Summary
The mechanisms that facilitate dissemination of the highly invasive spirochete, Treponema pallidum, are incompletely understood. Previous studies showed the treponemal metalloprotease pallilysin (Tp0751) possesses fibrin clot degradation capability, suggesting a role in treponemal dissemination. In the current study we report characterization of the functionally-linked protein Tp0750. Structural modelling predicts Tp0750 contains a von Willebrand factor type A (vWFA) domain, a protein-protein interaction domain commonly observed in extracellular matrix (ECM)-binding proteins. We report Tp0750 is a serine protease that degrades the major clot components fibrinogen and fibronectin. We also demonstrate Tp0750 cleaves a matrix metalloprotease (MMP) peptide substrate that is targeted by several MMPs, enzymes central to ECM remodelling. Through proteomic analyses we show Tp0750 binds the endothelial fibrinolytic receptor, annexin A2, in a specific and dose-dependent manner. These results suggest Tp0750 constitutes a multifunctional protein that is able to (1) degrade infection-limiting clots by both inhibiting clot formation through degradation of host coagulation cascade proteins and promoting clot dissolution by complexing with host proteins involved in the fibrinolytic cascade and (2) facilitate ECM degradation via MMP-like proteolysis of host components. We propose that through these activities Tp0750 functions in concert with pallilysin to enable T. pallidum dissemination.
Keywords: Treponema pallidum, syphilis, host interactions, protease, dissemination, pathogenesis
Introduction
The spirochete Treponema pallidum subsp. pallidum is the causative agent of syphilis, a chronic multistage disease that is primarily transmitted sexually or in utero. Treponema pallidum invades the tissue barrier and enters the bloodstream within minutes of treponemal exposure followed by widespread dissemination within hours of infection (Raiziss and Severac, 1937; Cumberland and Turner, 1949). Furthermore, T. pallidum traverses both the placental and blood-brain barriers and invades the central nervous system during early primary syphilis (Lukehart et al., 1988; Hollier and Cox, 1998; Radolf et al., 2006). Although T. pallidum dissemination via the bloodstream is central to the infection process, our understanding of the mechanics of this crucial aspect of treponemal pathogenesis is limited.
Under normal conditions the vascular endothelium exists in an anti-coagulant state for the purpose of maintaining blood fluidity. However, under conditions of infection pathogen-stimulated endothelial cell activation occurs, resulting in endothelial leukocyte adhesion (Takahashi et al., 1996; Keller et al., 2003). This induces a local inflammatory response which leads to loss of anti-coagulant factors from the endothelial surface and rapid production of endothelial cell-expressed pro-coagulant factors, including tissue factor (TF) and von Willebrand factor (vWF) (Keller et al., 2003). TF activates the host coagulation cascade which culminates in the formation of a clot via thrombin-mediated conversion of fibrinogen to fibrin while vWF mediates platelet adhesion and plug formation (Weisel, 2005). These pathogen-induced changes shift the endothelial environment from an anti-coagulant state to a pro-coagulant state (Keller et al., 2003) which causes fibrin clot formation in the vicinity of the invading pathogen to prevent pathogen dissemination via the vasculature (Keller et al., 2003; Levi et al., 2004; Delvaeye and Conway, 2009; Petaja, 2011). Studies focused on Escherichia coli, Streptococcus pyogenes, and Staphylococcus aureus have demonstrated that blood coagulation is induced as an early innate host immune response to entrap, localize, and promote killing of these invasive pathogens (Wang et al., 2010; Loof et al., 2011).
In the vasculature the coagulation cascade is intimately associated with the fibrinolytic system, which uses the serine protease plasmin to degrade fibrin clots in order to maintain the haemostatic balance. Plasmin is generated from plasminogen via the activity of tissue-type plasminogen activator (tPA) (Collen and Lijnen, 2005). During the process of vascular fibrinolysis, the endothelial cell surface-localized fibrinolytic receptor complex (comprised of annexin A2 and S100A10) binds both plasminogen and tPA, leading to plasmin deposition on the endothelial cell surface (Madureira et al., 2011; Flood and Hajjar, 2011; Bharadwaj et al., 2013). In response to infection the host concurrently activates the coagulation and fibrinolytic systems to avoid disseminated coagulation, with the endothelium playing a pivotal role in the initiation and regulation of these intertwined processes (Keller et al., 2003; Tanaka et al., 2009; Ait-Oufella et al., 2010).
Plasmin generation by the fibrinolytic receptor complex also results in extracellular matrix (ECM) component degradation via the activation of host matrix metalloproteases (MMPs) (Madureira et al., 2011). MMPs are a group of zinc-dependent proteases that routinely function in tissue remodelling and wound healing, although excessive MMP expression leads to uncontrolled host tissue destruction (Egeblad and Werb, 2002; Malemud, 2006). Multiple pathogenic bacteria, including S. pyogenes, S. aureus, Yersinia pestis, and the spirochetes Borrelia burgdorferi and Leptospira interrogans, exploit the host fibrinolytic and MMP systems to facilitate dissemination via plasmin-mediated clot degradation and plasmin- and MMP-mediated ECM component degradation, respectively (Coleman et al., 1995; Lahteenmaki et al., 2005; Fernandes et al., 2012). Tissue destruction is characteristic of syphilis infection, a phenomenon that may be partly explained by the capacity of T. pallidum to activate human vascular endothelial cells and the collagenase MMP-1 (Riley et al., 1992; Riley et al., 1994; Lee et al., 2000; Chung et al., 2002; LaFond and Lukehart, 2006).
Through previous studies our laboratory identified the T. pallidum metalloprotease pallilysin (Tp0751). This protein degrades the central components of the coagulation cascade, fibrinogen and fibrin. Pallilysin also degrades laminin, a major constituent of the basement membrane that lines the vascular endothelium and a barrier that T. pallidum must pass through during dissemination via the bloodstream (Raiziss and Severac, 1937). In the current study we demonstrate that the gene encoding pallilysin (tp0751) is co-transcribed with the immediate upstream gene, tp0750, suggesting possible functional linkage. We further show that Tp0750 co-associates with pallilysin, interacts with the fibrinolytic receptor complex protein annexin A2, exhibits MMP-like proteolytic activity and degrades the pro-coagulation protein fibrinogen. Our results are consistent with Tp0750 and pallilysin playing a role in treponemal dissemination via a multi-factorial, host-targeted mechanism involving coagulation inhibition, fibrinolysis promotion, and ECM component degradation.
Results
tp0750 and tp0751 (pallilysin) are co-transcribed
Genomic arrangement analysis of tp0750 and tp0751 using Artemis Genome Browser (Rutherford et al., 2000) shows the two genes are oriented in the same direction (forward strand), located in the same reading frame, separated by a 36 base pair intergenic region and flanked by the genes tp0749 and tp0752 on the opposite (reverse) strand (Fig. 1A). These genomic features are synonymous with prokaryotic operon arrangement (Chuang et al., 2012). Indeed, both the Database of Predicted Operons in Microbial Genomes (OperonDB) (Pertea et al., 2009) and the Prokaryotic Operon Database (ProOpDb) (Taboada et al., 2012) predict tp0750 and tp0751 comprise a two-gene operon (data not shown). To confirm co-transcription of tp0750/tp0751, we analyzed RNA isolated from T. pallidum by orientation-specific reverse transcriptase PCR (RT-PCR) using sense (tp0750) and antisense (tp0751) primers (Fig. 1A and Table 1). The primer pair amplified an 865 base pair product, matching a similarly sized amplicon generated from T. pallidum genomic DNA (Fig. 1B, lanes 1 and 3). An amplicon was not detected when reverse transcriptase was omitted from the RT-PCR reaction (Fig. 1B, lane 2), indicating that the 865 base pair product was amplified from RNA and not contaminating DNA. This demonstrates that tp0750 and tp0751 are co-transcribed, suggesting functional linkage.
Fig. 1. tp0750 and tp0751 comprise a two-gene operon.
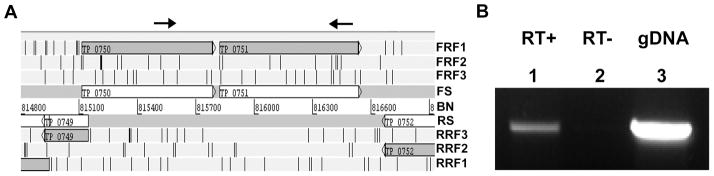
A. The potential for tp0750 and tp0751 to comprise a two-gene operon was investigated by analysis of the T. pallidum genome sequence using the Artemis Genome Browser. FS, forward strand; RS, reverse strand; BN, base pair number; FRF1-3, forward reading frames 1–3; RFR1-3, reverse reading frames 1–3. Arrows indicate the location and orientation of gene-specific primers used in the RT-PCR reaction. B. The two-gene operon organization of tp0750 and tp0751 was confirmed by analysing T. pallidum RNA by RT-PCR. The cDNA product amplified from the tp0750/tp0751 primer pair (RT+) and the PCR product of the same size amplified from T. pallidum genomic DNA (gDNA) are shown. A cDNA product was not detected when reverse transcriptase was omitted from the RT-PCR reaction (RT−).
Table 1.
ICP-MS quantitation of metal ions in purified recombinant Tp0750.
| Concentration in sample (nM) | Ratio of Tp0750:Metal Ion | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tp0750 | Ca | Mg | Zn | Co | Fe | Mn | Ca | Mg | Zn | Co | Fe | Mn |
| 86.0 | 74.8 | 16.0 | 7.6 | 1.0 | ND | ND | 1.0:0.87 | 1.0:0.19 | 1.0:0.09 | 1.0:0.01 | NA | NA |
ND: Not detected; NA: Not applicable; Detection limit: 1.0 part per billion.
