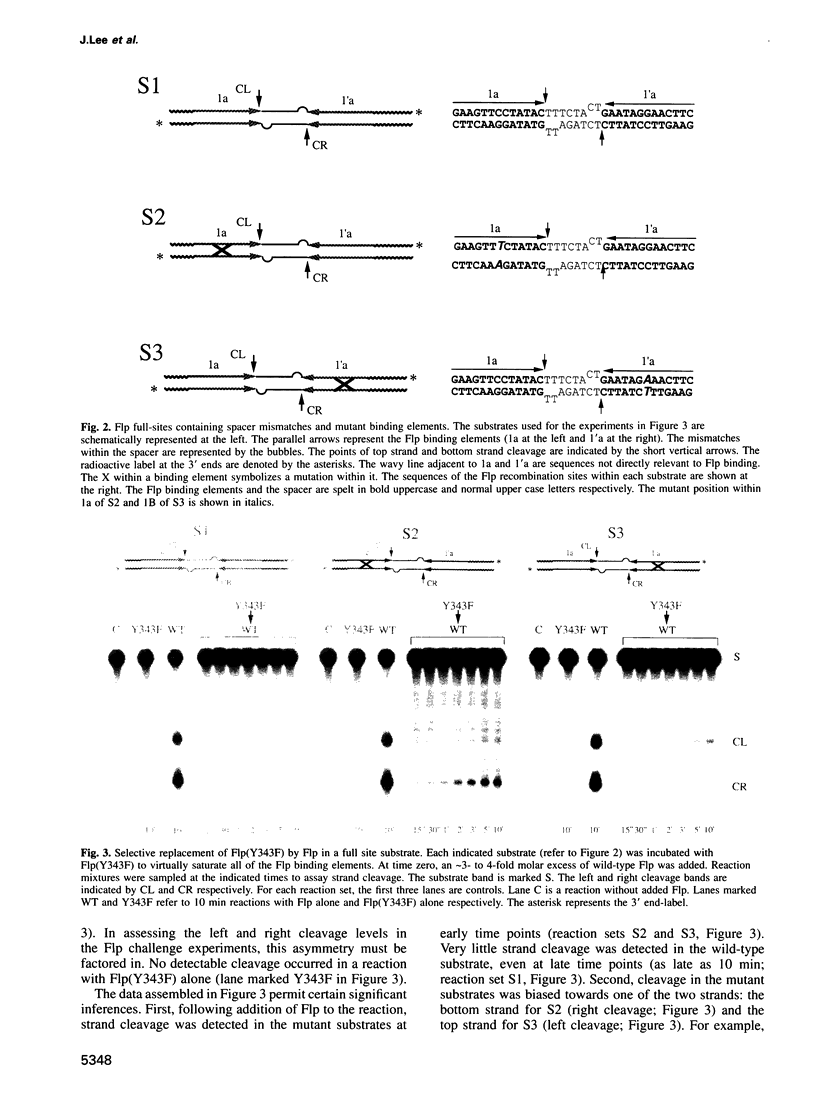
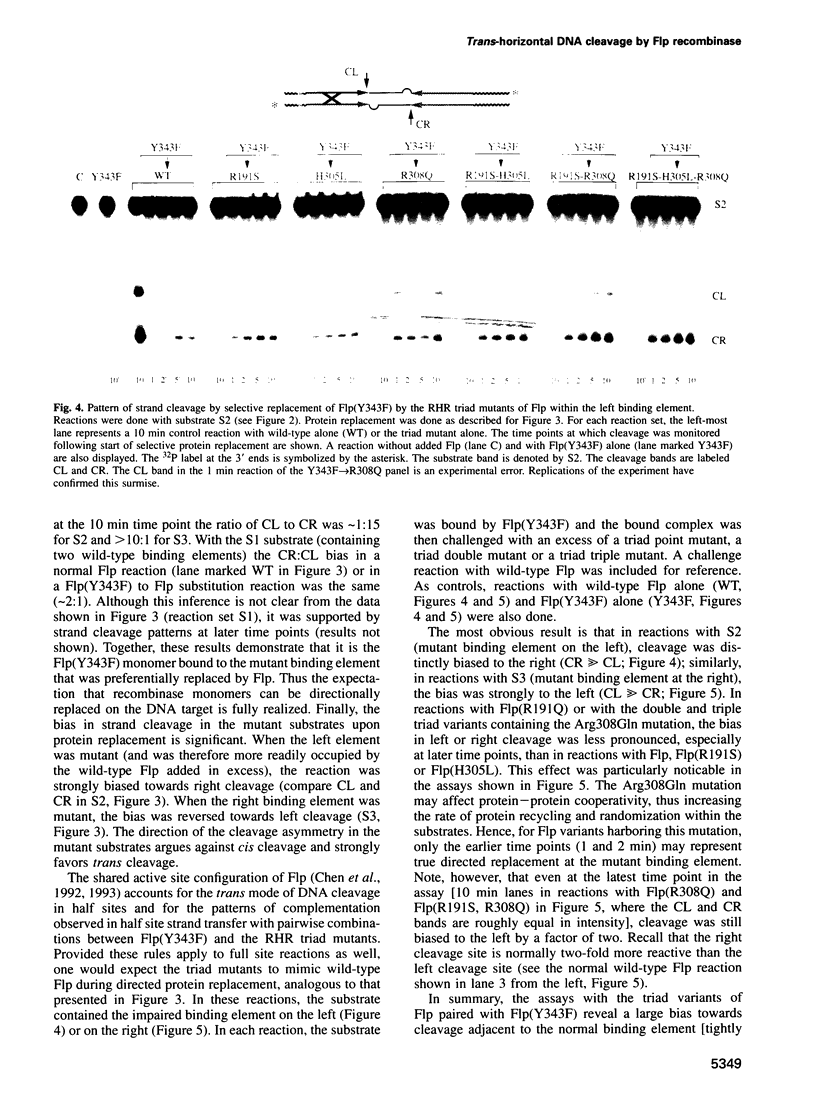
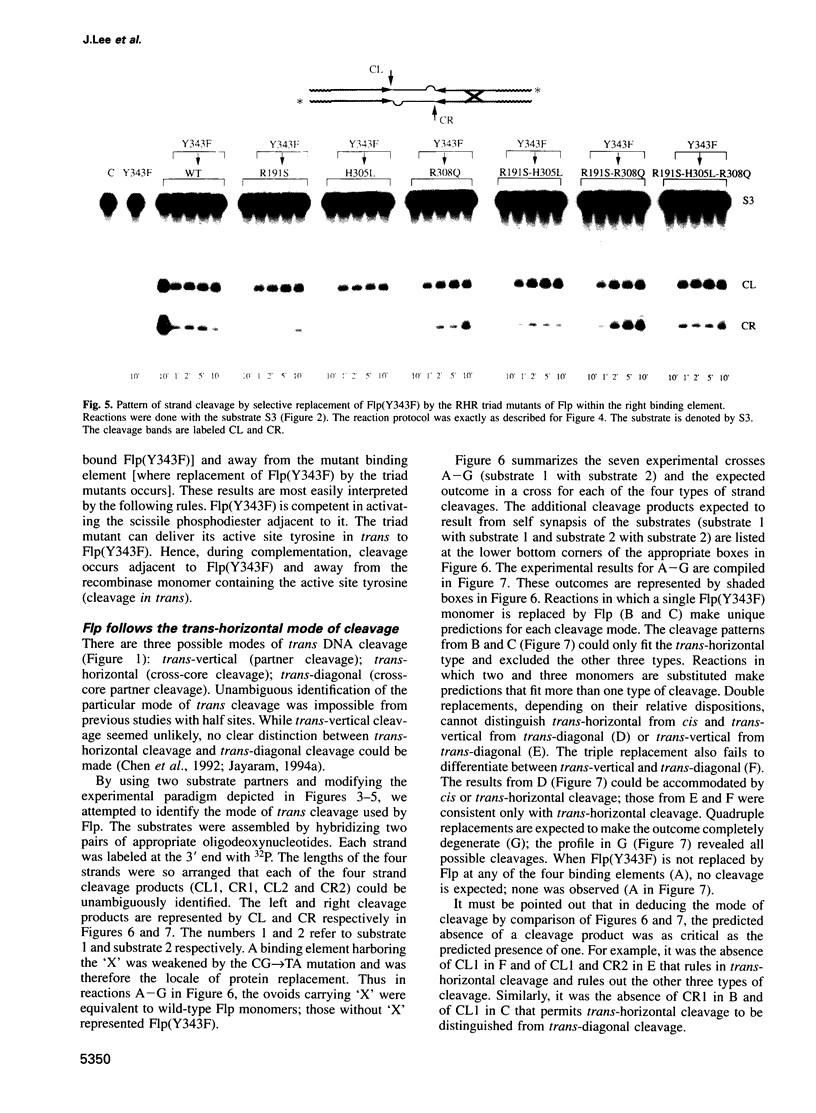
Abstract
One round of site-specific recombination between two DNA partners mediated by the Flp recombinase requires the breakage and reformation of four phosphodiester bonds. The reaction is accomplished by the combined action of four Flp monomers. Within the recombination complex, what is the relative disposition of a Flp monomer with respect to the target diester that it cleaves? To address this question, we have devised a strategy for the targeted orientation of Flp monomers within full-site recombination substrates. Our experimental design is not dependent on 'altered binding specificity' of the recombinase. Analysis of the pattern of DNA cleavage by this method reveals no evidence for DNA cleavage in cis. A Flp monomer bound to its recognition element within the full site does not cleave the scissile phosphodiester bond adjacent to it. Our results are most consistent with 'trans-horizontal cleavage'. Cleavage by Flp occurs at the scissile phosphodiester distal to it, but within the same full site. The general experimental design employed here will be of widespread utility in mechanistic analyses of nucleic acid transactions involving multimeric DNA-protein assemblies.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K. E., Hoess R. H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992 Jan;5(1):87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- Andrews B. J., McLeod M., Broach J., Sadowski P. D. Interaction of the FLP recombinase of the Saccharomyces cerevisiae 2 micron plasmid with mutated target sequences. Mol Cell Biol. 1986 Jul;6(7):2482–2489. doi: 10.1128/mcb.6.7.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Bruckner R. C., Cox M. M. Specific contacts between the FLP protein of the yeast 2-micron plasmid and its recombination site. J Biol Chem. 1986 Sep 5;261(25):11798–11807. [PubMed] [Google Scholar]
