Abstract
Background
Asthma is a common, chronic inflammatory airway disease of major public health importance with multiple genetic determinants. Previously, we found by positional cloning that PHD finger protein 11 (PHF11) on chromosome 13q14 modifies serum immunoglobulin E (IgE) concentrations and asthma susceptibility. No coding variants in PHF11 were identified.
Objective
Here we investigate the 3 single nucleotide polymorphisms (SNPs) in this gene most significantly associated with total serum IgE levels—rs3765526, rs9526569, and rs1046295—for a role in transcription factor binding.
Methods
We used electrophoretic mobility shift assays to examine the effect of the 3 SNPs on transcription factor binding in 3 cell lines relevant to asthma pathogenesis. Relative preferential expression of alleles was investigated by using the allelotyping method.
Results
Electrophoretic mobility shift assays show that rs1046295 modulates allele-specific binding by the octamer-binding transcription factor 1 (Oct-1). Analysis of the relative expression levels of the 2 alleles of this SNP in heterozygous individuals showed a modest, but highly significant (P = 6.5 × 10−16), preferential expression of the A allele consistent with a functional role for rs1046295.
Conclusion
These results suggest a mechanism by which rs1046295 may act as a regulatory variant modulating transcription at this locus and altering asthma susceptibility.
Keywords: Asthma genetics, PHF11, IgE, rs1046295, electrophoretic mobility shift assay, Oct-1, gene expression
Asthma is a common, chronic respiratory disorder, affecting an estimated 300 million people worldwide.1-6 Symptoms include coughing, wheezing, and breathlessness. Asthma is accompanied by bronchial hyperresponsiveness, excess mucus production, and raised IgE levels that in part respond to common environmental allergens.7-9 Twin studies have shown that a genetic component accounts for 36% to 79% of asthma heritability,10,11 and 6 novel susceptibility genes have been identified by positional cloning.12-17 More recently, several groups have also performed genome-wide association studies for asthma or asthma related phenotypes. These have identified new candidate genes and loci including ORM1-like 3 (ORMDL3),18 phosphodiesterase 4D cAMP-specific (PDE4D),19 chitinase 3-like 1 (CHI3L1),20 catenin alpha 3 (CTNNA3),21 Fc fragment of IgE high affinity 1 receptor for alpha polypeptide (FCER1A),22 chromosome 5q23.2,23 and chromosome 18.24 Although none of the 6 positionally cloned asthma genes has been identified in genome-wide studies, the genes remain of importance in identifying pathways underlying asthma susceptibility.
One of these positionally cloned asthma genes, PHF11, is located at chromosome 13q14 in the human genome and is flanked by SET domain bifurcated 2 (SETDB2) and regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 (RCBTB1).13 The region was original identified by Daniels et al7 as showing linkage with atopy, an IgE-mediated state. Wiltshire et al25 showed linkage of the same region with IgA levels, and 9 further studies have indicated the presence of a gene or genes influencing asthma susceptibility or its associated traits in this region in both Caucasian and Japanese subjects.26-35 Bhattacharyya et al36 created a saturated genetic map of the chromosome 13q14 region. Yeast artificial chromosome, bacterial artificial chromosome, and P1 artificial chromosome contigs of the resulting linkage region on chromosome 13 were created, covering 4 Mb, and 31 expressed sequence tags mapped within the contigs.37 Association between the USAT24G1 microsatellite marker in this region and total serum IgE was found in an Australian population and replicated in a British population.37 Because of the size of the subject panels used, the limit of detection of linkage disequilibrium was anticipated to be less than 100 Kb, indicating that a disease gene for asthma was likely to be located within 100 Kb of either side of USAT24G1.37
Zhang et al13 created a dense single nucleotide polymorphism (SNP) map consisting of 53 markers in the region of USAT24G1. Association of polymorphisms with total serum IgE levels in an Australian population was found to center on PHF11 and extend outward into the 2 flanking genes, SETDB2 and RCBTB1. The 3 SNPs most strongly associated with IgE levels (rs3765526 [formerly called b4_2], P = .002; rs9526569 [b5_2], P = .0005; rs1046295 [b5_3], P =.001) were located within PHF11, indicating that this was the most probable asthma susceptibility gene at the locus. Stepwise analysis indicated that these 3 SNPs had independent effects on total IgE levels. The association to PHF11 was confirmed by using haplotype analysis, including SNPs rs3765526 and rs1046295, in an extended Australian population, and replicated in an independent set of subjects from the United Kingdom. PHF11 was also shown to be expressed in B cells and immune-related tissues, and Northern blots identified a possible combined SETDB2-PHF11 transcript. It was predicted that both PHF11 and SETDB2 would regulate transcription.13 Despite extensive resequencing, no nonsynonymous polymorphisms have been identified within PHF11.