Tp0750 contains predicted vWFA and MIDAS domains and binds calcium
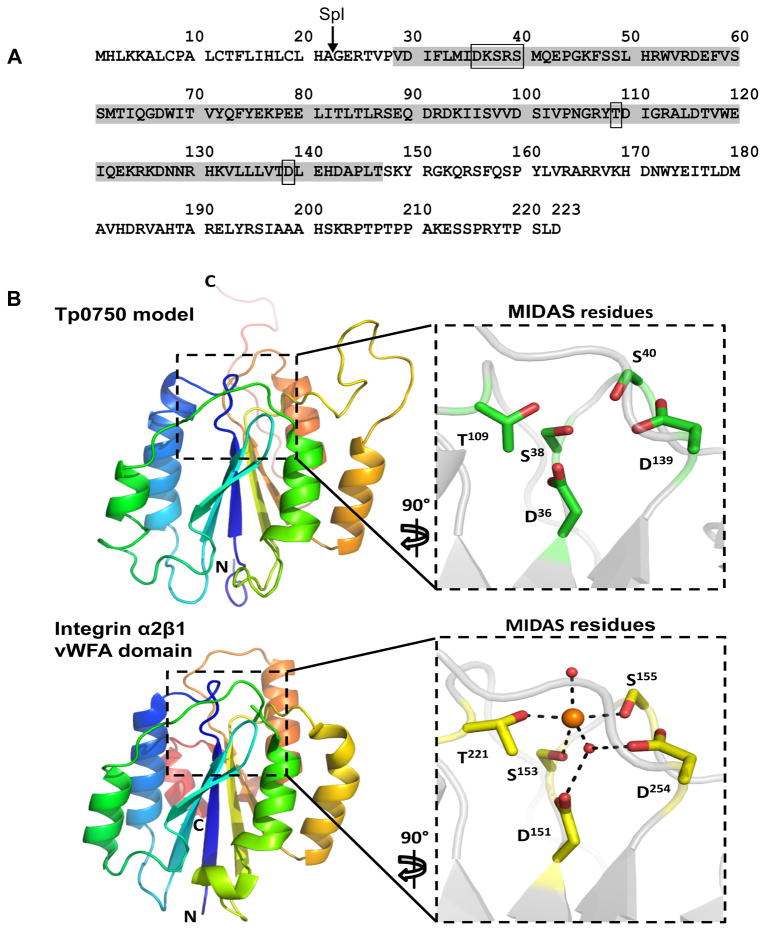
In order to investigate the function of Tp0750, we analyzed the amino acid sequence using multiple proteomic prediction software tools. SignalP (Version 4.1) (Petersen et al., 2011) analysis predicted the presence of an SpI (signal peptidase I) cleavage site between residues A22 and G23 (Fig. 2A). NCBI conserved domain database (Marchler-Bauer et al., 2011) analysis revealed significant similarity of residues V29-T147 (E value = 2.46 × 10−15) to the Metal Ion-Dependent Adhesion Site (MIDAS)-containing von Willebrand Factor type A (vWFA) domain with perfect conservation of the MIDAS motif (DXSXS........T.........D) (Fig. 2A). Tertiary structure prediction analyses using Modeller predicted structural similarity of Tp0750 with integrin alpha2beta1 (α2β1), which contains a vWFA domain and a MIDAS motif (Fig. 2B). vWFA domain-containing proteins often bind divalent metal ions via coordination to residues comprising the MIDAS motif (Springer, 2006). To determine whether Tp0750 binds divalent metal ions, recombinant Tp0750_G23-A199 was analyzed using quantitative inductively coupled plasma-mass spectrometry (ICP-MS), a highly sensitive mass spectrometry technique capable of measuring low concentrations of metal ions. ICP-MS analysis demonstrated that Tp0750 binds primarily to calcium (Table 1).
Fig. 2. Tp0750 molecular modelling predicts similarity to a MIDAS- and vWFA domain-containing protein.
A. Schematic representation of the results of SignalP and NCBI conserved domain database analyses performed on the Tp0750 amino acid sequence. Indicated are the predicted SpI (signal peptidase I) cleavage site (between A22 and G23, arrow) and Metal Ion-Dependent Adhesion Site (MIDAS)-containing vWFA (von Willebrand Factor type A) domain (V29-T147, grey shading; rectangles indicate MIDAS site residues). B. Model of Tp0750 generated by the Modeller program predicts a central 5-stranded beta sheet of mixed parallel and anti-parallel beta strands and 5 framing alpha helices (upper panel). The predicted structure of Tp0750 was modelled using integrin α2β1 (PDB 1DZI), which contains both a vWFA domain and a MIDAS motif (lower panel). Right (inset) panels: residues comprising the MIDAS motif within Tp0750 (upper panel) are conserved in sequence and structure with the cobalt-binding MIDAS motif of integrin α2β1 (lower panel), suggesting Tp0750 has metal ion-binding capability.
Tp0750 interacts with the fibrinolytic receptor complex protein annexin A2
Since the Tp0750 sequence includes a MIDAS-containing vWFA domain, we used an affinity chromatography-proteomics approach to determine if Tp0750 can interact with specific host proteins derived from a biologically relevant cell type. A recombinant Tp0750-cobalt chelate affinity column and a parallel control column (unconjugated cobalt chelate affinity resin) were incubated with a preparation enriched for human umbilical vein endothelial cell (HUVEC) membrane-associated proteins. Columns were extensively washed and Tp0750-interacting host proteins were eluted, separated by SDS-PAGE, subjected to in-gel trypsin digestion, and analyzed by tandem mass spectrometry. Data analysis using PEAKS, which provides protein identification as well as results validation in the form of a PEAKS score (0–100% protein/peptide identity confidence), identified annexin A2 as the highest scoring protein to be isolated using the Tp0750 affinity column (99.99% identity confidence/PEAKS score, 40.41% sequence coverage, 9 unique peptides identified). Also of note, one peptide unique to S100A10 was identified (96% identity confidence/PEAKS score, 26% sequence coverage). Importantly, annexin A2 and S100A10 peptides were not detected upon analysis of the parallel control affinity resin eluate.
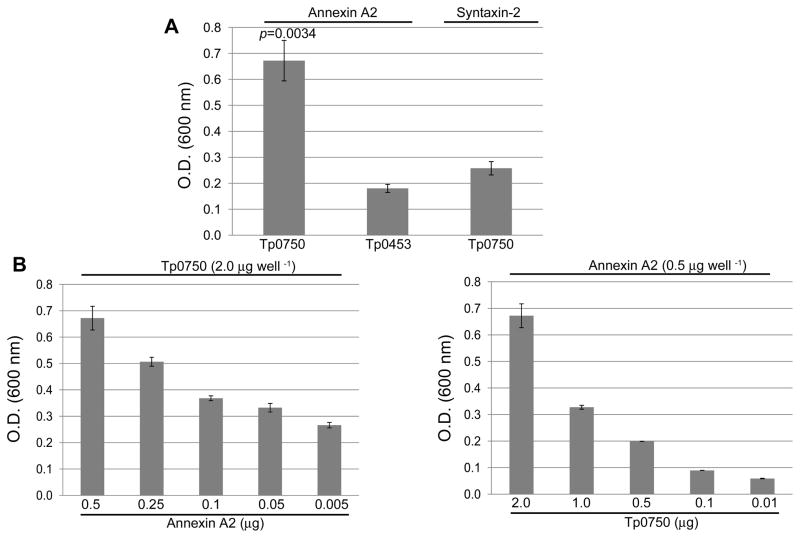
In order to confirm the specificity of the Tp0750-annexin A2 interaction, binding assays were performed using recombinant Tp0750 and annexin A2. Controls included testing the level of attachment of (1) Tp0750 to syntaxin-2, an endothelial membrane protein not identified in our proteomic studies and (2) a control recombinant protein Tp0453 to both annexin A2 and syntaxin-2. As shown in Fig. 3A, Tp0750 exhibited statistically significant level of binding to annexin A2 (p=0.0034) compared to the level of binding exhibited by the negative control protein to annexin A2. Tp0750 also exhibited statistically significant level of binding to annexin A2 (p=0.0072) compared to the level of binding exhibited by Tp0750 to syntaxin-2. Dose-dependent binding assays further validated the specificity of the Tp0750-annexin A2 interaction, with similar dose-dependent binding responses observed when increasing concentrations of immobilized annexin A2 (0.005–0.5 μg) were tested with a constant concentration of soluble Tp0750 (Fig. 3B, left panel) and a constant concentration of immobilized annexin A2 was tested with increasing concentrations of soluble Tp0750 (0.01–2.0 μg) (Fig. 3B, right panel).
Fig. 3. Tp0750 exhibits specific binding to annexin A2.
A. Binding assays were performed to compare the attachment of recombinant Tp0750 (2 μg) and a negative control protein (Tp0453; 2 μg) to immobilized annexin A2 (0.5 μg) and syntaxin-2 (0.5 μg). Tp0750 exhibited a statistically significant level of binding to annexin A2 (p≤0.0072) when compared to the binding of the negative control to annexin A2 and Tp0750 to syntaxin-2. B. Dose-dependent binding assays confirmed the specificity of the Tp0750-annexin A2 interaction. Similar dose-dependent binding responses were observed upon use of (i) varying concentrations of immobilized annexin A2 (0.005–0.5 μg per well) and a constant concentration of soluble Tp0750 (2 μg per well) (left panel), and (ii) a constant concentration of immobilized annexin A2 (0.5 μg per well) and varying concentrations of soluble Tp0750 (0.01–2.0 μg per well) (right panel). Average readings from triplicate measurements are presented with bars indicating standard error (SE), and the results are representative of three independent experiments. For statistical analyses, the Student’s two-tailed t test was used to compare the attachment of (i) Tp0750 to annexin A2 and the control host protein syntaxin-2 and (ii) Tp0750 and the control recombinant protein Tp0453 to annexin A2.
Tp0750 exhibits specific binding to human fibrinogen
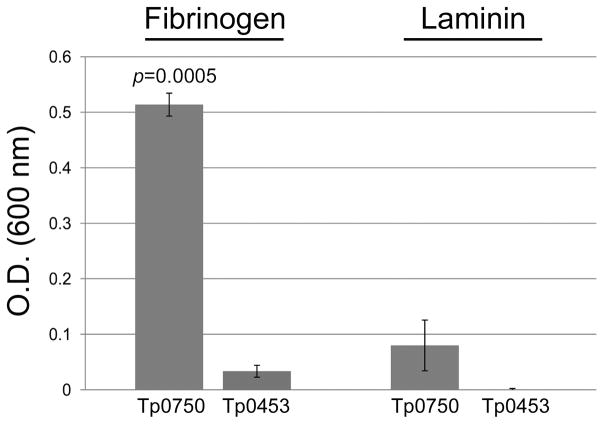
The observation that tp0751 (pallilysin) and tp0750 are co-transcribed, combined with our previous demonstration that pallilysin binds human fibrinogen (Houston et al., 2011), prompted us to investigate if Tp0750 also binds fibrinogen. Tp0750 exhibited statistically significant levels of binding to fibrinogen (p=0.0005) when compared to the level of binding by Tp0453, while exhibiting minimal levels of binding to the host component laminin (Fig. 4). This result suggests Tp0750 exhibits a narrower host component binding profile than pallilysin.
Fig. 4. Tp0750 binds human fibrinogen.
Attachment of Tp0750 and a negative control protein (Tp0453; 2 μg) to immobilized fibrinogen and laminin (0.5 μg) was investigated. Tp0750 exhibited statistically significant levels of binding to fibrinogen (p=0.0005) when compared to the level of binding by the negative control. Average readings of triplicate wells are presented with bars indicating standard error and the results are representative of three independent experiments. For statistical analyses, attachment to the host proteins by Tp0750 was compared to attachment by Tp0453 using the Student’s two-tailed t test.