- Chen J. W., Lee J., Jayaram M. DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell. 1992 May 15;69(4):647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- Chen J. W., Yang S. H., Jayaram M. Tests for the fractional active-site model in Flp site-specific recombination. Assembly of a functional recombination complex in half-site and full-site strand transfer. J Biol Chem. 1993 Jul 5;268(19):14417–14425. [PubMed] [Google Scholar]
- Doolittle R. F. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994 Jan;19(1):15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. Analysis of a nucleoprotein complex: the synaptosome of gamma delta resolvase. Science. 1993 Oct 29;262(5134):738–740. doi: 10.1126/science.8235593. [DOI] [PubMed] [Google Scholar]
- Han Y. W., Gumport R. I., Gardner J. F. Complementation of bacteriophage lambda integrase mutants: evidence for an intersubunit active site. EMBO J. 1993 Dec;12(12):4577–4584. doi: 10.1002/j.1460-2075.1993.tb06146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M. Phosphoryl transfer in Flp recombination: a template for strand transfer mechanisms. Trends Biochem Sci. 1994 Feb;19(2):78–82. doi: 10.1016/0968-0004(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Kim S., Moitoso de Vargas L., Nunes-Düby S. E., Landy A. Mapping of a higher order protein-DNA complex: two kinds of long-range interactions in lambda attL. Cell. 1990 Nov 16;63(4):773–781. doi: 10.1016/0092-8674(90)90143-3. [DOI] [PubMed] [Google Scholar]
- Lee J., Jayaram M. Mechanism of site-specific recombination. Logic of assembling recombinase catalytic site from fractional active sites. J Biol Chem. 1993 Aug 15;268(23):17564–17570. [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Heteroduplex substrates for bacteriophage lambda site-specific recombination: cleavage and strand transfer products. EMBO J. 1989 Nov;8(11):3523–3533. doi: 10.1002/j.1460-2075.1989.tb08518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G., Luetke K., Sadowski P. D. Mechanism of cleavage and ligation by FLP recombinase: classification of mutations in FLP protein by in vitro complementation analysis. Mol Cell Biol. 1993 Jun;13(6):3167–3175. doi: 10.1128/mcb.13.