Abstract
Self-assembly programmed by molecular structure and guided dynamically by energy dissipation is a ubiquitous phenomenon in biological systems that build functional structures from the nanoscale to macroscopic dimensions. This paper describes examples of one-dimensional self-assembly of peptide amphiphiles and the consequent biological functions that emerge in these systems. We also discuss here hierarchical self-assembly of supramolecular peptide nanostructures and polysaccharides, and some new results are reported on supramolecular crystals formed by highly charged peptide amphiphiles. Reflecting on presentations at this Faraday Discussion, the paper ends with a discussion of some of the future opportunities and challenges of the field.
1. Introduction to the Field
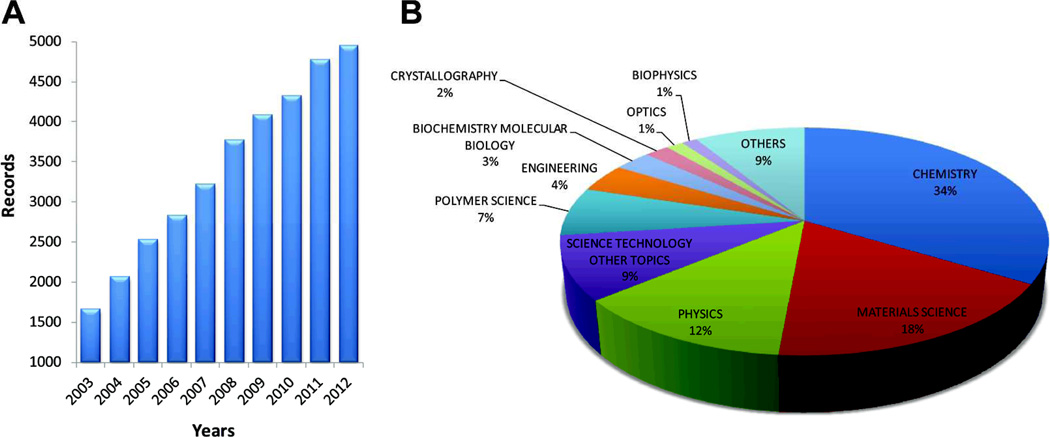
Self-assembly has fascinated a very large number of scientists over the past decade. If this bio-inspired strategy could be generally implemented in synthetic systems, it would have a profound impact on new materials and devices, as well as help discover new behaviors, even emergent ones, in abiotic systems. Figure 1 shows the number of papers dealing with self-assembly published over the past decade and the various fields of science associated with them. Chemistry, materials science, and physics dominate. In life sciences, not surprisingly, self-assembly as a strategy for fabrication of functional systems is taken for granted. To the physical scientist, the following phenomena are truly amazing examples of dynamic self-assembly involving molecular and supramolecular programming: protein folding, formation of receptor rafts for signaling on cell membranes, alignment of muscle fibers over macroscopic length scales, assembly of the ribosome as an efficient protein-making machine, reversible filaments of the cytoskeleton, hierarchical structure of articular cartilage with spatially varying orientations of collagen fibers to create a remarkably tough tissue with low coefficient of friction, and many others.
Fig. 1.
Analysis of publications using the term "self-assembly" over the past decade (2003–2012). This record was obtained from the Web of Science database using the keyword "self-assembly" as the search topic. (A) Bar graph reveals a continuous growth in publications per year over this period. (B) Pie chart showed the percentage of these publications categorized by research area.
Self-assembly of synthetic systems relies on components designed to spontaneously order into a “functional” structure with little or no intervention from humans or machines.1 As stated above, this process takes its inspiration from biology and can occur at molecular to macroscopic length scales. Noncovalent interactions such as hydrogen bonding, π–π stacking, metal–ligand interactions, electrostatic forces, dipole–dipole interactions, hydrophobic forces, and steric forces can now be used as part of a “supramolecular code” to design simple self-assembling materials, relative to biological systems.2 These so-called simple structures can be equilibrium structures and thus regarded as “static” at specific temperatures, pressures, or environments. However, they can also be highly metastable and exist in non-equilibrium states. In early stages of the self-assembly field, systems that could be regarded as nano-sized materials in which relatively small molecules aggregated through short range interactions were investigated. The assemblies created were nanoscale supramolecular objects such as non-centrosymmetric clusters of molecules,3 ribbons,4 tubes,5 helices,6, 7 among others. More complex systems in terms of structure and function could be accessed by dissipative systems which require energy input in the form of “fuels”8 or external forces. In these systems it may be possible, as observed in biology, to achieve higher complexity levels of self-organization. Other systems may require a structural template, such as in biomineralization processes. One of the grand challenges in the field of self-assembly is to develop strategies to create hierarchical structures, also commonly observed in biological systems. So far examples of hierarchical self-assembly have been discovered9 but the principles to design them rationally are effectively not known.
In the biopolymers space, the subject of this Faraday Discussion, the systems of interest are polypeptides, nucleic acids, polysaccharides, biologically synthesized condensation polymers, and supramolecular polymers built from biomolecular structural units such as peptides and oligonucleotides. Hybrid combinations of these systems could greatly expand the scope of the field. These macromolecules and supramolecular structures have potential for biocompatibility or bioactivity since they are built from biomolecular units. Peptides, polypeptides, and proteins naturally contain many self-assembling motifs and high potential for function.10, 11 New opportunities for functional materials have opened up with biosynthetic strategies which incorporate artificial amino acids or non-peptide backbones.12, 13 It is in fact now possible to express artificial proteins with non-canonical amino acids using bacteria.14 Self-assembly of designed oligonucleotides have recently become a highly active area initiated by Seeman and co-workers, yielding programmed structures of arbitrary shape driven by Watson–Crick pairing.15, 16 Very recently, these strategies have been used to design artificial DNA and RNA sequences for use in nanotechnology applications.15–18 Oligosaccharides, in contrast to oligonucleotides and peptides, are difficult to synthesize with specific sequences but offer great potential to create systems with chemically encrypted biological information and thus potential for many important biological functions. One example of utilizing polysaccharides in self-assembly is the formation of complex supramolecular pseudorotaxane polymers using cyclodextrins (cyclic oligosaccharides) and small molecules.19 Great progress has been made in oligosaccharide synthesis20, 21 but the preparation of more complex polysaccharide systems will require further synthetic innovation.
Many macromolecules with biological structural units have been synthesized and studied over the past few decades as covalent polymers, generating an enormous body of literature that is not covered here. However, the greatest potential for biomolecular soft matter with structural complexity and function lies with self-assembling systems in which supramolecular structure can be programmed. So far, this opportunity is on the horizon with peptide-based systems, which will eventually integrate with glycochemistry and oligonucleotides. This paper as well as many of the presentations at this Faraday Discussion are focused on peptide systems, particularly those that can create self-assembling structures across multiple scales.
Peptides offer a great structural tool for the science of self-assembly and for our understanding of proteins. At the same time, their synthesis is relatively simple and well developed (provided they are not too long) even though their purification can be challenging. It is also important that peptide-based systems can allow some degree of rational control based on current knowledge over molecular conformations and intermolecular interactions through the primary sequence of amino acids. This allows the possibility of interpreting the supramolecular complexity that often emerges in these systems. With regard to crafting function, the potential of peptides is enormous since they are the central signaling language of biology. This also provides the opportunity to create functional systems that are directly inspired by biology. The potential for biological interactions with other macromolecules, such as nucleic acids and polysaccharides, further augments the value of peptides. Thus, peptides can form the basis of systems programmed for useful applications in medicine, catalysis, energy related technologies, manipulation of microorganisms, and many other areas. On the structural side, the challenges include a deep understanding of the competition between inter-peptide vs. peptide-water interactions and also of the competition of hydrogen bonding, electrostatics, and hydrophobic contacts. These issues are critical for the development of a supramolecular code for peptides that can predict self-assembly of nanoscale and larger hierarchical architectures. Known architectures known so far include, cylindrical fibers, spheres, lamellae, twisted ribbons, flat ribbons/tapes/belts, tubes, and helical ribbons. On the functional side, the challenges include understanding the pathway dependence of peptide self-assembly towards equilibrium and non-equilibrium structures as well as optimizing strategies for incorporating bioactivity. This is particularly important in the context of scalability and robustness of functional peptide systems.
2. 1D Peptide Amphiphile Assemblies and their Biomedical Functions
Discussion of peptide self-assembly is perhaps best achieved by starting at the molecular level and progressing through increasing scales and complexity. The ultimate goal would be to encode the formation of nanoscopic, mesoscopic, and macroscopic structures based only on the design of primary sequence. Fibrous, filament-like nanostructures are commonly observed in self-assembly of peptides from water. It is difficult to predict their morphologies. Most importantly from a functional perspective, it is nearly impossible at this time to predict what segments of their sequences are displayed on the surfaces of these filamentous assemblies. Some early examples of peptide fibrils include those reported by Zhang and coworkers,22, 23 as well as the helical tapes and twisted ribbons reported by Aggeli and coworkers.24, 25 Inspired by amyloid formation, Collier and coworkers developed self-assembling peptides containing a glutamine-rich motif that drives β-sheet formation to stabilize nanofibers.26 The alkylation of peptides, molecules known as peptide amphiphiles, has offered a strategy to define supramolecular assemblies, particularly the nature of the surfaces since alkyl or other similar segments will always be more hydrophobic than any peptide segment. In this case, self-assembly in water would involve hydrophobic collapse of the alkyl segments, directing them into the interior of nanostructures and making it more likely that termini of peptide segments are displayed on surfaces of the aggregates. Peptide amphiphiles (PAs) were initially studied by Berndt and coworkers reporting on assemblies at the water-air interface.27 Formation of spherical micelles by PAs has been previously reviewed28 and will not be discussed further here. The remainder of this section will only consider one-dimensional filamentous structures..
One-dimensional nanostructures in which the length is orders of magnitude larger than the cross-sectional dimensions are functionally interesting since they can form aligned structures and networks.29, 30 These high aspect ratio supramolecular nanostructures can be considered as unconventional polymers in which monomer units are connected by strong secondary forces rather than covalent bonds. They can be described using classical terms such as polydispersity and degree of polymerization. A review by De Greef and coworkers draws analogies to covalent polymers and classifies “supramolecular polymerization” as isodesmic, ring-opening, or cooperative.31 However, depending on the nature of monomers, supramolecular polymers can be more complex than conventional polymers in that the process of monomer assembly in 1D can create structures with internal order.4, 32–35 Furthermore, because supramolecular polymers are held together by secondary interactions they can have dynamic structures given the finite time scales of their bond lifetimes. In this regard, they could exhibit self-healing behavior or have faster biodegradation rates than conventional covalent polymers. From a biomimetic and functional perspective, supramolecular polymers based on peptides or peptide amphiphiles as monomers are interesting since they mimic the fibrous components of natural extracellular matrices (ECMs). For example, both fibronectin and collagen proteins assemble into fibrils that comprise the 3D supportive matrix of cells. These ECM components additionally provide signals that regulate cell migration, proliferation, and differentiation. Artificial synthesis of ECM proteins and structure is challenging; however, self-assembling oligopeptides that can potentially capture their structures and functions are readily accessible.36–38 Furthermore, oligopeptides may be modified with biomolecular units such as sugars, lipids, or nucleic acids.