Tp0750 exhibits fibrinogenolytic and fibronectinolytic activity that is inhibited by serine protease inhibitors
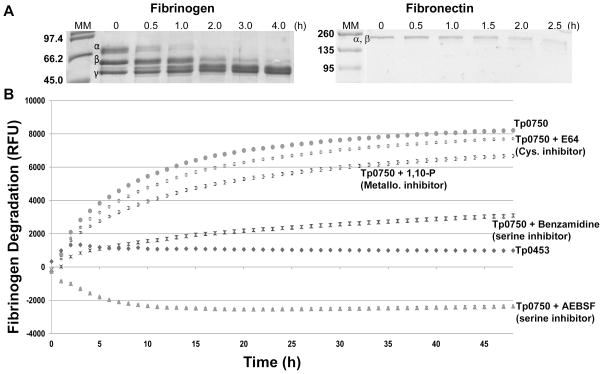
In vitro protein degradation assays were performed to determine if Tp0750 is capable of degrading the major structural protein present in clots, fibrinogen, and the clot-stabilizing protein, fibronectin. Purified recombinant proteins were incubated with plasminogen-free human fibrinogen or fibronectin for defined time intervals and degradation was analyzed by SDS-PAGE. Tp0750 initiated degradation of the fibrinogen α-chain within 0.5 h, with complete degradation of the fibrinogen α- and β-chains by 2 and 4 h post-incubation, respectively (Fig. 5A). Tp0750 also partially degraded the 220 kDa α- and β-chains of fibronectin (which migrate together in SDS-PAGE) within 2 h post-incubation (Fig. 5A). A negative control protein, Tp0453, failed to degrade either host protein and both fibrinogen and fibronectin were stable during the 4 and 2.5 h experiment durations, respectively (data not shown).
Fig. 5. Tp0750 mediates degradation of fibrinogen and fibronectin and is inhibited by serine protease inhibitors.
A. Protein degradation assays were performed to determine if Tp0750 is capable of degrading fibrinogen and fibronectin. Recombinant Tp0750_G23-A199 was incubated with human fibrinogen (20 μg Tp0750 and 100 μg fibrinogen) or fibronectin (2 μg Tp0750 and 30 μg fibronectin) for 4 h and 2.5 h, respectively. Samples (estimated to be 10 μg fibrinogen and 1.5 μg fibronectin in the absence of degradation) were removed at various time points and degradation of the fibrinogen α-, β-, and γ-chains (left panel) and co-migrating fibronectin α- and β-chains (right panel) was analyzed by SDS-PAGE. The negative control, Tp0453 (20 μg), failed to degrade fibrinogen or fibronectin (data not shown). Numbers to the left of the lanes indicate the size (kDa) of the corresponding molecular mass (MM) markers. B. Fluorescence-based degradation assays were performed to investigate the effect of protease inhibitors on the fibrinogenolytic activity of Tp0750. Recombinant proteins (Tp0453 and Tp0750_G23-A199) (1 μg) were pre-incubated for 1 h in the presence or absence of 1 mM 1,10 phenanthroline (1,10-P), 1 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 mM benzamidine, or 200 μM trans-epoxysuccinyl-L-leucylamido (4-guanidino) butane (E64) prior to incubation with FITC-labelled fibrinogen (10 μg) for 48 h. The degree of fibrinogen degradation was measured every hour by detecting the increase in relative fluorescence units (RFU) using standard fluorescein excitation/emission filters. The increase in RFU for the two recombinant proteins +/− inhibitors is shown. Average fluorescence intensity readings from triplicate measurements are presented with bars indicating standard error (SE) and the results are representative of three independent experiments.
To identify the catalytic type of the Tp0750 protease, fluorescence-based degradation assays were performed by incubating FITC-labelled fibrinogen with recombinant Tp0750 in the presence and absence of serine, cysteine, and metalloprotease inhibitors. Quenching occurs in heavily labelled FITC proteins due to the close proximity of the labels. However, upon digestion of the intact molecule by proteases, the FITC-labelled protein becomes dequenched, resulting in a measurable increase in fluorescence which is indicative of proteolysis. The extent of fibrinogen degradation was determined by measuring the level of fluorescence intensity over 48 h. As shown in Fig. 5B, Tp0750-mediated fibrinogenolysis was not inhibited by either a cysteine protease (E64, 200 μm) or metalloprotease (1,10 phenanthroline, 1 mM) inhibitor. However, incubation of Tp0750 with the reversible serine protease inhibitor, benzamidine (1 mM), significantly inhibited Tp0750-mediated fibrinogenolysis compared to the level observed in the absence of inhibitor (p≤0.0006 at 1 h post-incubation and beyond). Furthermore, incubation of Tp0750 with the irreversible serine protease inhibitor, AEBSF (1 mM), abolished fibrinogenolytic activity (Fig. 5B). A negative control recombinant protein, Tp0453, failed to degrade fibrinogen throughout the course of the experiment (Fig. 5B). Fibrin clot degradation assays showed Tp0750 could not degrade fibrin clots, in either the presence or absence of plasminogen (data not shown). These results demonstrate that Tp0750 is a serine protease that is capable of degrading the key host components involved in fibrin clot formation and stabilization but is incapable of degrading fibrin clots, either directly via Tp0750-mediated proteolysis or indirectly via plasminogen activation.
Tp0750 is capable of cleaving MMP substrate
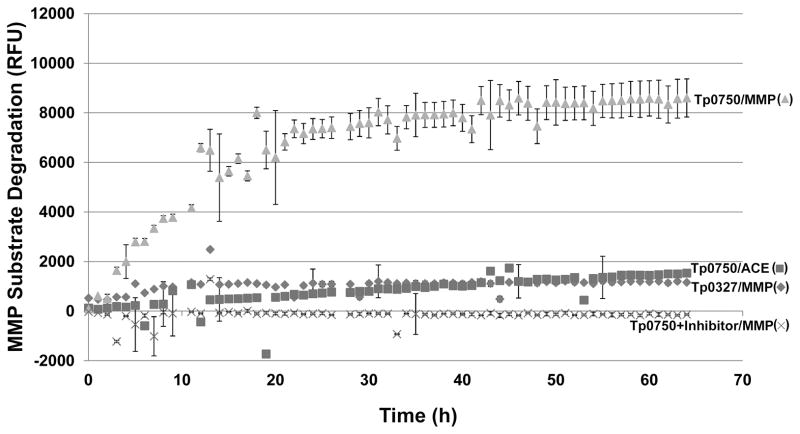
We investigated if Tp0750 exhibits MMP-like proteolytic activity by determining if Tp0750 can degrade a peptide substrate for MMPs 1–3, 7–9, and 13–14. Recombinant Tp0750 (in the presence and absence of the serine protease inhibitor AEBSF) and the positive control MMP-2 (400 ng each) were individually incubated with either the fluorescence-quenched peptide substrate (2.5 μg) or the negative control fluorescence-quenched angiotensin converting enzyme (ACE) peptide substrate. A negative control recombinant protein (Tp0327; 400 ng) was also incubated with the fluorescence-quenched peptide substrate (2.5 μg). Samples were gently mixed and MMP substrate cleavage was detected by measuring the increase in fluorescence intensities (excitation/emission=320 nm/420 nm) over 64 h. Tp0750 exhibited a statistically significant level of MMP substrate cleavage at 3 h post-incubation (p=0.003) and beyond (p=0.0008 at 64 h), compared to the absence of MMP substrate cleavage exhibited by the negative control (Fig. 6). Incubation of Tp0750 with its irreversible serine protease inhibitor, AEBSF, abolished Tp0750-mediated MMP substrate proteolysis (Fig. 6). As a positive control, MMP-2 exhibited rapid substrate cleavage, achieving maximum cleavage within 2 hours (data not shown). Tp0750 (Fig. 6) and MMP-2 (data not shown) failed to cleave the negative control peptide substrate. Proteases exhibit different preferences for substrate binding in the active site, which in turn dictate the proteases’ substrate cleavage rate. The synthetic peptide substrate used in the current study is composed of residues that are known to promote optimal MMP active site-substrate binding, and thus maximal proteolytic activity of the peptide substrate was observed with MMP-2. Although the peptide substrate was cleaved by Tp0750, the lower rate of proteolysis suggests the peptide sequence represents a suboptimal Tp0750 substrate.
Fig. 6. Tp0750-mediated MMP substrate degradation.
A fluorescence-based degradation assay was used to determine if recombinant Tp0750 is capable of cleaving a MMP substrate. A negative control protein (Tp0327; 400 ng) and Tp0750 (400 ng) +/− the serine protease inhibitor AEBSF were incubated with either the MMP substrate (MMP; 2.5 μg) or the negative control substrate (ACE; 2.5 μg) (Tp0750 only) for 0–64 h. MMP substrate cleavage was measured by detecting the increase in relative fluorescence units (RFU) using standard fluorescein excitation/emission filters (320 nm/420 nm). Average fluorescence intensity readings from triplicate measurements are presented with bars indicating standard error (SE) and the results are representative of three independent experiments.
Tp0750 interacts with pallilysin in vitro
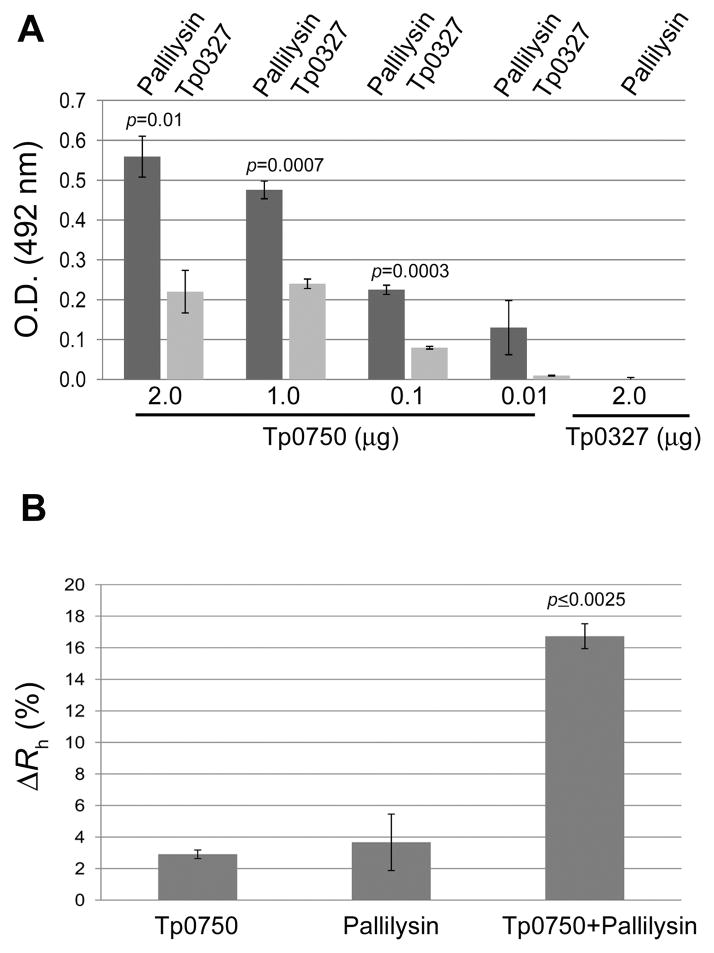
The presence of the vWFA protein-protein interaction domain within Tp0750, combined with the shared functionality of Tp0750 and pallilysin, prompted an investigation of the interaction capability of these two proteins using solid phase binding assays and dynamic light scattering. Dose-dependent binding showed pallilysin (2 μg) bound to increasing concentrations of immobilized Tp0750 in a statistically significant, dose-dependent manner (p<0.0001, Tp0750 concentration range of 0.01–2 μg) compared to minimal binding to an immobilized negative control recombinant protein (Tp0327; 2 μg). Furthermore, Tp0750 concentrations in the range of 0.1–2 μg exhibited statistically significant levels of binding to pallilysin (2 μg; p≤0.01) when compared to the corresponding levels of binding to immobilized Tp0750 by the negative control protein Tp0327 (2 μg) (Fig. 7A). The low levels of dose-dependent binding to Tp0750 exhibited by Tp0327 may be explained by the putative chaperone function that has been ascribed to this protein (Desrosiers et al., 2011).