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi G. B., Beatty L. G., Sadowski P. D. The FLP protein contacts both major and minor grooves of its recognition target sequence. Nucleic Acids Res. 1992 Nov 25;20(22):5927–5935. doi: 10.1093/nar/20.22.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R. L., Evans B. R., Zheng L., Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. Possible role of Arg-308 in coupling substrate binding to catalysis. J Biol Chem. 1990 Mar 15;265(8):4527–4533. [PubMed] [Google Scholar]
- Prasad P. V., Horensky D., Young L. J., Jayaram M. Substrate recognition by the 2 micron circle site-specific recombinase: effect of mutations within the symmetry elements of the minimal substrate. Mol Cell Biol. 1986 Dec;6(12):4329–4334. doi: 10.1128/mcb.6.12.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X. H., Inman R. B., Cox M. M. Protein-based asymmetry and protein-protein interactions in FLP recombinase-mediated site-specific recombination. J Biol Chem. 1990 Dec 15;265(35):21779–21788. [PubMed] [Google Scholar]
- Senecoff J. F., Rossmeissl P. J., Cox M. M. DNA recognition by the FLP recombinase of the yeast 2 mu plasmid. A mutational analysis of the FLP binding site. J Mol Biol. 1988 May 20;201(2):405–421. doi: 10.1016/0022-2836(88)90147-7. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Jayaram M. Generality of the shared active site among yeast family site-specific recombinases. The R site-specific recombinase follows the Flp paradigm [corrected]. J Biol Chem. 1994 Apr 29;269(17):12789–12796. [PubMed] [Google Scholar]
- Zhou Y., Busby S., Ebright R. H. Identification of the functional subunit of a dimeric transcription activator protein by use of oriented heterodimers. Cell. 1993 Apr 23;73(2):375–379. doi: 10.1016/0092-8674(93)90236-j. [DOI] [PubMed] [Google Scholar]