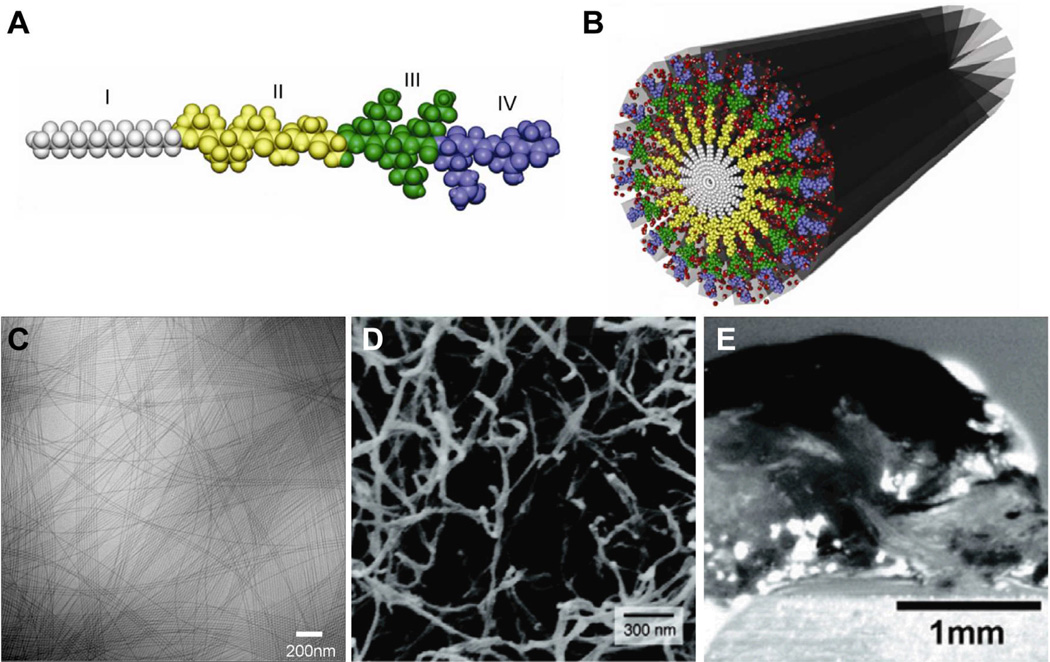
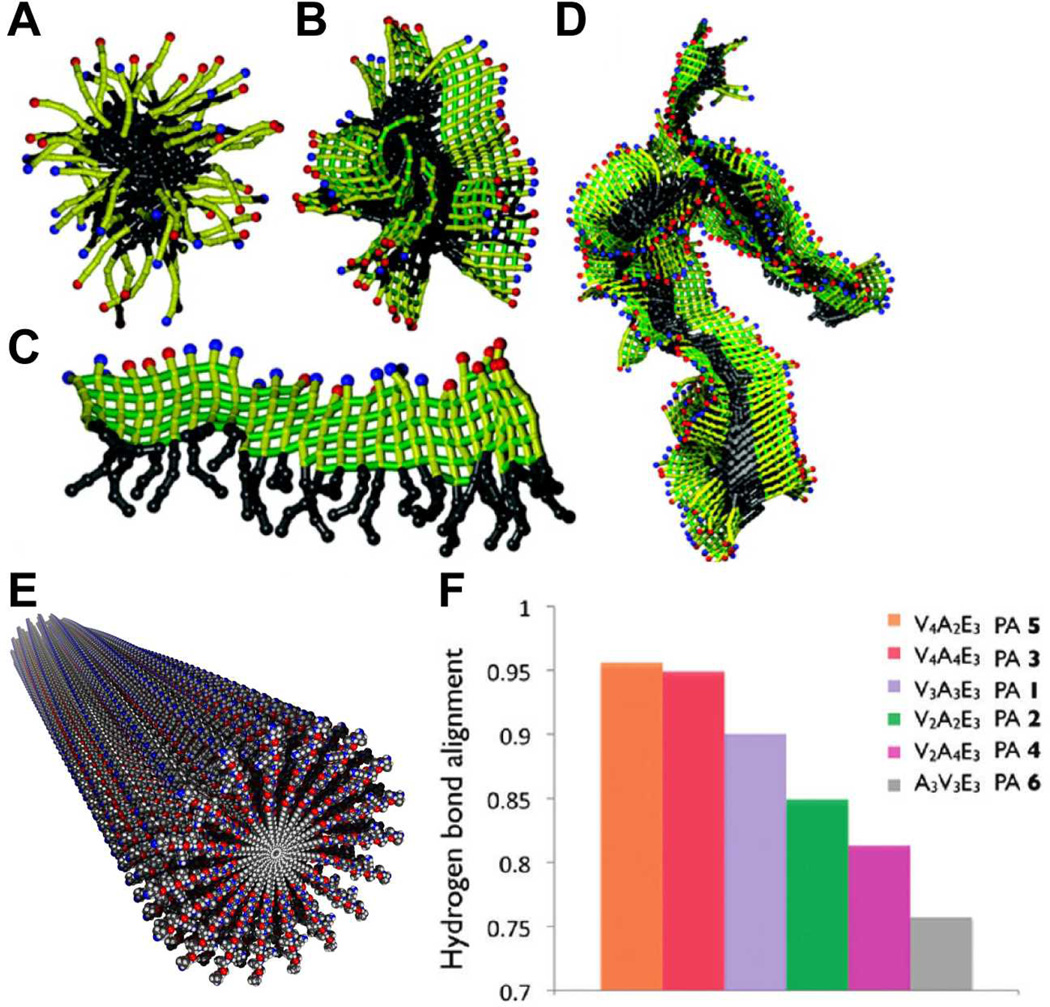
The Stupp Laboratory has developed a class of peptide amphiphiles capable of self-assembling into cylindrical nanofibers of high aspect ratio. This offered the opportunity to create mimic the architecture of extracellular fibers for biomedical functions. (Fig. 1).32, 39, 40 These PAs are generally composed of four domains: a hydrophobic moiety such as an alkyl group (I), a β--sheet forming peptide sequence (II), a charged peptide sequence which promotes solubility (III), and an optional sequence providing bioactive function (IV). The bioactive forms of these PAs therefore have the capacity to display biological signals in high density on the surfaces of the nanofibers as a result of the hydrophobic collapse of alkyl segments in water. The basis of one-dimensional assembly in the Stupp laboratory PAs has been hypothesized to be the formation of β-sheet secondary structure in the amino acids of the domain that is contiguous to the alkyl chain. This was supported by a coarse grained simulation in the authors’ laboratory, which showed that PAs without these intermolecular hydrogen bonding interactions assemble into spherical micelles with a hydrophobic core and hydrophilic corona (Fig. 2).41 In contrast, PAs modeled to have the β-sheet intermolecular hydrogen bonding assemble into β-sheet structures that collapse through hydrophobic interactions into larger 1D aggregates. We believe this to be the mechanism involved in the formation of the cylindrical nanofibers observed experimentally, which contain many β-sheets and a hydrophobic alkyl core. In this simulation, molecules lacking the hydrophobic alkyl segments assemble into polydisperse 1D β-sheets structures.
Fig. 2.
(A) Molecular graphics representation of a canonical peptide amphiphile (PA) molecule, showing the (I) hydrophobic alkyl tail, (II) β-sheet forming residues, (III) charged residues, and (IV) bioactive peptide sequence. (B) Molecular graphics representation of a cylindrical nanofiber formed by peptide amphiphiles in water. (C) Cryogenic TEM of PA nanofibers, (D) SEM micrograph of a nanofiber network, and (E) optical image of a hydrogel formed by the nanofiber network. Adapted with permission from reference 36. Copyright 2011 Elsevier Ltd.
The canonical PA nanofiber is 6–12 nm in diameter depending on the PA molecule and up to several microns in length (Fig. 1C).32 Typical PA nanofibers are highly charged, which helps with solubility in water. However, screening of PA nanofibers with ions, primarily divalent or higher valency, or reducing charge density through changes in pH results in the formation of viscous liquids or gels. However, the molecular mechanism of gelation is not clear and most likely involves changes in water structure around the nanofibers. (Fig. 1D–E).42, 43 Using spectroscopic techniques, studies have confirmed that cylindrical PA nanofibers contain β-sheets oriented parallel to the long axis of the fiber.44, 45 These structures exhibit a degree of twisting about the axis that can be tuned by altering the proportion of strong and weak β-sheet forming residues adjacent to the hydrophobic tail.46 The degree of twisting decreases and intermolecular hydrogen bond alignment increases when stronger β-sheet forming residues (i.e. valine as opposed to alanine) are positioned adjacent to the hydrophobic tail (Fig. 2F–G). This change in internal structure has direct consequences on the mechanical properties of the PA nanofibers, as gelled 3D nanofiber networks show increased stiffness with hydrogen bond alignment. These results are consistent with the work by Hartgerink and coworkers, which also suggests the importance of the first four amino acids adjacent to the alkyl tail in nanofiber assembly.45 Furthermore, changes to the β-sheet forming peptide sequence have been shown to affect gelation time of PAs, with more bulky and hydrophilic sequences (e.g. SLSLGGG versus AAAAGGG) requiring longer times to form self-supporting 3D networks.47 This feature allows the tuning of injectable scaffolds through molecular structure of the PA for use in biomedical applications. Surprisingly, internal order is found also within the hydrophobic nanofiber cores in PAs with linear alkyl chains as indicated by infrared spectroscopic data.44 Further evidence for this order was obtained by designing PAs containing diacetylene-derivatized hydrophobic tails.48 The topotactic polymerization of diacetylene was observed in these systems which is only possible if high degrees of internal order exist in the nanofiber core.
The peptide portion of PAs constitutes the hydrophilic corona of nanostructures formed and is highly hydrated. Stern–Volmer quenching analysis of PAs functionalized with tryptophan or pyrene at different locations within the peptide region indicates that even the PA nanofiber interior is well solvated.49 Chromophores located adjacent to the hydrophobic core respond significantly to aqueous quenchers, albeit weaker in comparison to those located on the nanostructure surface. This suggests that small molecules may access the nanofiber interior. Work by Webber et al. showed further that proteins also may penetrate the interior of PA nanofibers.50 In this previous work, a PA was designed to contain a consensus substrate sequence for protein kinase A (PKA), an enzyme important in intracellular signaling. Upon treatment of PA nanofibers with PKA, individual PA molecules become phosphorylated and the nanofiber consequently disassembles due to electrostatic repulsion. Dephosphorylation of the PA with alkaline phosphatase restores the nanofiber assembly. Since PKA is also a known cancer biomarker, this strategy has been investigated for the delivery of a hydrophobic chemotherapeutic drug, such as doxorubicin, encapsulated within the nanofiber core.