Fig. 7. Interaction capability of Tp0750 and pallilysin.
A. Dose-dependent binding assays were performed to determine the ability of His-tagged pallilysin (Tp0751_HAXXH mutant) (2 μg per well) and a His-tagged negative control protein (Tp0327) (2 μg per well) to bind varying concentrations of immobilized tagless Tp0750 (0.01 μg-2 μg per well). Pallilysin exhibited statistically significant levels of binding to immobilized Tp0750 (0.1–2 μg per well; p<0.01) when compared to the level of binding of the negative control (Tp0327) to immobilized Tp0750 and pallilysin to the immobilized tagless control protein (Tp0327). Tp0750 and the negative control protein Tp0327 exhibited similar coating efficiencies (data not shown). Average readings from triplicate measurements are presented with bars indicating standard error (SE) and the results are representative of three independent experiments. For statistical analyses, attachment capabilities were compared using the Student’s two-tailed t test. B. DLS analyses were performed to further confirm the Tp0750-pallilysin interaction. Tp0750 and pallilysin (0.9 mg ml−1) were incubated alone and together at room temperature for 60 min, centrifuged for 20 min and analyzed by DLS after total incubation times of 80 and 180 min. Average readings from triplicate measurements are presented as the percent difference between the hydrodynamic radius measurements recorded at 80 and 180 min (ΔRh [%]) with bars indicating standard error. The results are representative of three independent experiments. The Tp0750-pallilysin mixed sample exhibited a statistically significant higher increase in ΔRh when compared to Tp0750 or pallilysin alone (Student’s two-tailed t test: p≤0.0025).
To complement our binding study we also used Dynamic Light Scattering (DLS), a label-free spectroscopic technique that accurately detects protein-protein interactions by determining the size distribution of molecules in free solution. This technique measures the amount of input laser light scattered by molecules in solution, which is related to their hydrodynamic radius (Rh), with molecules of large Rh moving slowly and scattering more light than quickly rotating molecules with small Rh (Hanlon et al., 2010). DLS measurements of a mixture of recombinant Tp0750_G23-D223 (26.0 kDa) and pallilysin (Tp0751_C24-P237; 25.8 kDa) incubated for 180 minutes revealed an increase in Rh of 16.7% [± 0.79%] to 4.10 nm (± 0.03 nm) corresponding to a molecular mass of 45.2 (± 0.7 kDa; Fig. 7B). Importantly, following the 180-minute incubation period, the average Rh value for Tp0750 and pallilysin measured independently corresponded to a molecular weight of 24.9 kDa (± 0.7 kDa), with the two independent proteins maintaining Rh differences of less than 4% (Tp0750, 2.9% [± 0.27%]; pallilysin, 3.7% [± 1.79%]; Fig. 7B). These data are consistent with the formation of a Tp0750-pallilysin binary complex.
Discussion
Genomic and functional linkage of Tp0750 and pallilysin
In previous reports we established that the functional activity of the treponemal metalloprotease pallilysin is consistent with a role in T. pallidum dissemination, namely as a host component-specific adhesin/protease (Cameron, 2003; Cameron et al., 2005; Houston et al., 2011; Houston et al., 2012). The current study demonstrates the co-transcription of tp0750 and pallilysin, suggesting a potential functional linkage of these two treponemal proteins. Indeed, we have confirmed that both Tp0750 and pallilysin possess dual adhesive/protease activities and target host barriers encountered by T. pallidum during the course of infection, including the milieus of laminin-rich basement membranes and infection-limiting fibrin clots.
Significance of Tp0750-pallilysin heterodimeric complex formation
In the present study, we demonstrate that Tp0750 and pallilysin are capable of interacting in a dose-dependent manner, and results generated using the free-solution, label-free biophysical characterization technique DLS are consistent with Tp0750 and pallilysin forming a heterodimeric complex. Precedence exists for co-transcribed genes encoding proteins that form heteromers. In Mycobacterium tuberculosis members of the PE/PPE protein family are encoded by co-transcribed genes, exhibit ECM component-binding, and form hetero-oligomeric complexes on the bacterial surface (Espitia et al., 1999; Tundup et al., 2006). For these proteins, complex formation has been shown to be crucial for inducing proper protein folding, maintaining protein stability, and translocating the proteins to the bacterial surface (Tundup et al., 2006; Strong et al., 2006; Abdallah et al., 2006; Daleke et al., 2012).
With regards to T. pallidum, our previous investigations showed that pallilysin-specific antiserum promotes opsonophagocytosis of viable T. pallidum in the presence of rabbit macrophages, a result consistent with exposure of pallilysin on the surface of intact treponemes (Houston et al., 2012). Combined with our binding/DLS studies, this suggests the intriguing possibility that Tp0750 and pallilysin exist in a heterodimeric complex on the treponemal surface. However, similar phagocytosis assays conducted using Tp0750-sepcific antiserum did not demonstrate significant levels of opsonophagocytosis (S.A. Lukehart and B.J. Molini, University of Washington, unpublished observations). Interestingly, the protein-specific sera were generated against renatured pallilysin versus non-renatured insoluble Tp0750 (P.A. Cullen and C.E. Cameron, unpublished observations), suggesting the pallilysin protein preparation may have been more adept at generating antibodies specific for conformational epitopes that are likely to be key to the process of opsonophagocytosis. Due to the well-known labile nature of the T. pallidum outer membrane and the technical challenges associated with identification of its constituent proteins (Cameron, 2006), direct demonstration of the presence of co-associated Tp0750-pallilysin on the treponemal surface is not easily achievable and confirmation of the existence of such a heteroduplex must await future studies.
Significance of the observed multifunctional nature of Tp0750
(i) Coagulation cascade inhibition via Tp0750 serine protease activity
In vitro studies have demonstrated that virulent T. pallidum activates human vascular endothelial cells, resulting in surface TF expression/pro-coagulant activity and production of extensive fibrin networks on the endothelium surface (Riley et al., 1992; Riley et al., 1994; Lee et al., 2000). Heat-killed T. pallidum and T. phagedenis (which is non-invasive) failed to induce these host changes, suggesting the observed coagulation cascade activation is a specific host response to T. pallidum that may function to contain infection (Riley et al., 1992; Riley et al., 1994). In the present study, we have shown Tp0750 exhibits fibrinogenolytic and MMP-like proteolytic activity that is abolished by AEBSF, an irreversible inhibitor of serine proteases. We also demonstrate that Tp0750 has fibronectinolytic activity. During the final steps of blood coagulation, the structure of the fibrin clot is stabilized by incorporation of fibronectin (Pankov and Yamada, 2002; Midwood et al., 2004). Furthermore, during clot formation fibronectin promotes platelet adhesion via interaction of the arginine-glycine-aspartic acid (RGD) cell binding motif present within fibronectin with the major platelet integrin, αIIbβ3, and is essential for stabilization of platelet aggregates (Gardner and Hynes, 1985; Thurlow et al., 1990; Cho and Mosher, 2006). Following endothelial cell traversal and activation, the capacity of Tp0750 to degrade both fibrinogen and fibronectin in the vicinity of T. pallidum could eliminate key clot constituents, which may in turn inhibit local clot formation and stabilization and aid in treponemal spread.
(ii) Fibrinolysis facilitation through Tp0750 interaction with annexin A2
Using protein structure modeling we demonstrated that Tp0750 contains a predicted MIDAS-containing vWFA domain. This domain is the prototype for a protein superfamily involved in protein-protein interactions and is commonly found in plasma, cell adhesion, and extracellular matrix proteins (Whittaker and Hynes, 2002). Surface-exposed MIDAS motifs, which are present in the majority of vWFA-containing proteins, use divalent metal cation coordination to mediate and stabilize protein-protein interactions (Lee et al., 1995; Whittaker and Hynes, 2002; Craig et al., 2004). vWFA domains frequently coordinate calcium via their MIDAS motif (Whittaker and Hynes, 2002; Loewen and Forsyth, 2005; Dolphin, 2012), which is in agreement with our finding that calcium associates with Tp0750.
To identify Tp0750-interacting host proteins we used a proteomics approach to isolate Tp0750-interacting HUVEC membrane-associated proteins. This identified the endothelial protein annexin A2 as a Tp0750 binding partner. One peptide from S100A10 was also identified; previous studies report the attainment of low peptide coverage (10%) upon mass spectrometry analysis of S100A10 (Suzuki et al., 2011), suggesting this protein may be inherently difficult to analyze by mass spectrometry methodologies and possibly explaining why a single peptide was observed for this protein. Regardless of whether S100A10 serves as an additional protein interaction partner for Tp0750 or was co-isolated with annexin A2, the isolation of both annexin A2 and S100A10 is of paramount interest. Annexin A2 and S100A10 form a heterotetramer on the endothelial cell membrane (Bharadwaj et al., 2013), and in the absence of S100A10 annexin A2 is not translocated to the endothelial cell surface. Further, binding of annexin A2 stabilizes S100A10 by masking a polyubiquitination site on S100A10; in the absence of annexin A2, S100A10 is polyubiquitinated and targeted to the proteosome for degradation (Hedhli et al., 2012). Thus, the fact that we detected both annexin A2 and S100A10 in our proteomic analyses indicates interaction of Tp0750 with endothelial membrane-associated annexin A2.
