende A., Teertstra R., Weisbeek P. J. n' Protein activator sites of plasmid pBR322 are not essential for its DNA replication. J Mol Biol. 1983 Jul 5;167(3):751–756. doi: 10.1016/s0022-2836(83)80108-9. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Yamaguchi M., Tomizawa J. Incompatibility of plasmids containing the replication origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5347–5351. doi: 10.1073/pnas.79.17.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavitz K. H., Marians K. J. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J Biol Chem. 1992 Apr 5;267(10):6933–6940. [PubMed] [Google Scholar]
- Zipursky S. L., Marians K. J. Escherichia coli factor Y sites of plasmid pBR322 can function as origins of DNA replication. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6111–6115. doi: 10.1073/pnas.78.10.6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Meyenburg K., Boye E., Skarstad K., Koppes L., Kogoma T. Mode of initiation of constitutive stable DNA replication in RNase H-defective mutants of Escherichia coli K-12. J Bacteriol. 1987 Jun;169(6):2650–2658. doi: 10.1128/jb.169.6.2650-2658.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]