Because PA nanofibers can readily interact with cells and proteins via peptide motifs displayed on their surfaces,51 they have been extensively investigated for use in applications such as neural regeneration,51–55 cartilage regeneration,56 bone regeneration,57–60 enamel regeneration,61, 62 angiogenesis to improve pancreatic islet transplantation63,treatment of myocardial infarction64 and peripheral arterial disease,65 and cancer therapies,66–68, among others. The role of PA nanostructures in some of these applications is to act as a bioactive vehicle for exogenous therapeutic proteins, small molecule drugs, or biopolymers.. For example, PA nanofibers have been designed to bind and display heparin on their surfaces for the purpose of capturing and displaying growth factors containing heparin binding domains with greater efficacy.69 More recently, a PA molecule with a ruthenium tricarbonyl group was designed to locally release the signaling gas carbon monoxide to improve the viability of stress cardiomyocytes.70 These PAs exhibit prolonged release kinetics 8-fold compared to soluble carbon monoxide donors. In another example, PAs have been functionalized with drugs at the terminus of the peptide segment conjugated through a labile bond such as a hydrazine linkage,71 leading to useful controlled release patterns. Another approach to the design of bioactive PAs involves the use of peptides that have been discovered by phage display methodology to bind specific growth factors. One recent example was the design of a PA that displays at its terminus a given concentration of a peptide that binds the growth factor TGFβ-1, which is important in differentiation of stem cells into chondrocytic lineage for cartilage regeneration. The high density of peptides displayed by the nanofibers offers the possibility to recruit endogeneous growth factor and thus rendering the fiber bioactive without adding any exogenous protein to the therapy.72 There are also PA nanofibers that are designed molecularly to interact directly with cells for signaling and this way induce a desired behavior. For example, PAs have been designed to display RGDS,32, 42, 73 a cell adhesion/survival epitope derived from fibronectin, or IKVAV,51, 74, 75 a laminin-derived oligopeptide that directs neurite growth. Similarly, PAs have been designed to mimic the activity of proteins such as vascular endothelial growth factor65 and glucagon-like peptide 176 by displaying sequences in high density on the nanofiber surface that activates their cell surface receptors. Epitopes displayed in this manner are stabilized in the active conformation, whereas the corresponding free peptide generally adopts random coil conformations.65, 76 Additionally, the density of epitopes on the nanofiber surface can be tuned by co-assembly of the bioactive PA with a non-bioactive diluent PA, allowing for optimization of epitope presentation.74, 77 Thus, the basic design of bioactive PAs developed in the Stupp laboratory to self-assemble into ECM-like nanofibers have multiple mechanisms as bioactive nanostructures for specific therapies.
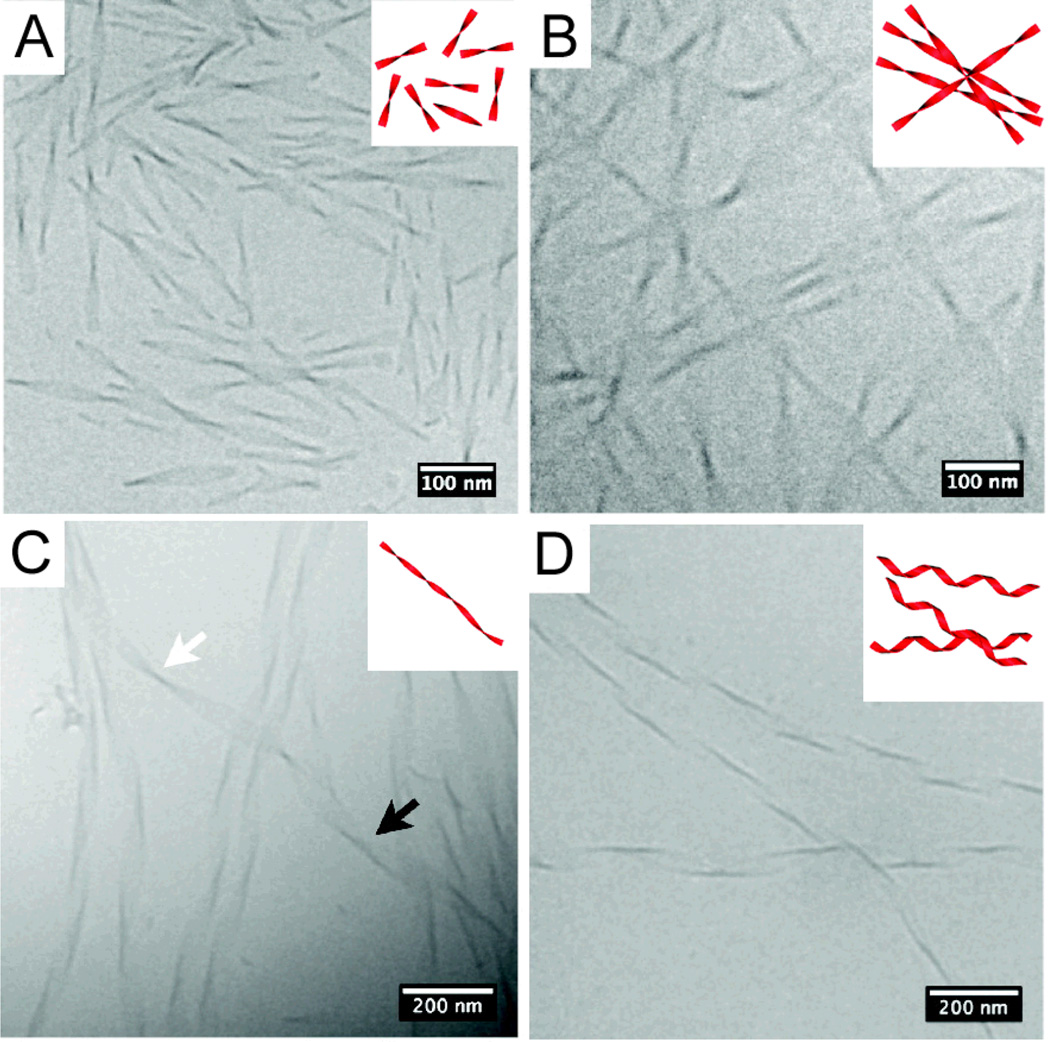
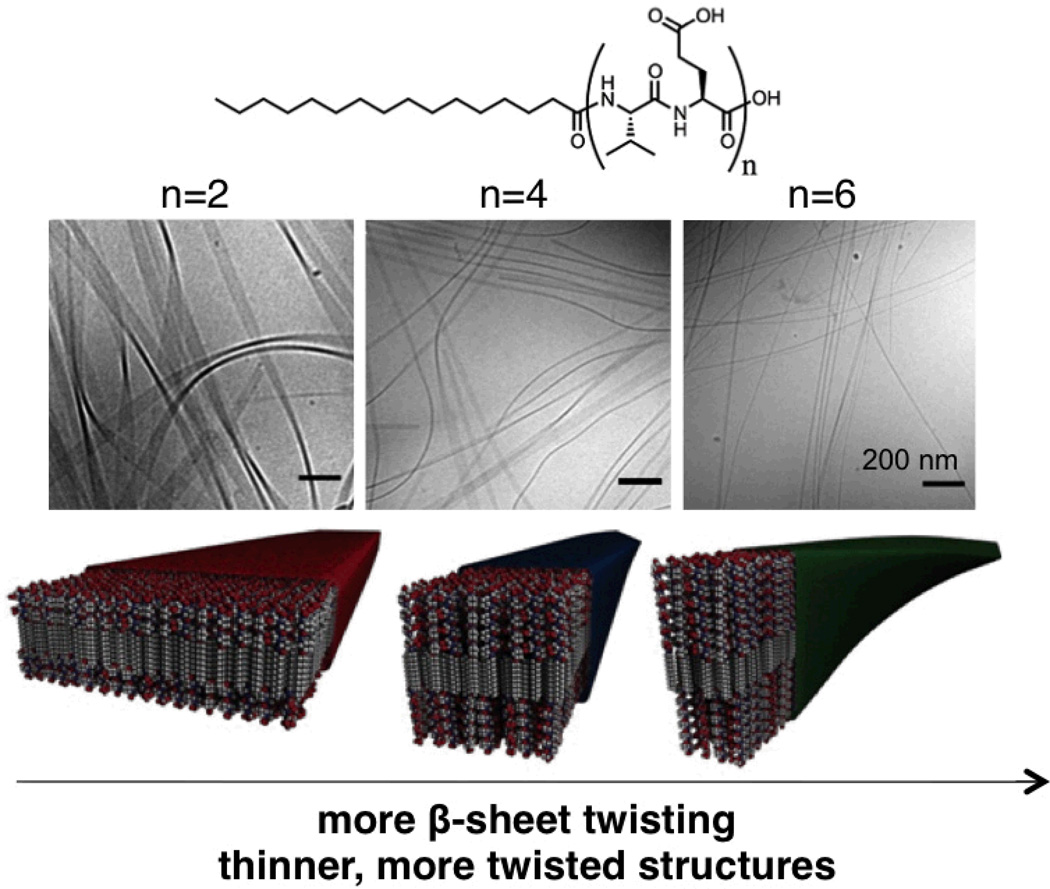
PA self-assembly kinetics and dimensions of resulting nanostructures can generally be predictably tuned by altering molecular structure. For example, the length of the alkyl tail may be changed, the propensity for β-sheet formation in the peptide segment may be adjusted, or the charge of the molecule may be varied. Cylindrical morphology typically persists when there are β-sheet forming residues in which side chain interactions are not too strong. The current view is that interactions between aggregates with partial β-sheet secondary structure and water internal to the fiber lead to cylindrical aggregates collapsed by hydrophobic interactions. On the other hand, strong side chain interactions among β-sheet aggregates could displace water and the curvature of the entire supramolecular ensemble is lost. For example, PAs have been designed that self-assemble into flat nanobelts of large width78, and others have ribbon-like, often twisted, morphologies.79 The PAs that form wide and flat nanobelts contain a sequence of alternating hydrophobic and hydrophilic residues and seem to dimerize. Specifically, these nanobelts were discovered with the peptide sequence VEVE, which exhibits hydrophobic and hydrophilic side chains on opposite sides of the peptide backbone. Thus, hydrophobic valine side chains can associate and PA molecules can dimerize. These dimers pack with interdigitated tails into a flat bilayer 5 nm thick and several microns long. Thus, strong side chain interactions can be a function of sequence, as observed in these systems. In contrast, control PAs bearing a VVEE motif exhibit the more canonical core-shell cylindrical nanostructure. Moyer et al also demonstrated that the lateral width of nanobelts can be tuned between 10 – 100 nm by decreasing the number of VE dimeric repeats from 6 to 2 (Fig. 3).80 Circular dichroism and cryo-TEM studies show that increasing the number of dimeric repeats increases β-sheet twisting and that the resultant twisting of nanobelts limits lateral growth. Cylinders become the dominant morphology over flat nanobelts above a threshold number of dimeric repeats. Pashuck et. al. have also observed a morphological transition in flat, non-cylindrical PA nanostructures occurring over time. In this work, PAs bearing phenylalanine residues adjacent to the hydrophobic tail rather than β-sheet forming residues assemble into short twisted ribbons immediately after dissolution in water (Fig. 4).81 Upon aging for several minutes, these short, twisted ribbons elongate. Further aging (i.e. up to 4 weeks) reveals a transition to helical ribbons, thus suggesting that the supramolecular packing behind the twisting of ribbons is energetically unfavorable. The cylindrical curvature of helical ribbons allows for more ideal supramolecular packing when compared to the saddle-like curvature of twisted ribbons.