During the process of vascular fibrinolysis, the cell surface-associated annexin A2-S100A10 fibrinolytic receptor complex binds both tPA and plasminogen and induces rapid plasmin generation via the tPA-mediated conversion of plasminogen to plasmin (Madureira et al., 2011; Flood and Hajjar, 2011). If our in vitro findings mirror what occurs during an in vivo T. pallidum infection, the Tp0750-annexin A2 interaction could have functional significance in three inter-related ways. First, T. pallidum has long been known to associate with perivascular areas, a localization that may be facilitated by the Tp0750-annexin A2 interaction. Second, the Tp0750-annexin A2 interaction would allow T. pallidum to exploit, through proximal location, the highly efficient host fibrinolysis system which would promote treponemal spread through clot degradation. And third, previous studies have shown that virulent T. pallidum, but not heat-killed T. pallidum or non-pathogenic treponemes, can invade the intercellular tight junctions of endothelial cells (Thomas et al., 1988; Riley et al., 1992). Since annexin A2 co-localizes with tight junction markers along the lateral plasma membrane of endothelial cells, it has been proposed to play a role in remodeling of tight junctions (Lee et al., 2004). This raises the intriguing possibility that the annexin A2-Tp0750 interaction may also position the bacteria at the site of endothelial cell tight junction remodelling and in this way assist T. pallidum entry into, and exit out of, the bloodstream.
(iii) Tp0750 MMP-like proteolysis
In the current study we show that Tp0750 is capable of cleaving a peptide substrate targeted by MMPs-1, -2, -3, -7, -8, -9, -13, and -14, suggesting that Tp0750 has MMP-like activity. Studies have shown that MMPs -2, -3, -7, -9, and -13 degrade the abundant basement membrane protein collagen IV (Fessler et al., 1984; Nguyen et al., 1993; Knauper et al., 1997; Sternlicht et al., 1999; Monaco et al., 2006), and that MMPs -3, -7, and -14 cleave laminin and fibronectin (Imai et al., 1995; Ohuchi et al., 1997; Sternlicht et al., 1999), major components of basement membranes and blood clots, respectively. Degradation of fibrinogen by the concerted action of MMPs -8, -12, -13, and -14 results in significantly impaired clotting (Hiller et al., 2000). Furthermore, MMPs -2 and -9 degrade the tight junction proteins occludin, ZO-1, and claudin-5, which causes blood-brain barrier (BBB) disruption (Feng et al., 2011; Liu et al., 2012). The observed MMP-like activity of Tp0750 suggests this treponemal protein may be able to degrade a broad range of host components that serve as critical barriers to T. pallidum dissemination.
Potential model of the contribution of Tp0750 and pallilysin to T. pallidum invasion and dissemination
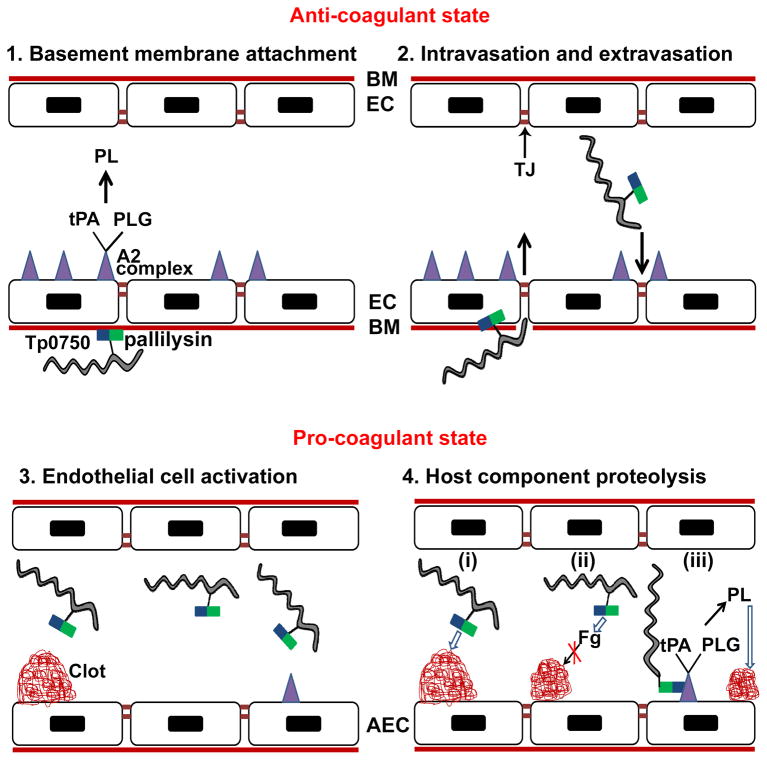
In light of the findings of the current and previous studies (Cameron, 2003; Houston et al., 2011; Houston et al., 2012), we propose a model to summarize the possible roles of Tp0750 and its protein partner pallilysin in T. pallidum invasion and dissemination (Fig. 8). In this model, Tp0750 and pallilysin associate on the treponemal surface, with membrane anchorage occurring through lipidation of pallilysin. Pallilysin mediates attachment to, and degradation of, the laminin-rich basement membrane underlying vascular endothelial cells, thereby promoting treponemal invasion into the bloodstream (Fig. 8 panels 1–2). Endothelial cell activation occurs, resulting in decreased expression of anti-coagulant factors including the annexin A2-associated fibrinolytic complex. Conversely, fibrin clot formation is up-regulated to contain the infection and curtail pathogen spread, thereby shifting the endothelial haemostatic balance from an anti- to a pro-coagulant state (Fig. 8 panel 3). Degradation of fibrinogen, fibrin clots, and the clot-stabilizer fibronectin by the concerted action of Tp0750 and pallilysin results in degradation of existing clots and a decrease in nascent clot formation (Fig. 8 panel 4). The Tp0750-annexin A2 interaction on the endothelial membrane is proposed to enhance the process of clot dissolution in the immediate vicinity of T. pallidum and localize T. pallidum to areas of tight junction remodelling, thus enabling extravasation and tissue invasion (Fig. 8 panels 2 and 4). Overall our model suggests T. pallidum is capable of interacting with key components of the host coagulation/fibrinolytic systems via the coordinated action of the physically- and functionally-linked Tp0750/pallilysin proteins, a pathogenic mechanism we predict is central to treponemal dissemination.
Fig. 8. Proposed model of the role of Tp0750/pallilysin in T. pallidum dissemination via the bloodstream.
(1) Surface-exposed pallilysin (
 ) mediates attachment of Treponema pallidum (
) mediates attachment of Treponema pallidum (
 ) to the laminin-rich basement membrane (BM) that underlies the endothelial cells (EC) of blood vessels. The unperturbed endothelium exists primarily in an anti-coagulant state due to the expression of profibrinolytic factors including the fibrinolytic complex comprising annexin A2 and S100A10 (
) to the laminin-rich basement membrane (BM) that underlies the endothelial cells (EC) of blood vessels. The unperturbed endothelium exists primarily in an anti-coagulant state due to the expression of profibrinolytic factors including the fibrinolytic complex comprising annexin A2 and S100A10 (
 ) which binds tissue plasminogen activator (tPA) and plasminogen (PLG) resulting in plasmin (PL)-mediated clot dissolution; (2) T. pallidum intravasation and extravasation: pallilysin degrades the basement membrane. Tp0750 (
) which binds tissue plasminogen activator (tPA) and plasminogen (PLG) resulting in plasmin (PL)-mediated clot dissolution; (2) T. pallidum intravasation and extravasation: pallilysin degrades the basement membrane. Tp0750 (
 ), through interaction with annexin A2, localizes T. pallidum to areas of tight junction (TJ) remodeling; (3) Activation of endothelial cells occurs upon infection with T. pallidum leading to the formation of a pro-coagulant endothelial surface. On the surface of activated endothelial cells (AEC), the coagulation cascade is initiated resulting in the generation of vascular clots composed of fibrin, fibronectin, and platelets; (4) Tp0750/pallilysin-mediated host component proteolysis: (i) Direct clot degradation via pallilysin-mediated fibrinolysis and Tp0750-mediated fibronectinolysis, (ii) clot formation inhibition via Tp0750/pallilysin-mediated fibrinogen (Fg) degradation, and (iii) Tp0750-annexin A2 interaction localizes T. pallidum to the immediate vicinity of plasmin-mediated vascular fibrinolysis. Open arrows indicate direct proteolysis of host components. Closed arrows indicate an intermediate pro-protease activation step prior to direct proteolysis of host components.
), through interaction with annexin A2, localizes T. pallidum to areas of tight junction (TJ) remodeling; (3) Activation of endothelial cells occurs upon infection with T. pallidum leading to the formation of a pro-coagulant endothelial surface. On the surface of activated endothelial cells (AEC), the coagulation cascade is initiated resulting in the generation of vascular clots composed of fibrin, fibronectin, and platelets; (4) Tp0750/pallilysin-mediated host component proteolysis: (i) Direct clot degradation via pallilysin-mediated fibrinolysis and Tp0750-mediated fibronectinolysis, (ii) clot formation inhibition via Tp0750/pallilysin-mediated fibrinogen (Fg) degradation, and (iii) Tp0750-annexin A2 interaction localizes T. pallidum to the immediate vicinity of plasmin-mediated vascular fibrinolysis. Open arrows indicate direct proteolysis of host components. Closed arrows indicate an intermediate pro-protease activation step prior to direct proteolysis of host components.
Experimental procedures
Ethics statement
All animal studies were approved by the local institutional review boards at the University of Victoria and the University of Washington, and were conducted in strict accordance with standard accepted principles as set forth by the Canadian Council on Animal Care (CCAC), National Institutes of Health (NIH) and the United States Department of Agriculture (USDA) in a facility accredited by the American Association for the Accreditation of Laboratory Animal Care.
Bacteria
T. pallidum subsp. pallidum (Nichols strain) was propagated in, and extracted from, New Zealand White rabbits as described elsewhere (Lukehart and Marra, 2007).
Host proteins
Plasminogen-depleted human fibrinogen was purchased from VWR International (Mississauga, ON). Fibronectin isolated from human plasma and laminin isolated from Engelbreth-Holm-Swarm murine sarcoma were purchased from Sigma Aldrich (Oakville, ON). Recombinant human annexin A2 (S2-D339) and recombinant human syntaxin-2 (M1-R188) were purchased from Cedarlane Labs (Burlington, ON).
RNA extraction and RT-PCR
RNA was isolated from T. pallidum subsp. pallidum (Nichols strain) as described elsewhere (Witchell et al., 2006). The orientation-specific RT-PCR sense and antisense primers for tp0750 and tp0751 (pallilysin) are indicated in Table 2. RT-PCR reactions were performed as previously described (Boucher et al., 2005), with reverse transcription performed at 42°C for 2 h, and denaturation at 70°C for 15 min. Negative control RT-PCR reactions did not include reverse transcriptase. PCR products were electrophoresed on agarose gels and visualized with ethidium bromide staining.
Table 2.