Fig. 3.
(A–D) Snapshots from molecular simulations of peptide amphiphiles with increasing degrees of hydrogen bonding strength in their β domains. (A) A spherical micelle forms through hydrophobic collapse; (B) a more elliptical micelle is predicted as β-sheets form; (C) an extended supramolecular aggregate formed by an extended β-sheet of peptide amphiphiles; (D) aggregating β-sheets through hydrophobic interactions. (E) molecular graphics representation of a cylindrical nanofiber in which β-sheets are twisted about its long axis. (F) The degree of twist decreases with increasing hydrogen bond alignment, which is controlled by the first amino acids adjacent to the alkyl tail. (A–D) are adapted with permission from reference 41. Copyright 2010 American Chemical Society. (F) is adapted with permission from reference 46. Copyright 2010 American Chemical Society.
Fig. 4.
Chemical structure of a series of amphiphilic peptide amphiphiles (PAs) with different numbers of dimeric repeats of hydrophobic (valine) and hydrophilic (glutamic acid) amino acids (top). Transmission electron micrographs of ribbon-like assemblies of the PAs containing two, four, and six dimeric units (middle), and their respective molecular graphics representations. As the number of dimeric repeats is increased from two to six, ribbon twisting increases and lateral width of the assemblies is reduced from 100 nm down to 10 nm. Adapted with permission from reference 80. Copyright 2013 American Chemical Society.
3. Hierarchical Structures
The use of hierarchical self-assembly as a strategy to synthesize materials is a bio-inspired idea driven by the enormous potential of novel properties and functions when structure is controlled across scales, from the nanometers to macroscopic dimensions. Whereas self-assembly can be seen as a spontaneous organization of matter from a disordered state into an ordered state, hierarchical self-assembly additionally entails generation of multiple levels of organization where, at each level of organization, properties emerge that may be informed by lower levels but do not exist at lower levels.82 As an example, collagen is the primary component of connective tissue and is comprised of three polypeptide strands twisted into a right-handed triple helix. The association of many triple helices results in microfibrils, which further aggregate into larger fibers. Such bundling of 1D structures is a ubiquitous phenomenon in nature and helps mediate events such as cell mitosis, protein transport, and signal transduction. Furthermore, regions of the central nervous system (e.g. brain and spinal cord) show areas of long-range alignment,83 which is crucial for directional transport of electrical signals.
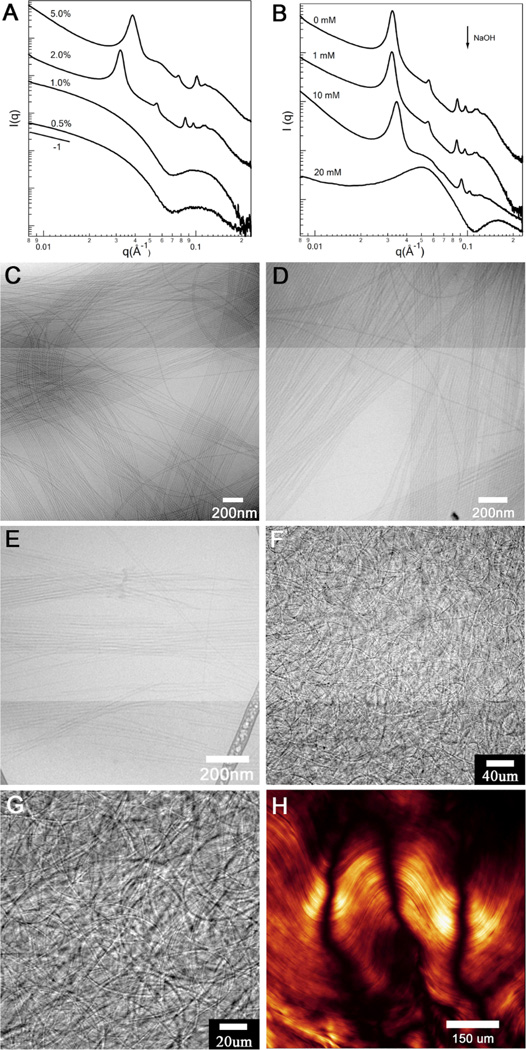
We previously showed that charged PA cylindrical nanofibers containing the peptide A6E3 can self-assemble into crystallizing arrays with surprisingly large d-spacing (~32 nm) in water (Fig. 5).84 This arrangement occurs spontaneously at high PA concentrations (2 wt% or above) or can be induced reversibly by ionizing X-ray irradiation at lower concentrations. Figure 5A shows that synchrotron small angle X-ray scattering (SAXS) on 2 wt% peptide aqueous solution displays a series of Bragg peaks that correspond to a two dimensional hexagonal lattice. The hexagonal stacking with significant long-range order can be identified by seven clear reflections at q/q* ratios of 1:√3:√4:√7:√9:√12:√13 (q* is the position of the first peak). The √4 peak is slightly depressed possibly due to its overlapping with the first minima of the nanofiber form factor. Considering the low volume fraction of peptide amphiphile molecules, the assembled nanofibers must exist as bundles. Experiments with NaCl and CaCl2 clearly suggested that the observed crystalline ordering among highly-charged PA nanofibers are mediated by electrostatic repulsion.84 The crystalline ordering was gradually suppressed with the increase of the salt concentration due to Manning condensation weakening electrostatic interactions among like-charge objects. We report here the influence of nanofiber surface charge on their bundling behaviour by varying the solution pH. Figure 5B displays the scattering profiles of 2 wt% peptide solutions with different amounts of NaOH, and cryo-TEM was used as a complementary tool to characterize the system (Figures 5C–5E). Presumably, NaOH would cause nanofibers to carry more negative charge by deprotonating –COOH groups located on nanofiber surface, leading to strengthened electrostatic repulsions. In the presence of 1 mM NaOH, little or no peak shift was observed. Surprisingly, with 10 mM NaOH added, instead of observing loosely packed bundles with increased spacing between nanofibers, we found that nanofibers tend to pack more closely inside the bundles. Accordingly, Bragg peaks that correspond to a hexagonal lattice were shifted to higher q (Figure 5B). With addition of 20 mM NaOH, the dramatic changes in the scattering profile are attributable to a decrease in nanofiber lengths. Strong electrostatic interactions within peptide shell eventually break up nanofibers into smaller aggregates, as supported by cryo-TEM imaging (Figure 5E). The diffuse peak around 0.0520 Å−1 corresponds to the structural factor for interactions among nanofibers of reduced sizes. In contrast to the bundling behaviour of F-actin and microtubules, where the bundles are constructed in the presence of multivalent counterions85, 86, hexagonal stacking of nanofibers reported here occurs in water solution lacking multivalent counterions. We suggest that the resulting bundles are kinetically trapped hierarchical structures that are polydisperse in size. Interestingly, we were able to visualize directly the bundles by optical microscopy (Figures 5F and 5G). We interpret the micrograph as a network of micron scale filamentous crystals that contain hundreds to thousands of nanofibers. If a small shear is applied to the network by slightly pressing the cover slip, we observe the formation of nematic domains across several hundred micrometers (figure 5H).
Fig. 5.
Cryo-TEM of C16-FFFEEE PA assemblies forming (A) short twisted ribbons after 30 seconds dissolution of the PA in water, (B) elongated twisted ribbons after 10 minutes dissolved in water, (C) twisted ribbons (white arrow) transforming into helical ribbons (black arrow) after 2 weeks in water, and (D) helical ribbons after aging in water at 25 °C for 1 month. Adapted with permission from reference 81. Copyright 2010 American Chemical Society.
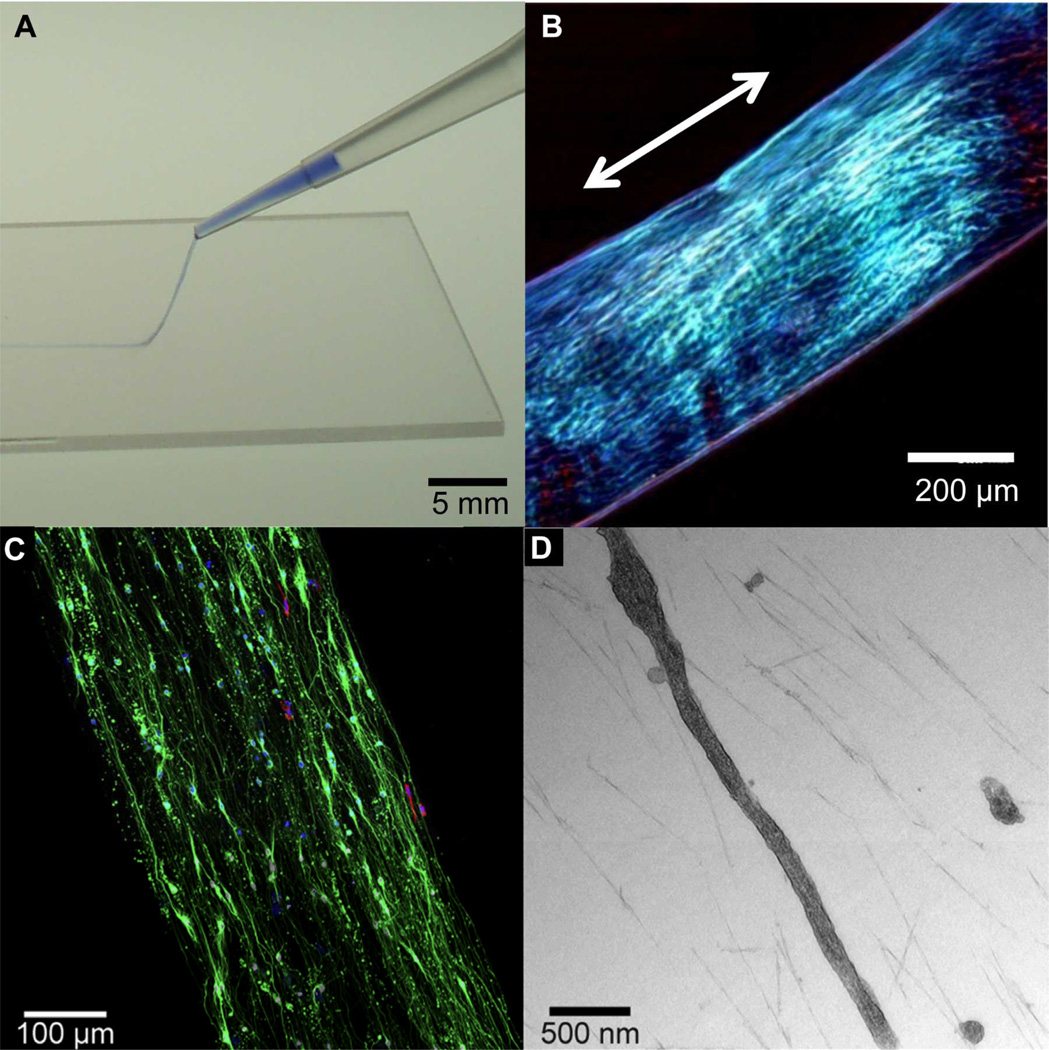
Bundling and 1D alignment of PA nanofibers can also be induced by thermal treatments.87 We previously reported that cylindrical nanofibers formed by relatively non-bioactive simple anionic PAs when heated to 80°C for 30 min organize into large flat plaque-like aggregates. This is presumably due to dehydration of water bound or trapped within the nanofibers. Upon cooling, large domains of bundled nanofibers emerge, resulting in a lyotropic liquid crystal with noticeable birefringence even in dilute solutions (~1 wt%) (Fig. 6). Domains of aligned nanofiber bundles several centimeters long can further be created by extruding the solution from a pipette tip in a bath containing divalent cations that screen the negative charges on the nanofiber surface. Due to their peptide-based components and their gentle alignment process, these monodomain aligned gels are compatible with living cells, which can be introduced after heating and cooling the PA solution.55, 88 Mesenchymal stem cells incorporated within monodomain aligned gels using this method show good viability. Moreover, cell bodies and filopodia align in the direction of extracellular PA nanofibers. By incorporating PA molecules bearing the IKVAV epitope in this hierarchical assembly process, these monodomain, aligned gels are further able to induce directional neurite outgrowth (Fig. 7C). Thus, this material shows promise in preliminary rodent studies as a transplantable scaffold for delivering neural progenitor cells in spinal cord repair applications.88 It is worth noting that these aligned gels are extremely hydrated (~99 wt% water), thus exhibiting a highly porous, open architecture that facilitates cell growth and migration. Electron microscopy shows the space between PA nanofibers into which cells extend (Fig. 7D).88 It is possible that the structuring of water by aligned PA nanofibers aid in directing neurite orientation in this seemingly open structure and also contributes to the gelation process itself in the presence of divalent ions.
Fig. 6.
(A) Small angle x-ray scattering (SAXS) scans at different peptide amphiphile concentrations (PA with A6E3 peptide sequence) and (B) SAXS scans of PA solutions at different concentrations of added NaOH. (C–E) Cryo transmission electron micrographs of aqueous 2 wt% PA solutions without added NaOH (C), containing 10mM NaOH (D), and 20mM NaOH (E). (G–F) Optical micrographs of aqueous 2 wt% PA solution, and (H) optical micrograph of the same solution under cross polars after pressing the cover slip (artificial color is used for clarity). (A) is adapted with permission from reference 84. Copyright AAAS 2010.
Fig. 7.
(A) Photograph of a thermally treated (see text) peptide amphiphile (PA) solution colored with trypan blue, dispensed manually from a pipette onto a thin layer of aqueous CaCl2 to form a noodle-like gel string. (B) Optical micrograph of a single string between cross polars indicating macroscopic alignment along the string axis. (C) Hippocampal neurons cultured for 7 days within a noodle-like aligned gel containing a PA with the 20% bioactive laminin epitope IKVAV and 80% of a non-bioactive diluent PA. Neurons were stained for β-tubulin III- (green) (neuron marker), glial fibrillary acidic protein (GFAP) (red) (marker for astrocytes), and 4’,6-diamino-2-phenylindole (DAPI) (blue) (nuclear stain) revealing extensive, aligned neurite growth. (D) TEM micrograph of a hippocampal neurite inside the aligned gel showing its parallel orientation to the aligned PA nanofibers. (A) and (B) are adapted with permission from reference 87. Copyright 2010 Nature Publishing Group. (C) and (D) are adapted with permission from reference 88. Copyright 2014 Elsevier.
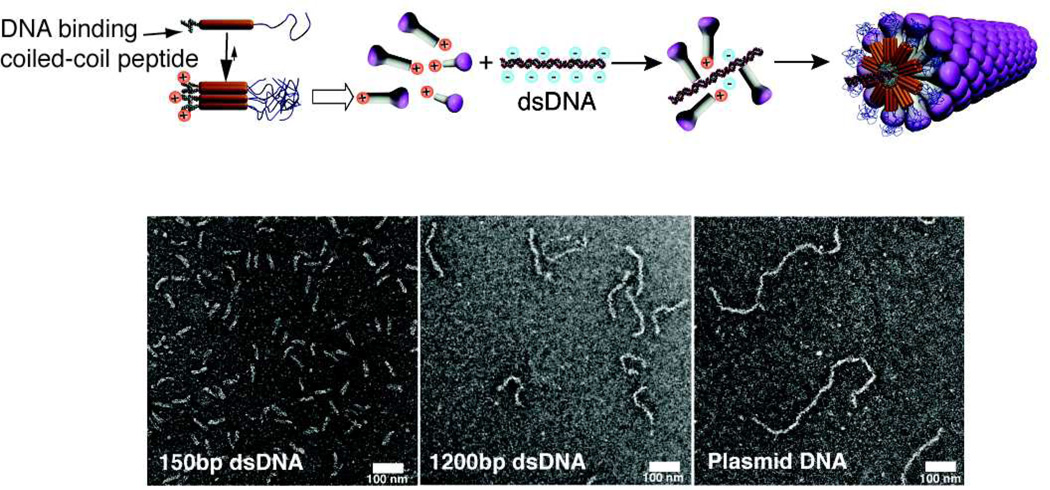
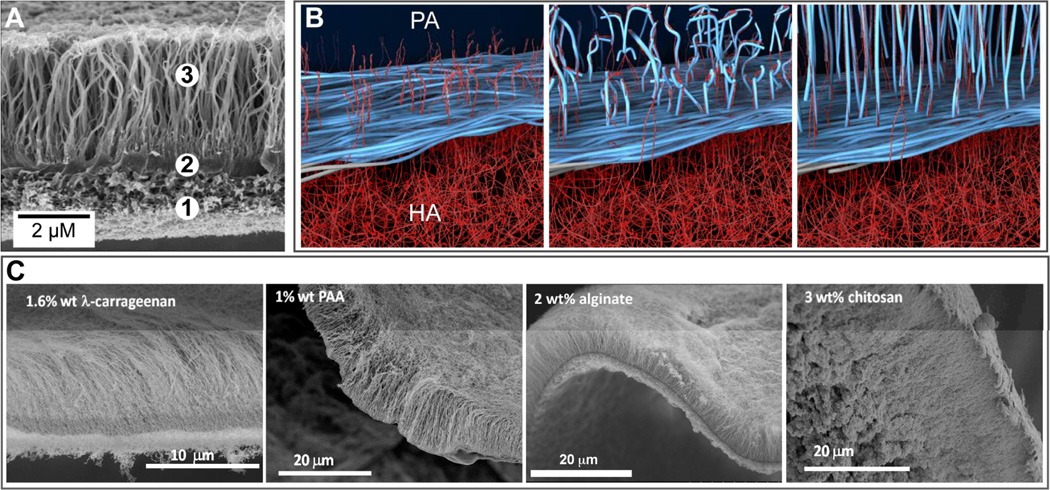
Hierarchical self-assembly can be utilized as a strategy to overcome some challenges faced by simple molecular self-assembly. One such challenge is the formation of one-dimensional nanostructures with monodisperse length. Inspired by the assembly of filamentous viruses one may consider the use of templates; for example the tobacco mosaic virus uses a single RNA strand to template the self-assembly of capsid proteins into a cylinder of precise length.89 Recently, a system has been designed in the Stupp laboratory to mimic this process using double-stranded DNA to template pre-assembled mushroom-shaped nanoparticles into monodisperse nanofibers (Fig. 7).90 The mushroom-shaped nanoparticle is a heptamer comprised of self-assembled coil-coil oligopeptides with a PEG chain on one terminus and a cationic spermine moiety on the other terminus. These heptamers interact with the DNA backbone via electrostatic interaction between spermine and phosphate groups but require the steric crowding of PEG chains to prevent buckling of the DNA strand. Without these steric forces, the hierarchical nanofibers formed are heterogeneous in shape and size. The lesson here is that multiple orthogonal interactions were necessary to build the hierarchical structure. As it is often observed in biological systems, hierarchical self-assembly can readily incorporate multiple components in order to create enhanced function. Common hybrid systems include aggrecan, which consists of chondroitin sulfate and keratan sulfate attached to a protein core to form a bottlebrush structure.91 Many aggrecan complexes then associate with a hyaluronic acid backbone by linker proteins in order to form a major component of ECM, particularly in shock absorbing tissue such as cartilage. Also, multi-component self-assembly systems can be used to access dynamic processes that may result in complex structures with order on multiple length scales. In 2008, Capito and coworkers reported on the formation of a robust membrane at the interface between aqueous solutions of PAs and high molecular weight hyaluronic acid (HA).9 Membrane self-assembly is initiated in this system by the rapid electrostatic complexation of the two oppositely charged components to form a contact layer at the liquid-liquid interface that acts as a barrier to diffusion, preventing chaotic mixing of PA and HA (Fig. 8). Subsequently, excess osmotic pressure in the HA solution promotes the diffusion of HA molecules through the diffusion barrier. Reptation and outward diffusion of HA chains into the PA solution over the course of minutes to days templates growth of nanofiber bundles aligned perpendicular to the interface. Thus, longer incubation time results in membranes with thicker regions of perpendicularly aligned nanofibers. Further studies showed that the mechanical properties and water permeability of membranes also depend on incubation time as well as solution concentrations.92 Since the membrane self-assembly mechanism is sensitive to osmotic pressure and ion flux, electric fields have been used to control membrane structure and growth rate.93
Fig. 8.
Schematic representation of the assembly of molecules containing a coiled coil segment, flanked by a polyethylene oxide segment and cationic spermine segment into mushroom-shaped nanostructures which then assemble into a hierarchical structure on a double stranded DNA template (top). (Bottom) Transmission electron micrographs of the filamentous virus-like particles templated by double stranded DNA (dsDNA) with 150 base pairs (left), 1200 base pairs (middle), and pbr322 plasmid DNA approximately 721 nm long (right). Adapted with permission from reference 90. Copyright 2013 American Chemical Society.
PA-HA hierarchical self-assembly differs from previously studied interactions between simple surfactants and polyelectrolytes94–96 in that a cohesive material with nanoscale and microscale structure, rather than complexes and precipitates in solution, can be formed without post-processing. Previous experiments suggest that the presence of high aspect ratio PA nanofibers in solution are required for diffusion barrier assembly and aligned nanofiber growth.97 In contrast, membranes formed with PAs that form only spherical micelles exhibit an afibrous morphology with cubic phase symmetry and nanoscale periodicities.98 The difference between these two morphologies impacts significantly on membrane properties. To illustrate this point, membranes have been prepared using nanostructures containing two PAs, one containing a peptide sequence for cell lysis as a potential cancer therapy (KLAKLAK)2 and the second a non-bioactive diluent.97 When nanostructures with a low proportion of anti-cancer PA to diluent PA are used, membranes exhibit the previously described aligned-fiber morphology. When nanostructures with a high proportion of anti-cancer PA to diluent PA are used, membranes exhibit the afibrous morphology. The spherical vs. nanofiber aggregation state in these two different systems is the result of the high positive charge and steric bulkiness of (KLAKLAK)2 sequences. In the context of anti-cancer activity, it has been found that cytotoxic compounds are only released upon enzymatic degradation in the case of afibrous membranes formed by PA spherical micelles. Aligned-fiber membranes formed by PA nanofibers do not release anti-cancer products upon enzymatic degradation but can kill cancer cells through direct contact. This example highlights the cascading effects that molecular structure in relatively simple molecules can have on nanoscale morphology, microscale morphology, and bulk properties in hierarchical structures formed by self-assembly.