Primers used in RT-PCR and recombinant protein expression.
| ORF1 | Primer Orientation | Sequence2 |
|---|---|---|
| Tp0750 (RT-PCR) | Sense | 5′-TCAAAGTACCGTGGCAAACA |
| Tp0751 (RT-PCR) | Antisense | 5′-CCTCGTGGGATTCAAATGTT |
| Tp0750_G23-A199 (pET28a) | Sense | 5′-CTAGACCATATGGGTGAACGCACTGTCC |
| Tp0750_G23-A199 (pET28a) | Antisense | 5′-GTCAGCTCGAGTCAAGCTGCGATGCTGCGATAC |
| Tp0750_G23-A199 (pET32a) | Sense | 5′-CTACTACCATGGCTGGTGAACGCACTGTCC |
| Tp0750_G23-A199 (pET32a) | Antisense | 5′-CTACTACTCGAGTTAAGCTGCGATGCTGCGATAC |
| Tp0750_G23-D223 (pDEST17) | Sense | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGCGAACGCACTGTCCCCG |
| Tp0750_G23-D223 (pDEST17) | Antisense | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAGTCAAGCGAAGGGG |
| Tp0750_G23-D223 (pAcGP67B) | Sense | 5′-CTACTACCATGGCTGGTGAACGCACTGTCCCC |
| Tp0750_G23-D223 (pAcGP67B) | Antisense | 5′-CTACTAGCGGCCGCGTCAAGCGAAGGGGTATAGCG |
| Tp0327_I23-S172 (pET28a) | Sense | 5′-CATGACCATATGATCACGCGCTTTGCCGTC |
| Tp0327_I23-S172 (pET28a) | Antisense | 5′-GTCAGCTCGAGTCACGAGCTGCTCAGCTC |
Shown in brackets is either the methodology that the primers were used for [RT-PCR] or the expression vector used for recombinant expression of each protein.
Restriction sites are highlighted in bold.
Molecular modeling and homology analyses
The potential two-gene operon comprising tp0750 and tp0751 (pallilysin) was analysed for features synonymous with prokaryotic operon arrangement using Artemis Genome Browser (http://www.sanger.ac.uk/resources/software/artemis/). The Tp0750 amino acid sequence was analysed using SignalP (Version 4.1) (http://www.cbs.dtu.dk/services/SignalP/), which predicts the presence and location of signal peptide cleavage sites in Gram-negative protein sequences. Amino acid sequence homology analysis of Tp0750 was performed using the National Center for Biotechnology Information (NCBI) conserved domain database search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). For tertiary modelling of Tp0750, the program Modeller was used with a manually edited sequence alignment based off the conserved domain assignment from NCBI using integrin α2β1 (PDB 1DZI) as the template.
Construct cloning
DNA fragments encoding G23-A199 and G23-D223 of Tp0750 and I23-S172 of Tp0327 were PCR amplified from T. pallidum subsp. pallidum (Nichols strain) genomic DNA using the primers listed in Table 2. All cloning procedures were conducted according to the manufacturer’s instructions for the particular destination vector. The Tp0750 (G23-A199) and Tp0327 (I23-S172) amplicons were cloned into the NdeI/Xhol sites of the E. coli expression vectors pET28a (Tp0750 and Tp0327) and pET32a (Tp0750) (VWR International, Mississauga, ON). The Tp0750 amplicon comprising residues G23-D223 was cloned into the T7 expression vector pDEST-17 (Invitrogen, Carlsbad, CA). Constructs encoding wild-type pallilysin (C24-P237), the active site mutant pallilysin_HAXXH (C24-P237) and Tp0453 (A32-S287) were generated as previously described (Houston et al., 2011; Houston et al., 2012). For insect recombinant protein expression, a DNA fragment encoding G23-D223 of Tp0750 was PCR amplified using the primers listed in Table 2 and cloned into the NotI/NcoI sites of a pAcGP67B baculovirus transfer vector (BD Biosciences, Mississauga, ON) modified to incorporate a C-terminal hexahistidine tag and thrombin cleavage site. The sequence and reading frames of all constructs were verified by DNA sequencing.
Recombinant protein expression and purification
pET constructs were transformed into the E. coli expression strain BL21 Star (DE3) (Invitrogen). Transformants were cultured at 37°C in LB media containing 50 μg ml−1 kanamycin (pET28a) or 100 μg ml−1 ampicillin (pET32a) until the A600 reached between 0.8 and 1.0. Cultures were induced with 0.4 mM IPTG for 16 h at 16–23°C and harvested by centrifugation at 4°C. Cell pellets were resuspended in buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole, 1% glycerol), flash frozen in liquid nitrogen and stored at −80°C. Recombinant His-tagged proteins were purified using immobilized metal ion affinity chromatography followed by size exclusion chromatography, as previously described (Houston et al., 2011). The N-terminal thioredoxin (Trx)/Histidine (His) tag of pET32a-expressed Tp0750 was removed by cleavage with enterokinase (New England Biolabs, Whitby, ON), and enterokinase was immediately removed following cleavage using anti-enterokinase-conjugated agarose beads (Sigma Aldrich). Following enterokinase removal, the Trx/His tag was removed from Tp0750 by incubation with Nickel-NTA agarose beads (Qiagen, Toronto, ON) in closed poly-prep chromatography columns (BioRad, Mississauga, ON) for 16 h at 4°C. Purified tagless Tp0750 was eluted in 20 ml buffer (20 mM HEPES pH 7.5, 150 mM NaCl, and 1% glycerol), concentrated and stored at −80°C as previously described (Houston et al., 2011). The pDEST17 constructs (wild-type pallilysin, active site mutant pallilysin [HAXXH], and Tp0750_G23-D223) and insect recombinant Tp0750 were expressed and purified as previously described (Crawford et al., 2010; Houston et al., 2011; Houston et al., 2012). Mass spectrometry, SDS-PAGE analyses, and western blotting were performed to confirm that Tp0750 proteins were purified to homogeneity (Fig. S1).
Inductively coupled plasma-mass spectrometry
Tp0750_G23-A199 and the corresponding buffer used for size exclusion chromatography (20mM HEPES pH 7.5, 150 mM NaCl, 1% glycerol) were analyzed for divalent metal ions at Cantest Ltd. (Burnaby, BC) using Inductively Coupled Plasma-Mass Spectrometry (ICP-MS), as previously described (Houston et al., 2011).
HUVEC tissue culture and membrane-associated protein purification
HUVECs were purchased from ATCC (American Type Culture Collection, Manassas, VA) and cultured according to the manufacturer’s instructions. HUVEC membrane-associated proteins were extracted using the Thermo Scientific Mem-PER eukaryotic membrane protein extraction kit (Fisher Scientific, Ottawa, ON) according to the manufacturer’s instructions, with the following modifications: Thermo Scientific halt protease inhibitor cocktail was added immediately after the addition of the Mem-PER cell lysis detergent (reagent A), samples were centrifuged at 10,000 × g for 5 min at 4°C after adding the Mem-PER reagents B and C (membrane protein solubilisation reagents), and the membrane protein fraction was dialyzed 3 times at 4°C against 150-fold volumes of PBS/0.05% CHAPS buffer.
Tp0750-HUVEC membrane-associated protein pull-down assay
The ProFound Pull-Down PolyHis Protein:Protein Interaction kit (Fisher Scientific) was used to conduct the Tp0750-HUVEC pull-down assay according to the manufacturer’s instructions, with the following modifications. The ‘bait’ protein, recombinant hexahistidine-tagged Tp0750_G23-A199, was dialysed against PBS/0.05% CHAPS buffer at 4°C, followed by addition of CHAPS to a final concentration of 0.5%. During equilibration of the immobilized cobalt chelate resin, 20 μl of 4M imidazole and CHAPS (final concentration 0.5%) was added to 8 ml of ProFound wash solution for each column/sample. For immobilization of the bait protein, 150 μg of Tp0750 was diluted in PBS wash buffer containing 0.5% CHAPS, and immobilized Tp0750 was washed 3 times with PBS/0.5% CHAPS. The mammalian ‘prey’ sample, comprising the sample of enriched HUVEC membrane-associated proteins, was dialysed against PBS/0.05% CHAPS buffer at 4°C, followed by addition of CHAPS and imidazole to final concentrations of 0.5% and 40 mM, respectively. During the prey protein capture steps, 375 μl of enriched HUVEC membrane-associated proteins were added to a Tp0750-conjugated cobalt chelate resin column (‘sample’) and in parallel to an unconjugated cobalt chelate affinity column (prey only control). In addition, wash buffer (PBS/0.5% CHAPS) alone was added to a Tp0750-conjugated cobalt chelate resin column (bait only control). Columns were incubated at 4°C for 16 h to allow for maximal binding and then washed 3 times with wash buffer. Bait-prey elution steps were performed according to manufacturer’s instructions.
Mass spectrometry sample preparation
To prepare Tp0750-HUVEC eluates for mass spectrometry, samples were acetone precipitated at −20°C for 16 h, centrifuged for 15 min at 16,000 × g at 4°C, and excess acetone removed. Proteins were separated by SDS-PAGE using 15% tris-glycine polyacrylamide gels, stained for 16 h with colloidal Coomassie G250 stain, and destained in sterile distilled water. Protein bands were excised from the gels, de-stained in 1M ammonium bicarbonate/20% acetonitrile solution followed by 50% methanol/5% acetic acid, reduced using 10mM dithiothreitol, alkylated in 100mM iodoacetamide, and digested with trypsin (20 ng μl−1 in 50mM ammonium bicarbonate) for 16 h. Peptides were extracted in successive washes of 50% acetonitrile, 10% formic acid, and 50mM ammonium bicarbonate.
Liquid chromatography-electrospray ionization tandem mass spectrometry
Protein samples were subjected to LC-MS/MS analysis using an integrated Famos autosampler, Switchos II switching pump, and UltiMate micro pump (LC Packings, Amsterdam, Netherlands) system with a QStar Pulser i Hybrid Quadrupole-TOF LC/MS/MS Mass Spectrometer (Applied Biosystems, Carlsbad, CA) equipped with a nanoelectrospray ionization source (Proxeon, Odense, Denmark) and fitted with a 10μm fused-silica emitter tip (New Objective, Woburn, MA). LC-MS/MS set-up, chromatographic separation, and data acquisition were performed as previously described (Eshghi et al., 2009).
LC-MS/MS data analysis
Analyst QS 1.1 software (ABI Sciex LP, Concord, ON) was used to view the information dependent acquisition (IDA) file and Mascot script 1.6b23 (Matrix Science Limited, London, UK) was used to generate peak lists using the following parameters: ion charge states of +2 to +4 were used; spectra were discarded if they contained less than 10 peaks. Peak lists were then sent to a local Mascot search engine V 2.0 (Matrix Science Limited). The generated Mascot generic format files were searched against the Uniprot-SwissProt ‘all species’, ‘homo sapiens’, and ‘Eubacteria’ databases and the software used to search the MS data against the databases was PEAKS client and PEAKS online (Bioinformatics Solutions Inc., Waterloo, ON). PEAKS proteomics mass spectrometry software analyzes raw mass spectrometry data and performs validated peptide and protein identification in the form of PEAKS scores (0–100% protein/peptide identity confidence) and indicates the number and location of peptides within a protein that were identified following mass spectrometry. The following PEAKS search parameters were used; enzyme specificity = trypsin; maximum missed cleavages = 2; precursor mass = monoisotopic; merge tolerance ± 0.0 Da; parent tolerance ± 0.5 Da; fragment tolerance 0.3 Da; maximum variable PTM = 3; variable modifications = carbamidomethyl (C), deamidated (NQ), and oxidation (M).