The dynamic self-assembly of aligned-fiber membranes is surprisingly tolerant towards different molecular components, provided that certain conditions are met. As described previously, formation of this membrane morphology requires the use of PAs capable of nanofiber assembly. Because charge complexation initiates membrane self-assembly, both PA and polyelectrolyte components should have zeta potentials above a threshold level.9 Furthermore, the osmotic pressure of the polyelectrolyte solution should initially be higher than that of the PA solution in order to drive growth of the aligned nanofibers oriented perpendicular to the membrane plane. When these essential requirements are met, aligned-fiber membranes have been fabricated using different PAs and polyelectrolytes (Fig. 8C). For example, alginate and λ-carrageenan have both been used to fabricate such membranes. However, these membranes show slower perpendicular fiber growth as compared to membranes formed with HA. Because studies show massive absorption of released counterions by HA after initiation of membrane self-assembly,92 slower growth rate in the case of alginate and λ-carrageenan is likely due to a lesser increase in osmotic pressure. Aligned-fiber membranes have also been formed using cationic chitosan and anionic nanofiber-forming PAs. Furthermore, aligned-fiber membranes have been successfully fabricated using a heparin-binding PA (HBPA), which does not form nanofibers in solution on its own.99 Here, incorporation of heparin in the self-assembly process facilitates PA aggregation into nanofibers. These HBPA-containing membranes were found to significantly increase angiogenesis in vitro due to enhanced growth factor retention, thus making them potentially attractive for wound healing applications. Structural investigation of these membranes revealed that a critical concentration of heparin is required to promote diffusion barrier assembly and aligned nanofiber growth. Membranes formed with lower concentration of heparin are non-fibrous and contain a nanoscale phase with cubic symmetry.
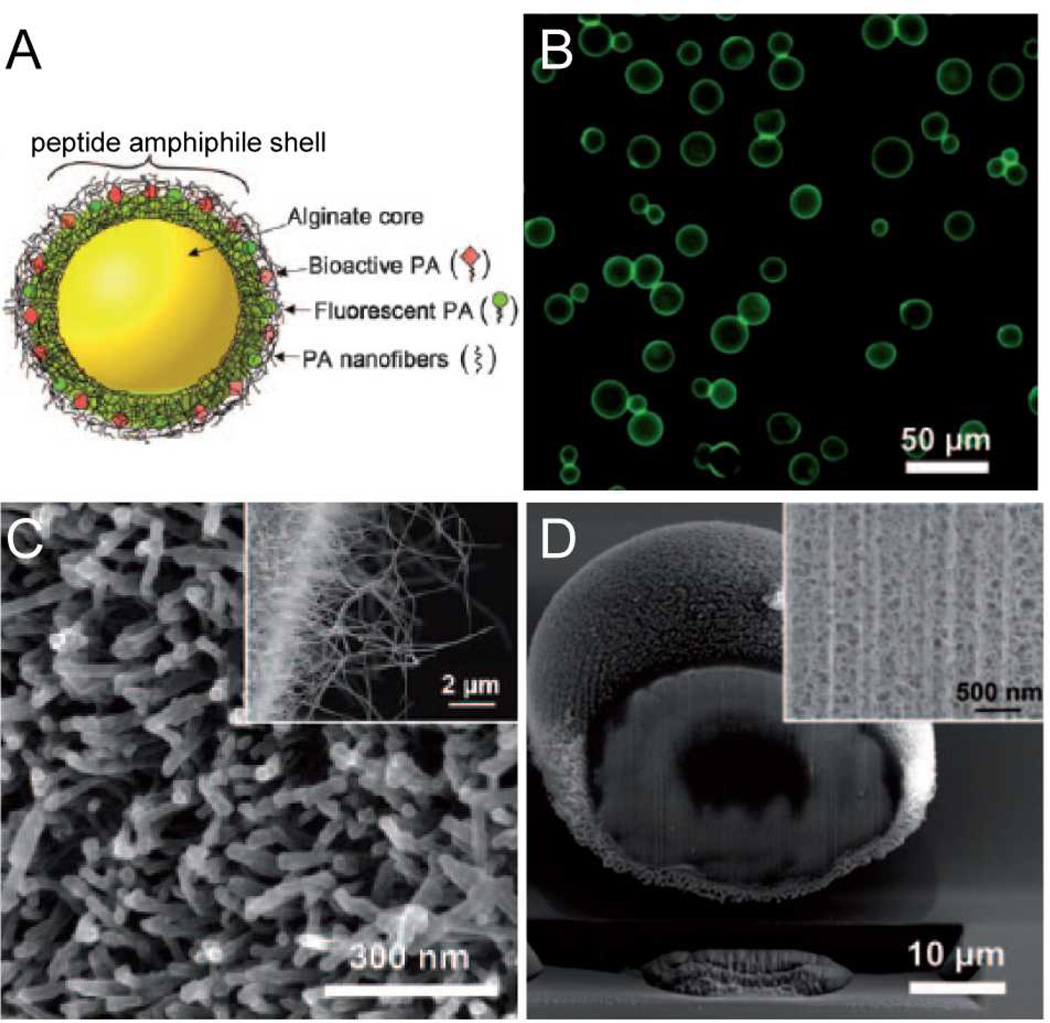
Hybrid hierarchical self-assembly between PA and polyelectrolytes has been also explored for fabrication of multifunctional drug delivery vehicles (Fig. 9). For this purpose, microcapsules less than 100 µm have been produced when nebulized microdroplets of alginate under high nitrogen pressure were are introduced into a PA solution.100 The resulting particles consisted of an alginate gel core covered on their surface with PA nanofibers. This nanofibrous shell exhibits high surface area for potential interaction with cells and can be produced with bioactive PAs. Furthermore, the nanofibrous shell is porous and allows release of compounds encapsulated within the alginate core.
Fig. 9.
(A) Scanning electron micrograph (SEM) of a membrane formed at the liquid-liquid interface between an aqueous solution of the peptide amphiphile (PA) C16VVVAAAKKK and hyaluronic acid. The SEM shows the (1) amorphous region, (2) dense diffusion barrier, and (3) nanofibers aligned perpendicular to the interface. (B) Schematic representation illustrating the mechanism of diffusion barrier formation between the PA and HA aqueous compartments and the subsequent growth of aligned nanofibers over time. (C) From left to right, SEMs of hierarchical membranes formed with the PA in (A) and aqueous solutions of 1.6 wt% of the polysaccharide λ carrageenan, 1 wt% poly(acrylic acid) (PAA), 2 wt% alginate, or 3 wt% chitosan. (B) is adapted with permission from reference 9. Copyright 2008 AAAS.
4. Reflections on the Discussion
There was broad range of topics covered by this Faraday Discussion covering peptides, peptide amphiphiles, proteins, self-assembling drug molecules, copolymers, and microtubules, among others. It was interesting to conclude from the Discussion that many common principles apply to these widely varying systems. Also one gleans from naturally occurring systems possible bio-inspired strategies that could be useful in functional synthetic systems. Safinya and co-workers, for example, reported very interesting ion specific effects on depolymerization of taxol-stabilized microtubules.101 It is fascinating that specific interactions with physiologically relevant ions can depolymerize these one-dimensional supramolecular structures even though they are stabilized by molecules well known to prevent their disassembly. Could there be similar strategies that massively disassemble amyloids? There was also the paper by Cornelissen and co-workers reporting on photo-driven cargo release from virus-like assemblies, describing the depolymerization with light of a self-immolative polymer encapsulated in a virus-like particle with proteins from the cowpea chlorotic mottle virus.102 This offers ideas on therapy delivery using vehicles built with naturally occurring proteins pre-programmed for self-assembly. From the paper by Mezzenga and co-workers, it was interesting to find that proteins such as bovine serum albumin can create the types of fibrillar structures observed in very simple short peptides, for example ribbon-like fibrils wrapping into closed tubes pointing to common basic supramolecular principles.103 At the same time Lecommandoux and co-workers reported on disc-like self-assembly of rodcoil branched biopolymers formed by a rigid poly(amino acid) and flexible oligosaccharides that have been observed in small synthetic molecules.104 Biofunctionality on such discs could be interesting in biomedical applications. Also the systems described by Cui and co-workers demonstrate the possibility of integrating drugs into self-assembling molecules that create nanostructures similar to those one might obtain from biomolecular structures.105 This way the vehicle is the drug and not cargo. Also in the context of functional systems, Guler and co-workers showed how peptide amphiphiles can expand the capabilities of the liposome.106
From the Discussion it was clear that self-assembly of peptides and peptide-containing molecules is presently at a pivotal position in the field of self-assembly of biopolymers. Papers from the groups of Jan van Hest/Lowik,107 Mazza/Kostarelos,108 Saiani,109 Adams,110 and Hamley111 demonstrated the richness of these molecules given their programmable secondary structure and also their potential biomedical translation. Strong evidence was presented from solid state NMR, x-ray diffraction, and circular dichroism for the internal arrangement of peptide amphiphiles in flat nanostructures.107 It was also clear from these papers how complex is the assembly of peptides into gels even when molecules are as simple as dipeptides and octapeptides,109, 110 and intriguing how a single amino acid in a terminal bioactive epitope of a peptide amphiphile can change the supramolecular energy landscape yielding different periodicities in the assembly.111 On the functional side, the paper by Mazza/Kostarelos showed at the Discussion that cationic peptide amphiphile nanofibers can be internalized by neurons and can also degrade in the brain.108 One intriguing point linked to this paper is the question of whether or not the supramolecular nanofibers could cross the blood brain barrier.
There are both scientific and translational challenges in the bio-inspired field of peptide self-assembly which is both fascinating and potentially of high impact on human welfare. During the Discussion a number of issues surfaced that are of fundamental importance for the field to move forward. One of them is the challenge of understanding the role of water, the solvent of living systems, in both the static and dynamic aspects of self-assembly. Water in this context is a complex solvent which must be regarded as an integral, strongly bound part of the biomolecular structures being investigated. The many configurations of water that must exist in these systems must mediate the kinetics and pathways of self-assembly, defining the stability of the resulting structures. This is related to another large issue of interest to the community gathered at the Discussion, namely the scalability and functional robustness of self-assembled systems. This aspect is clearly connected to the possible non-equilibrium states in which these systems may exist depending on self-assembly pathways and their complex energy landscapes. This connects to the use of these systems in biomedical applications as they need to be for this purpose reproducible in supramolecular structure. To make further progress along these lines, the community will need to specifically target these questions experimentally, and continue to work closely with computation and modeling to explore the energy landscapes. The paper by Aleman and co-workers at the Discussion illustrated how computational methods can contribute to the advancement of the field.112
One of the questions that intrigued participants in the Discussion is the use of peptide systems in targeted drug delivery in which the biomolecular structures could be either the therapy itself or the targeting information to a specific tissue, parts of the brain, tumors, and specific organs or connective tissues. Considering the enormous number of drug targets that are bioactive peptides, this is a particularly rich area for research. The nature of the assembly in which the therapy is delivered in such systems could be strategies to protect the bioactive peptide from rapid degradation or avoid immune response. Other areas of interest in the Discussion were the manipulation of cell functions with self-assembled systems through receptor signaling and generally their use in the regeneration of tissues and organs, an area covered in the keynote lecture.