Binding assays
To test for binding of recombinant Tp0750 (G23-A199 and G23-D223 constructs) and a negative control (Tp0453) to the host proteins annexin A2, syntaxin 2, fibrinogen, and laminin, and to test for binding of recombinant Tp0750 and a negative control (Tp0327) to pallilysin, binding assays were performed as previously described (Cameron, 2003). Investigation of the coating efficiency of Tp0750 and Tp0327 was done by coating wells with 2 μg of each protein and detecting the level of bound protein with Ni-HRP, as previously described (Cameron, 2003). In all assays, the background binding level of proteins to wells coated with buffer alone was subtracted from the absorbance values. Plates were read at 600 nm or 492 nm with a BioTek ELISA plate reader (Fisher Scientific). All statistical analyses were performed using the Student’s two-tailed t test.
Host protein degradation and protease inhibition assays
SDS-PAGE-based in vitro host protein degradation assays were performed as previously described (Houston et al., 2011) with the following modifications. To assess Tp0750 fibrinogenolytic activity, E. coli-expressed recombinant tagless Tp0750_G23-A199 (20 μg) and a negative control (Tp0453; 20 μg) were independently incubated with 100 μg of plasminogen-free human fibrinogen in protease activation buffer (20 mM HEPES pH 7.5, 25 mM CaCl2). To assess Tp0750 fibronectinolytic activity, tagless Tp0750_G23-A199 (2 μg) and Tp0453 (2 μg) were independently incubated with 30 μg of fibronectin in the same protease activation buffer. The reactions were incubated at 37°C for 4 h or 2.5 h, respectively, samples were removed at various time points, and analysed for degradation of the three fibrinogen chains (α, β, and γ) or two fibronectin chains (α and β) by SDS-PAGE and Coomassie Brilliant Blue staining. A fluorescence- based degradation assay was performed to investigate the effect of broad range protease inhibitors on the fibrinogenolytic activity of Tp0750, using methodology as previously described (Houston et al., 2011) with the following modifications. Recombinant proteins (Tp0453 and Tp0750_G23-A199) were diluted in protease activation buffer (20 mM HEPES pH 7.5, 25 mM CaCl2) and added in triplicate to sterile Corning Costar 96-well black plates (Fisher Scientific). Recombinant proteins were pre-incubated at 37°C for 1 h in the presence and absence of 1 mM 1,10 phenanthroline, 1 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 mM benzamidine, or 200 μM trans-epoxysuccinyl-L-leucylamido (4-guanidino) butane (E64).
Generic MMP peptide substrate degradation
Pro-MMP-2 (AnaSpec Inc., San Jose, CA) was activated by incubation with 1 mM APMA (p-aminophenylmercuric acetate) (AnaSpec Inc.) at 37°C for 45 min. The positive control MMP-2 (400 ng), a negative control recombinant protein (Tp0327; 400 ng), and tagless Tp0750_G23-A199 (400 ng) in the presence/absence of 10 mM AEBSF, were added in triplicate to sterile Corning Costar 96-well black plates (Fisher Scientific). The MMP substrate (Mca - Pro - Leu - Gly - Leu – Dap[Dnp] - Ala - Arg - NH2) (2.5 μg)or angiotensin-converting enzyme-2 (ACE-2) substrate (Mca - Ala - Pro – Lys[Dnp] – OH) (2.5 μg) (AnaSpec Inc.) were then added to the wells, gently mixed, and incubated at 37°C in the dark for 0–64 h. Fluorescence-based degradation assays were performed as previously described (Houston et al., 2011) using a standard fluorescein excitation/emission filter set at 320 nm/420 nm. Mean blank fluorescence readings (fluorescence from wells containing assay buffer and substrate) were subtracted from the mean sample fluorescence readings and the increase in relative fluorescence units (RFU) was plotted against time. Statistical analyses were performed using the Student’s two-tailed t test.
Dynamic Light Scattering (DLS)
Tp0750_G23-D223 (0.9 mg ml−1) in 20 mM HEPES pH 7.5, 150 mM NaCl, and 1% glycerol and Tp0751_C24-P237 (0.9 mg ml−1) in the same buffer at pH 7.0 were incubated alone and together at room temperature for 60 min. Samples were centrifuged at 16,000 × g for 20 min at 8°C and 25 μl of each sample was added to Corning 384-well optical bottom plates (Fisher Scientific). Initial measurements were performed after a total incubation time of 80 min using a DynaPro plate reader (Wyatt Technology, Santa Barbara, CA) with the following parameters: temperature stabilized at 25°C, wavelength of 834 nm, automatically attenuated laser power, solvent refractive index of 1.33 and solvent viscosity of 0.91 to account for 1% glycerol in the buffers, and 10 acquisitions of 5 sec each. The samples were then incubated in the assay plate at 16°C with shaking at 300 rpm for 100 min using an Eppendorf Thermomixer, following which a second set of measurements was performed after a total incubation time of 180 min using the same parameters. All data were collected and analyzed using the Dynamics V7.1 software and an elongated polymer model for molecular weight determination. The analyzed data are an average of triplicate measurements, and presented as a percent difference between the hydrodynamic radius measurements recorded at 80 and 180 min.
Supplementary Material
Acknowledgments
We thank Azad Eshghi for his knowledge and technical assistance regarding mass spectrometry and Kellie Brown for assistance with protein production. This work was funded by Public Health Service Grant AI-051334 from the National Institutes of Health, as well as by awards from the Canada Foundation for Innovation, the Michael Smith Foundation for Health Research, and the British Columbia Knowledge Development Fund. Support for this research was also provided in part by the BC Proteomics Network. C.E.C. is a Canada Research Chair in Molecular Pathogenesis and a Michael Smith Foundation for Health Research Scholar.
References
- Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, Eisenberg D, et al. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol. 2006;62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- Ait-Oufella H, Maury E, Lehoux S, Guidet B, Offenstadt G. The endothelium: physiological functions and role in microcirculatory failure during severe sepsis. Intensive Care Med. 2010;36:1286–1298. doi: 10.1007/s00134-010-1893-6. [DOI] [PubMed] [Google Scholar]
- Bharadwaj A, Bydoun M, Holloway R, Waisman D. Annexin A2 heterotetramer: structure and function. Int J Mol Sci. 2013;14:6259–6305. doi: 10.3390/ijms14036259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher DJ, Adler B, Boyce JD. The Pasteurella multocida nrfE gene is upregulated during infection and is essential for nitrite reduction but not for virulence. J Bacteriol. 2005;187:2278–2285. doi: 10.1128/JB.187.7.2278-2285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE. Identification of a Treponema pallidum laminin-binding protein. Infect Immun. 2003;71:2525–2533. doi: 10.1128/IAI.71.5.2525-2533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Brouwer NL, Tisch LM, Kuroiwa JMY. Defining the interaction of the Treponema pallidum adhesin Tp0751 with laminin. Infect Immun. 2005;73:7485–7494. doi: 10.1128/IAI.73.11.7485-7494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE. T. pallidum outer membrane and outer membrane proteins. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponema, molecular and cellular biology. Norfolk, England: Caister Academic Press; 2006. pp. 237–266. [Google Scholar]
- Cho J, Mosher DF. Role of fibronectin assembly in platelet thrombus formation. J Thromb Haemost. 2006;4:1461–1469. doi: 10.1111/j.1538-7836.2006.01943.x. [DOI] [PubMed] [Google Scholar]
- Chuang LY, Chang HW, Tsai JH, Yang CH. Features for computational operon prediction in prokaryotes. Brief Funct Genomics. 2012;11:291–299. doi: 10.1093/bfgp/els024. [DOI] [PubMed] [Google Scholar]
- Chung KY, Kim KS, Lee MG, Chang NS, Lee JB. Treponema pallidum induces up-regulation of interstitial collagenase in human dermal fibroblasts. Acta Derm Venereol. 2002;82:174–178. doi: 10.1080/00015550260132442. [DOI] [PubMed] [Google Scholar]
- Coleman JL, Sellati TJ, Testa JE, Kew RR, Furie MB, Benach JL. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect Immun. 1995;63:2478–2484. doi: 10.1128/iai.63.7.2478-2484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collen D, Lijnen HR. Thrombolytic agents. Thromb Haemost. 2005;93:627–630. doi: 10.1160/TH04-11-0724. [DOI] [PubMed] [Google Scholar]
- Craig D, Gao M, Schulten K, Vogel V. Structural insights into how the MIDAS ion stabilizes integrin binding to an RGD peptide under force. Structure. 2004;12:2049–2058. doi: 10.1016/j.str.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Crawford J, Lamb E, Wasmuth J, Grujic O, Grigg ME, Boulanger MJ. Structural and functional characterization of SporoSAG: a SAG2-related surface antigen from Toxoplasma gondii. J Biol Chem. 2010;285:12063–12070. doi: 10.1074/jbc.M109.054866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberland MC, Turner TB. Rate of multiplication of Treponema pallidum in normal and immune rabbits. Amer J Syph. 1949;33:201–212. [PubMed] [Google Scholar]
- Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, et al. Specific chaperones for the type VII protein secretion pathway. J Biol Chem. 2012;287:31939–31947. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvaeye M, Conway EM. Coagulation and innate immune responses: can we view them separately? Blood. 2009;114:2367–2374. doi: 10.1182/blood-2009-05-199208. [DOI] [PubMed] [Google Scholar]
- Desrosiers DC, Anand A, Luthra A, Dunham-Ems SM, LeDoyt M, Cummings MAD, et al. Tp0326, a Treponema pallidum β-Barrel Assembly Machinery A (BamA) ortholog and rare outer membrane protein. Mol Microbiol. 2011;80:1496–1515. doi: 10.1111/j.1365-2958.2011.07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolphin AC. Calcium channel auxiliary alpha2delta and beta subunits: trafficking and one step beyond. Nat Rev Neurosci. 2012;13:542–555. doi: 10.1038/nrn3311. [DOI] [PubMed] [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE. Global proteome analysis of Leptospira interrogans. J Proteome Res. 2009;8:4564–4578. doi: 10.1021/pr9004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espitia C, Laclette JP, Mondragon-Palomino M, Amador A, Campuzano J, Martens A, et al. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology. 1999;145:3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- Feng S, Cen J, Huang Y, Shen H, Yao L, Wang Y, Chen Z. Matrix metalloproteinase-2 and -9 secreted by leukemic cells increase the permeability of blood-brain barrier by disrupting tight junction proteins. PLoS ONE. 2011;6:e20599. doi: 10.1371/journal.pone.0020599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes LG, Vieira ML, Kirchgatter K, Alves IJ, de Morais ZM, Vasconcellos SA, et al. OmpL1 is an extracellular matrix- and plasminogen- interacting protein of Leptospira spp. Infect Immun. 2012;80:3679–3692. doi: 10.1128/IAI.00474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler LI, Duncan KG, Fessler JH, Salo T, Tryggvason K. Characterization of the procollagen IV cleavage products produced by a specific tumor collagenase. J Biol Chem. 1984;259:9783–9789. [PubMed] [Google Scholar]