5. Conclusions and Future Prospects
The field described here has developed rapidly over the past decade and clearly offers many opportunities for translation in biomedical or bio-inspired technologies. Among the exciting challenges ahead is to discover strategies to create dynamic systems that can respond to stimuli. This could generate biomimetic functions such as adaptability to the environment, structural transformations that change physical properties, spatio-temporal changes in biological signaling, energy transduction that mimics muscle tissue, self-healing behavior, and self-replication, among many others. The capacity for structural changes is great, for example, Hamley and coworkers very recently showed that a peptide amphiphile that self-assembles into nanotubes and ribbons can be cleaved with the enzyme α-chymotrypsin to give shorter peptides that form only spherical micelles.113 In our own recent work, we have shown that we can induce temporal changes in peptide bioactivity of a scaffold using host–guest interactions.114 Self-assembly systems that dissipate energy could also provide access to unique transformations in supramolecular architecture and chemical structure.8, 115 The future of the field in the context of biomolecular soft matter will ultimately require integrating all the capabilities of the four basic structural chemistry pillars in biological systems: peptides, sugars, lipids, and nucleobases. There are also great opportunities for creating systems that use interactions synergistically or orthogonally along with external stimuli to create hierarchically organized materials.
Fig. 10.
(A) Schematic representation of hybrid peptide amphiphile (PA) -alginate microcapsules containing a gel core and a shell of nanofibrous PA. (B) Optical micrograph showing the fluorescence of PA shells, SEM images showing the nanofibrous PA surface of the microcapsule (C) and the cross-section of the microcapsule (D). Adapted with permission from reference 100. Copyright 2011 Wiley.
Acknowledgments
Research in the authors’ laboratory on hierarchical self-assembling systems was supported by a grant from the U. S. Department of Energy, Basic Energy Sciences, DE-FG02-00ER45810, and research on bioactive peptide amphiphiles by National Institutes of Health grants NIBIB 5R01EB003806-04 and NIDCR 2R01DE015920-06. The authors are also grateful for the use of experimental facilities at the Institute for BioNanotechnology in Medicine (IBNAM), the Biological Imaging Facility (BIF), the Integrated Molecular Structure Education and Research Center (IMSERC), the Northwestern University Atomic- and Nanoscale Characterization Experimental Center (NUANCE, EPIC, NIFTI, and Keck-II) and Keck Biophysics Facilities, all at Northwestern University.
References
- 1.Aida T, Meijer EW, Stupp SI. Science. 2012;335:813–817. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer LC, Velichko YS, Olvera de la Cruz M, Stupp SI. Phil. Trans. R. Soc. A. 2007;365:1417–1433. doi: 10.1098/rsta.2007.2024. [DOI] [PubMed] [Google Scholar]
- 3.Stupp SI, LeBonheur V, Walker K, Li LS, Huggins KE, Keser M, Amstutz A. Science. 1997;276:384–389. doi: 10.1126/science.276.5311.384. [DOI] [PubMed] [Google Scholar]
- 4.Zubarev ER, Pralle MU, Sone ED, Stupp SI. J. Am. Chem. Soc. 2001;123:4105–4106. doi: 10.1021/ja015653+. [DOI] [PubMed] [Google Scholar]
- 5.Hill J, Jin W, Kosaka A, Fukushima T, Ichihara H, Shimomura T, Ito K, Hashizume T, Ishii N, Aida T. Science. 2004;304:1481–1483. doi: 10.1126/science.1097789. [DOI] [PubMed] [Google Scholar]
- 6.Hirschberg JH, Brunsveld L, Ramzi A, Vekemans JA, Sijbesma RP, Meijer EW. Nature. 2000;407:167–170. doi: 10.1038/35025027. [DOI] [PubMed] [Google Scholar]
- 7.Messmore BW, Sukerkar PA, Stupp SI. J. Am. Chem. Soc. 2005;127:7992–7993. doi: 10.1021/ja051183y. [DOI] [PubMed] [Google Scholar]
- 8.Boekhoven J, Poolman JM, Maity C, Li F, van der Mee L, Minkenberg CB, Mendes E, van Esch JH, Eelkema R. Nat. Chem. 2013;5:433–437. doi: 10.1038/nchem.1617. [DOI] [PubMed] [Google Scholar]
- 9.Capito RM, Azevedo HS, Velichko YS, Mata A, Stupp SI. Science. 2008;319:1812–1816. doi: 10.1126/science.1154586. [DOI] [PubMed] [Google Scholar]
- 10.van Hest JCM, Tirrell DA. Chem. Commun. 2001:1897–1904. doi: 10.1039/b105185g. [DOI] [PubMed] [Google Scholar]
- 11.Stephanopoulos N, Ortony JH, Stupp SI. Acta Mater. 2013;61:912–930. doi: 10.1016/j.actamat.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Schultz PG. Angew. Chem. Int. Ed. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 14.Ngo JT, Schuman EM, Tirrell DA. Proc. Natl. Acad. Sci. U.S.A. 2013 doi: 10.1073/pnas.1216375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 16.Zheng J, Birktoft JJ, Chen Y, Wang T, Sha R, Constantinou PE, Ginell SL, Mao C, Seeman NC. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng J, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinheiro AV, Han D, Shih WM, Yan H. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson HW, Yamaguchi N, Jones JW. J. Am. Chem. Soc. 2003;125:3522–3533. doi: 10.1021/ja020900a. [DOI] [PubMed] [Google Scholar]
- 20.Plante OJ, Palmacci ER, Seeberger PH. Science. 2001;291:1523–1527. doi: 10.1126/science.1057324. [DOI] [PubMed] [Google Scholar]
- 21.Calin O, Eller S, Seeberger PH. Angew. Chem. Int. Ed. 2013;52:5862–5865. doi: 10.1002/anie.201210176. [DOI] [PubMed] [Google Scholar]
- 22.Zhang S, Lockshin C, Cook R, Rich A. Biopolymers. 1994;34:663–672. doi: 10.1002/bip.360340508. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Biomaterials. 1995;16:1385–1393. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- 24.Aggeli A, Nyrkova IA, Bell M, Harding R, Carrick L, McLeish TC, Semenov AN, Boden N. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11857–11862. doi: 10.1073/pnas.191250198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maude S, Ingham E, Aggeli A. Nanomedicine. 2013;8:823–847. doi: 10.2217/nnm.13.65. [DOI] [PubMed] [Google Scholar]
- 26.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berndt P, Fields GB, Tirrell M. J. Am. Chem. Soc. 1995;117:9515–9522. [Google Scholar]
- 28.Trent A, Marullo R, Lin B, Black M, Tirrell M. Soft Matter. 2011;7:9572–9582. [Google Scholar]
- 29.Sayar M, Stupp SI. Phys. Rev. E. 2005;72:011803. doi: 10.1103/PhysRevE.72.011803. [DOI] [PubMed] [Google Scholar]
- 30.Palmer LC, Stupp SI. Acc. Chem. Res. 2008;41:1674–1684. doi: 10.1021/ar8000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Greef TFA, Smulders MMJ, Wolffs M, Schenning APHJ, Sijbesma RP, Meijer EW. Chem. Rev. 2009;109:5687–5754. doi: 10.1021/cr900181u. [DOI] [PubMed] [Google Scholar]
- 32.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 33.Schenning APHJ, Peeters E, Meijer EW. J. Am. Chem. Soc. 2000;122:4489–4495. [Google Scholar]
- 34.Zubarev ER, Sone ED, Stupp SI. Chem. Eur. J. 2006;12:7313–7327. doi: 10.1002/chem.200600619. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Fukushima T, Suna Y, Ishii N, Saeki A, Seki S, Tagawa S, Taniguchi M, Kawai T, Aida T. Science. 2006;314:1761–1764. doi: 10.1126/science.1134441. [DOI] [PubMed] [Google Scholar]
- 36.Matson JB, Zha RH, Stupp SI. Curr. Opin. Solid State Mater. Sci. 2011;15:225–235. doi: 10.1016/j.cossms.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fairman R, Akerfeldt KS. Curr. Opin. Struct. Bio. 2005;15:453–463. doi: 10.1016/j.sbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Webber MJ, Kessler JA, Stupp SI. J. Intern. Med. 2010;267:71–88. doi: 10.1111/j.1365-2796.2009.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui H, Webber MJ, Stupp SI. Pept. Sci. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webber MJ, Berns EJ, Stupp SI. Isr. J. Chem. 2013;53:530–554. doi: 10.1002/ijch.201300046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velichko YS, Stupp SI, Olvera de la Cruz M. J. Phys. Chem. B. 2008;112:2326–2334. doi: 10.1021/jp074420n. [DOI] [PubMed] [Google Scholar]
- 42.Hartgerink JD, Beniash E, Stupp SI. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5133–5138. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenfield MA, Hoffman JR, Olvera de la Cruz M, Stupp SI. Langmuir. 2009;26:3641–3647. doi: 10.1021/la9030969. [DOI] [PubMed] [Google Scholar]
- 44.Jiang H, Guler MO, Stupp SI. Soft Matter. 2007;3:454–462. doi: 10.1039/b614426h. [DOI] [PubMed] [Google Scholar]
- 45.Paramonov SE, Jun HW, Hartgerink JD. J. Am. Chem. Soc. 2006;128:7291–7298. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 46.Pashuck ET, Cui H, Stupp SI. J. Am. Chem. Soc. 2010;132:6041–6046. doi: 10.1021/ja908560n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niece KL, Czeisler C, Sahni V, Tysseling-Mattiace V, Pashuck ET, Kessler JA, Stupp SI. Biomaterials. 2008;29:4501–4509. doi: 10.1016/j.biomaterials.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu L, Cvetanovich GL, Stupp SI. J. Am. Chem. Soc. 2008;130:3892–3899. doi: 10.1021/ja076553s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tovar JD, Claussen RC, Stupp SI. J. Am. Chem. Soc. 2005;127:7337–7345. doi: 10.1021/ja043764d. [DOI] [PubMed] [Google Scholar]