- Flood EC, Hajjar KA. The annexin A2 system and vascular homeostasis. Vascul Pharmacol. 2011;54:59–67. doi: 10.1016/j.vph.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JM, Hynes RO. Interaction of fibronectin with its receptor on platelets. Cell. 1985;42:439–448. doi: 10.1016/0092-8674(85)90101-1. [DOI] [PubMed] [Google Scholar]
- Hanlon AD, Larkin MI, Reddick RM. Free-solution, label-free protein-protein interactions characterized by dynamic light scattering. Biophys J. 2010;98:297–304. doi: 10.1016/j.bpj.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedhli N, Falcone DJ, Huang B, Cesarman-Maus G, Kraemer R, Zhai H, et al. The annexin A2/S100A10 system in health and disease: emergine paradigms. J Biomed Biotechnol. 2012;2012:406273. doi: 10.1155/2012/406273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller O, Lichte A, Oberpichler A, Kocourek A, Tschesche H. Matrix metalloproteinases collagenase-2, macrophage elastase, collagenase-3, and membrane type 1-matrix metalloproteinase impair clotting by degradation of fibrinogen and factor XII. J Biol Chem. 2000;275:33008–33013. doi: 10.1074/jbc.M001836200. [DOI] [PubMed] [Google Scholar]
- Hollier LM, Cox SM. Syphilis. Semin Perinatol. 1998;22:323–331. doi: 10.1016/s0146-0005(98)80021-9. [DOI] [PubMed] [Google Scholar]
- Houston S, Hof R, Francescutti T, Hawkes A, Boulanger MJ, Cameron CE. Bifunctional role of the Treponema pallidum extracellular matrix binding adhesin Tp0751. Infect Immun. 2011;79:1386–1398. doi: 10.1128/IAI.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston S, Hof R, Honeyman L, Hassler J, Cameron CE. Activation and proteolytic activity of the Treponema pallidum metalloprotease, pallilysin. PLoS Pathog. 2012;8:e1002822. doi: 10.1371/journal.ppat.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Yokohama Y, Nakanishi I, Ohuchi E, Fujii Y, Nakai N, Okada Y. Matrix metalloproteinase 7 (matrilysin) from human rectal carcinoma cells. Activation of the precursor, interaction with other matrix metalloproteinases and enzymic properties. J Biol Chem. 1995;270:6691–6697. doi: 10.1074/jbc.270.12.6691. [DOI] [PubMed] [Google Scholar]
- Keller TT, Mairuhu AT, de Kruif MD, Klein SK, Gerdes VE, ten Cate H, et al. Infections and endothelial cells. Cardiovasc Res. 2003;60:40–48. doi: 10.1016/s0008-6363(03)00354-7. [DOI] [PubMed] [Google Scholar]
- Knauper V, Cowell S, Smith B, Lopez-Otin C, O’Shea M, Morris H, et al. The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J Biol Chem. 1997;272:7608–7616. doi: 10.1074/jbc.272.12.7608. [DOI] [PubMed] [Google Scholar]
- LaFond RE, Lukehart SA. Biological basis for syphilis. Clin Microbiol Rev. 2006;19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahteenmaki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 2005;13:79–85. doi: 10.1016/j.tim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Lee DB, Jamgotchian N, Allen SG, Kan FW, Hale IL. Annexin A2 heterotetramer: role in tight junction assembly. Am J Physiol Renal Physiol. 2004;287:F481–F491. doi: 10.1152/ajprenal.00175.2003. [DOI] [PubMed] [Google Scholar]
- Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- Lee KH, Choi HJ, Lee MG, Lee JB. Virulent Treponema pallidum 47 kDa antigen regulates the expression of cell adhesion molecules and binding of T-lymphocytes to cultured human dermal microvascular endothelial cells. Yonsei Med J. 2000;41:623–633. doi: 10.3349/ymj.2000.41.5.623. [DOI] [PubMed] [Google Scholar]
- Levi M, van der Poll T, Buller HR. Bidirectional relation between inflammation and coagulation. Circulation. 2004;109:2698–2704. doi: 10.1161/01.CIR.0000131660.51520.9A. [DOI] [PubMed] [Google Scholar]
- Liu J, Jin X, Liu KJ, Liu W. Matrix metalloproteinase-2-mediated occludin degradation and caveolin-1-mediated claudin-5 redistribution contribute to blood-brain barrier damage in early ischemic stroke stage. J Neurosci. 2012;32:3044–3057. doi: 10.1523/JNEUROSCI.6409-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev. 2005;85:1061–1092. doi: 10.1152/physrev.00016.2004. [DOI] [PubMed] [Google Scholar]
- Loof TG, Morgelin M, Johansson L, Oehmcke S, Olin AI, Dickneite G, et al. Coagulation, an ancestral serine protease cascade, exerts a novel function in early immune defense. Blood. 2011;118:2589–2598. doi: 10.1182/blood-2011-02-337568. [DOI] [PubMed] [Google Scholar]
- Lukehart SA, Hook EW, Baker-Zander SA, Collier AC, Critchlow CW, Handsfield HH. Invasion of the central nervous system by Treponema pallidum: implications for diagnosis and treatment. Ann Intern Med. 1988;109:855–62. doi: 10.7326/0003-4819-109-11-855. [DOI] [PubMed] [Google Scholar]
- Lukehart SA, Marra CM. Isolation and laboratory maintenance of Treponema pallidum. Curr Protoc Microbiol Chapter. 2007;12:12A.1.1–12A.1.18. doi: 10.1002/9780471729259.mc12a01s7. [DOI] [PubMed] [Google Scholar]
- Madureira PA, Surette AP, Phipps KD, Taboski MA, Miller VA, Waisman DM. The role of the annexin A2 heterotetramer in vascular fibrinolysis. Blood. 2011;118:4789–4797. doi: 10.1182/blood-2011-06-334672. [DOI] [PubMed] [Google Scholar]
- Malemud CJ. Matrix metalloproteinases (MMPs) in health and disease: an overview. Front Biosci. 2006;11:1696–1701. doi: 10.2741/1915. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Weese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midwood KS, Williams LV, Schwarzbauer JE. Tissue repair and the dynamics of the extracellular matrix. Int J Biochem Cell Biol. 2004;36:1031–1037. doi: 10.1016/j.biocel.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Monaco S, Sparano V, Gioia M, Sbardella D, Di Pierro D, Marini S, Coletta M. Enzymatic processing of collagen IV by MMP-2 (gelatinase A) affects neutrophil migration and it is modulated by extracatalytic domains. Protein Sci. 2006;15:2805–2815. doi: 10.1110/ps.062430706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q, Murphy G, Hughes CE, Mort JS, Roughley PJ. Matrix metalloproteinases cleave at two distinct sites on human cartilage link protein. Biochem J. 1993;295:595–598. doi: 10.1042/bj2950595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y. Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J Biol Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37:D479–D482. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petaja J. Inflammation and coagulation. An overview. Thromb Res. 2011;127(Suppl 2):S34–S37. doi: 10.1016/S0049-3848(10)70153-5. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Radolf JD, Hazlett KRO, Lukehart SA. Pathogenesis of syphilis. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponema, molecular and cellular biology. Norfolk, England: Caister Academic Press; 2006. pp. 197–236. [Google Scholar]
- Raiziss GW, Severac M. Rapidity with which Spirochaeta pallida invades the bloodstream. Arch Dermatol Syphilol. 1937;35:1101–1109. [Google Scholar]
- Riley BS, Oppenheimer-Marks N, Hansen EJ, Radolf JD, Norgard MV. Virulent Treponema pallidum activates human vascular endothelial cells. J Infect Dis. 1992;165:484–493. doi: 10.1093/infdis/165.3.484. [DOI] [PubMed] [Google Scholar]
- Riley BS, Oppenheimer-Marks N, Radolf JD, Norgard MV. Virulent Treponema pallidum promotes adhesion of leukocytes to human vascular endothelial cells. Infect Immun. 1994;62:4622–4625. doi: 10.1128/iai.62.10.4622-4625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Springer TA. Complement and the multifaceted functions of VWA and integrin I domains. Structure. 2006;14:1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–146. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Yamayoshi Y, Nishimuta A, Tanigawara Y. S100A10 protein expression is associated with oxaliplatin sensitivity in human colorectal cancer cells. Proteome Sci. 2011;9:76–87. doi: 10.1186/1477-5956-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: Prokaryotic Operon DataBase. Nucleic Acids Res. 2012;40:D627–D631. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Ikeda U, Masuyama J, Kitagawa S, Kasahara T, Shimpo M, et al. Monocyte-endothelial cell interaction induces expression of adhesion molecules on human umbilical cord endothelial cells. Cardiovasc Res. 1996;32:422–429. doi: 10.1016/0008-6363(96)00085-5. [DOI] [PubMed] [Google Scholar]
- Tanaka KA, Key NS, Levy JH. Blood coagulation: hemostasis and thrombin regulation. Anesth Analg. 2009;108:1433–1446. doi: 10.1213/ane.0b013e31819bcc9c. [DOI] [PubMed] [Google Scholar]
- Thomas DD, Navab M, Haake DA, Fogelman AM, Miller JN, Lovett MA. Treponema pallidum invades intercellular junctions of endothelial cell monolayers. Proc Natl Acad Sci U S A. 1988;85:3608–3612. doi: 10.1073/pnas.85.10.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow PJ, Kenneally DA, Connellan JM. The role of fibronectin in platelet aggregation. Br J Haematol. 1990;75:549–556. doi: 10.1111/j.1365-2141.1990.tb07797.x. [DOI] [PubMed] [Google Scholar]
- Tundup S, Akhter Y, Thiagarajan D, Hasnain SE. Clusters of PE PPE genes of Mycobacterium tuberculosis are organized in operons: evidence that PE Rv. 2431c is co-transcribed with PPE Rv2430c and their gene products interact with each other. FEBS Lett. 2006;580:1285–1293. doi: 10.1016/j.febslet.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, et al. Pathogen entrapment by transglutaminase--a conserved early innate immune mechanism. PLoS Pathog. 2010;6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchell TD, Coutts SA, Bulach DM, Adler B. Differential expression of the Bhmp39 major outer membrane proteins of Brachyspira hyodysenteriae. Infect Immun. 2006;74:3271–3276. doi: 10.1128/IAI.02000-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.