- 50.Webber MJ, Newcomb CJ, Bitton R, Stupp SI. Soft Matter. 2011;7:9665–9672. doi: 10.1039/c1sm05610g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 52.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. J. Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sur S, Pashuck ET, Guler MO, Ito M, Stupp SI, Launey T. Biomaterials. 2012;33:545–555. doi: 10.1016/j.biomaterials.2011.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tysseling VM, Sahni V, Pashuck ET, Birch D, Hebert A, Czeisler C, Stupp SI, Kessler JA. J. Neurosci. Res. 2010;88:3161–3170. doi: 10.1002/jnr.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angeloni NL, Bond CW, Tang Y, Harrington DA, Zhang S, Stupp SI, McKenna KE, Podlasek CA. Biomaterials. 2011;32:1091–1101. doi: 10.1016/j.biomaterials.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah RN, Shah NA, Lim DRMM, Hsieh C, Nuber G, Stupp SI. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3293–3298. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee SS, Huang BJ, Kaltz SR, Sur S, Newcomb CJ, Stock SR, Shah RN, Stupp SI. Biomaterials. 2012;34:452–459. doi: 10.1016/j.biomaterials.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mata A, Geng Y, Henrikson KJ, Aparicio C, Stock SR, Satcher RL, Stupp SI. Biomaterials. 2010;31:6004–6012. doi: 10.1016/j.biomaterials.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Chem. Rev. 2008;108:4754–4783. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spoerke ED, Murray NG, Li H, Brinson LC, Dunand DC, Stupp SI. Acta Biomater. 2005;1:523–533. doi: 10.1016/j.actbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 61.Huang Z, Newcomb CJ, Bringas P, Jr, Stupp SI, Snead ML. Biomaterials. 2010;31:9202–9211. doi: 10.1016/j.biomaterials.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Newcomb CJ, Bitton R, Velichko YS, Snead ML, Stupp SI. Small. 2012;8:2195–2202. doi: 10.1002/smll.201102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stendahl JC, Wang L-J, Chow LW, Kaufman DB, Stupp SI. Transplantation. 2008;86:478–481. doi: 10.1097/TP.0b013e3181806d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webber MJ, Han X, Murthy SNP, Rajangam K, Stupp SI, Lomasney JW. J. Tissue Eng. Regen. Med. 2010;4:600–610. doi: 10.1002/term.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webber MJ, Tongers Jr, Newcomb CJ, Marquardt K-T, Bauersachs J, Losordo DW, Stupp SI. Proc. Natl. Acad. Sci. U.S.A. 2011 doi: 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soukasene S, Toft DJ, Moyer TJ, Lu H, Lee H-K, Standley SM, Cryns VL, Stupp SI. ACS Nano. 2011;5:9113–9121. doi: 10.1021/nn203343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Standley SM, Toft DJ, Cheng H, Soukasene S, Chen J, Raja SM, Band V, Band H, Cryns VL, Stupp SI. Cancer Res. 2010;70:3020–3026. doi: 10.1158/0008-5472.CAN-09-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toft DJ, Moyer TJ, Standley SM, Ruff Y, Ugolkov A, Stupp SI, Cryns VL. ACS Nano. 2012;6:7956–7965. doi: 10.1021/nn302503s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajangam K, Behanna HA, Hui MJ, Han X, Hulvat JF, Lomasney JW, Stupp SI. Nano Lett. 2006;6:2086–2090. doi: 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- 70.Matson JB, Webber MJ, Tamboli VK, Weber B, Stupp SI. Soft Matter. 2012;8:2689–2692. doi: 10.1039/C2SM25785H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Webber MJ, Matson JB, Tamboli VK, Stupp SI. Biomaterials. 2012;33:6823–6832. doi: 10.1016/j.biomaterials.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Proc. Natl. Acad. Sci. U.S.A. 2010 doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guler MO, Hsu L, Soukasene S, Harrington DA, Hulvat JF, Stupp SI. Biomacromolecules. 2006;7:1855–1863. doi: 10.1021/bm060161g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niece KL, Hartgerink JD, Donners JJJM, Stupp SI. J. Am. Chem. Soc. 2003;125:7146–7147. doi: 10.1021/ja028215r. [DOI] [PubMed] [Google Scholar]
- 75.Beniash E, Hartgerink JD, Storrie H, Stendahl JC, Stupp SI. Acta Biomater. 2005;1:387–397. doi: 10.1016/j.actbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 76.Khan S, Sur S, Newcomb CJ, Appelt EA, Stupp SI. Acta Biomater. 2012;8:1685–1692. doi: 10.1016/j.actbio.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Behanna HA, Donners JJJM, Gordon AC, Stupp SI. J. Am. Chem. Soc. 2005;127:1193–1200. doi: 10.1021/ja044863u. [DOI] [PubMed] [Google Scholar]
- 78.Cui H, Muraoka T, Cheetham AG, Stupp SI. Nano Lett. 2009;9:945–951. doi: 10.1021/nl802813f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goldberger JE, Berns EJ, Bitton R, Newcomb CJ, Stupp SI. Angew. Chem. Int. Ed. 2011;50:6292–6295. doi: 10.1002/anie.201100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moyer TJ, Cui H, Stupp SI. J. Phys. Chem. B. 2013;117:4604–4610. doi: 10.1021/jp3087978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pashuck ET, Stupp SI. J. Am. Chem. Soc. 2010;132:8819–8821. doi: 10.1021/ja100613w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lehn J-M. Angew. Chem. Int. Ed. 2013;52:2836–2850. doi: 10.1002/anie.201208397. [DOI] [PubMed] [Google Scholar]
- 83.Wedeen VJ, Rosene DL, Wang R, Dai G, Mortazavi F, Hagmann P, Kaas JH, Tseng W-YI. Science. 2012;335:1628–1634. doi: 10.1126/science.1215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui H, Pashuck ET, Velichko YS, Weigand SJ, Cheetham AG, Newcomb CJ, Stupp SI. Science. 2010;327:555–559. doi: 10.1126/science.1182340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Needleman DJ, Ojeda-Lopez MA, Raviv U, Ewert K, Jones JB, Miller HP, Wilson L, Safinya CR. Physical Review Letters. 2004;93 doi: 10.1103/PhysRevLett.93.198104. [DOI] [PubMed] [Google Scholar]
- 86.Needleman DJ, Ojeda-Lopez MA, Raviv U, Miller HP, Wilson L, Safinya CR. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16099–16103. doi: 10.1073/pnas.0406076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang S, Greenfield MA, Mata A, Palmer LC, Bitton R, Mantei JR, Aparicio C, de La Cruz MO, Stupp SI. Nat. Mater. 2010;9:594–601. doi: 10.1038/nmat2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berns EJ, Sur S, Pan L, Goldberger JE, Suresh S, Zhang S, Kessler JA, Stupp SI. Biomaterials. 2014;35:185–195. doi: 10.1016/j.biomaterials.2013.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klug A. Philos. Trans. R. Soc. Lond., B. 1999;354:531–535. doi: 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruff Y, Moyer T, Newcomb CJ, Demeler B, Stupp SI. J. Am. Chem. Soc. 2013;135:6211–6219. doi: 10.1021/ja4008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kiani C, Chen L, Wu YJ, Yee AJ, Yang BB. Cell Res. 2002;12:19–32. doi: 10.1038/sj.cr.7290106. [DOI] [PubMed] [Google Scholar]
- 92.Carvajal D, Bitton R, Mantei JR, Velichko YS, Stupp SI, Shull KR. Soft Matter. 2010;6:1816–1823. [Google Scholar]
- 93.Velichko YS, Mantei JR, Bitton R, Carvajal D, Shull KR, Stupp SI. Adv. Funct. Mater. 2012;22:369–377. doi: 10.1002/adfm.201101538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Langevin D. Adv. Colloid Interface Sci. 2009;147–148:170–177. doi: 10.1016/j.cis.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 95.Thalberg K, Lindman B. J. Phys. Chem. 1989;93:1478–1483. [Google Scholar]
- 96.Zhou SQ, Chu B. Adv. Mater. 2000;12:545–556. [Google Scholar]
- 97.Zha RH, Sur S, Stupp SI. Adv. Healthcare Mater. 2013;2:126–133. doi: 10.1002/adhm.201200118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bitton R, Chow LW, Zha RH, Velichko YS, Pashuck ET, Stupp SI. Small. 2013 doi: 10.1002/smll.201300254. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chow LW, Bitton R, Webber MJ, Carvajal D, Shull KR, Sharma AK, Stupp SI. Biomaterials. 2011;32:1574–1582. doi: 10.1016/j.biomaterials.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rożkiewicz DI, Myers BD, Stupp SI. Angew. Chem. Int. Ed. 2011;50:6324–6327. doi: 10.1002/anie.201100821. [DOI] [PubMed] [Google Scholar]
- 101.Needleman DJ, Ojeda-Lopez MA, Raviv U, Miller HP, Li Y, Song C, Feinstein SC, Wilson L, Choi MC, Safinya CR. Faraday Discussions. 2013 doi: 10.1039/c3fd00063j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brasch M, Voets IK, Koay MST, Cornelissen JJLM. Faraday Discussions. 2013 doi: 10.1039/c3fd00088e. [DOI] [PubMed] [Google Scholar]
- 103.Usov I, Adamcik J, Mezzenga R. Faraday Discussions. 2013 doi: 10.1039/c3fd00083d. [DOI] [PubMed] [Google Scholar]
- 104.Bonduelle C, Mazzaferro S, Huang J, Lambert O, Heise A, Lecommandoux S. Faraday Discussions. 2013 doi: 10.1039/c3fd00082f. [DOI] [PubMed] [Google Scholar]
- 105.Lock LL, LaComb M, Schwarz K, Cheetham AG, Lin Y-a, Zhang P, Cui H. Faraday Discussions. 2013 doi: 10.1039/c3fd00099k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sardan M, Kilinc M, Genc R, Tekinay AB, Guler MO. Faraday Discussions. 2013 doi: 10.1039/c3fd00058c. [DOI] [PubMed] [Google Scholar]
- 107.Nieuwland M, Ruizendaal L, Brinkmann A, Kroon-Batenburg L, van Hest JCM, Lowik DWPM. Faraday Discussions. 2013 doi: 10.1039/c3fd00055a. [DOI] [PubMed] [Google Scholar]
- 108.Mazza M, Patel A, Pons R, Bussy C, Kostarelos K. Faraday Discussions. 2013 doi: 10.1039/c3fd00100h. [DOI] [PubMed] [Google Scholar]
- 109.Boothroyd S, Miller AF, Saiani A. Faraday Discussions. 2013 doi: 10.1039/c3fd00097d. [DOI] [PubMed] [Google Scholar]
- 110.Cardoso AZ, Alvarez Alvarez AE, Cattoz BN, Griffiths PC, King SM, Frith WJ, Adams DJ. Faraday Discussions. 2013 doi: 10.1039/c3fd00104k. [DOI] [PubMed] [Google Scholar]
- 111.Castelletto V, Gouveia RM, Connon CJ, Hamley IW. Faraday Discussions. 2013 doi: 10.1039/c3fd00064h. [DOI] [PubMed] [Google Scholar]
- 112.Bertran O, Curco D, Zanuy D, Aleman C. Faraday Discussions. 2013 doi: 10.1039/c3fd00079f. [DOI] [PubMed] [Google Scholar]
- 113.Dehsorkhi A, Hamley IW, Seitsonen J, Ruokolainen J. Langmuir. 2013;29:6665–6672. doi: 10.1021/la401025r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Boekhoven J, Rubert Pérez CM, Sur S, Worthy A, Stupp SI. Angew. Chem. Int. Ed. 2013;52:12077–12080. doi: 10.1002/anie.201306278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fialkowski M, Bishop KJM, Klajn R, Smoukov SK, Campbell CJ, Grzybowski BA. J. Phys. Chem. B. 2006;110:2482–2496. doi: 10.1021/jp054153q. [DOI] [PubMed] [Google Scholar]