Recently, an association was reported by Jang et al38 for 2 SNPs in PHF11 and childhood atopic dermatitis. The work was performed in a population of Australian children by using 7 SNPs located in SETDB2 and PHF11. The 4 PHF11 polymorphisms genotyped, rs2031532, rs2247119, rs2274276, and rs1046295, were previously referred to as 154016_2R, ren_in2, b4_3, and b5_3, respectively, by Zhang et al.13 The SNPs showing association to childhood atopic dermatitis were rs2247119 (P = .029) and rs1046295 (P = .007).38 In the work of Zhang et al,13 1 of these SNPs, rs1046295, also showed significant association to total serum IgE levels (P = .001). A second study investigating association to asthma susceptibility in the Chinese Han population also found significant association with 2 PHF11 SNPs, including rs1046295 (P = .0096).39 These studies confirm the association of PHF11 with atopic disease.
We sought to characterize the functional significance of the 3 SNPs in PHF11 most strongly associated with total serum IgE levels in our populations (rs3765526, rs9526569, and rs1046295). These SNPs are located in intron 5, intron 9, and the 3′ untranslated region (UTR) of PHF1113 (Fig 1). In the current investigation, we used electrophoretic mobility shift assay (EMSA) experiments to investigate whether transcription factor binding is affected by rs3765526, rs9526569, and rs1046295 and show that this is the case for rs1046295. Subsequently, by using this SNP, we show evidence of significant allele-specific differences in gene expression consistent with rs1046295 acting as a regulatory variant modulating PHF11 transcription.
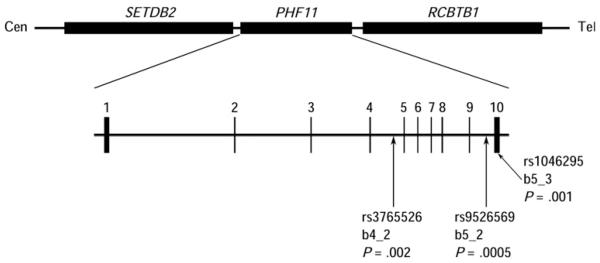
FIG 1. Gene structure of PHF11.
Figure drawn approximately to scale. The locations of the genes according to the Human March 2006 assembly are as follows: SETDB2: chr13:48,916,511-48,964,299; PHF11: chr13:48,967,802-49,001,118; RCBTB1: chr13:49,004,083-49,057,720. The detailed structure of PHF11 is shown with exons numbered. The positions of SNPs rs3765526, rs9526569, and rs1046295 are indicated, together with their names and P values from the study by Zhang et al.13
METHODS
Subjects
Allele-specific expression analysis was performed in a subset of the MRC-A population, MRC-A/RNA. The MRC-A panel consists of 200 families of British descent recruited through a proband between the ages of 5 and 18 years with severe asthma, defined as 1 or more hospital admissions caused by asthma and taking maintenance-inhaled steroids. Both parents and at least 1 sibling were included for each family, regardless of disease status. EBV immortalized lymphoblastoid cell lines were created from blood taken from all the children in the population. The MRC-A/RNA cohort is a subset of the MRC-A population. It is made up of 94 children from the 49 sibling pairs selected from the 200 families on the basis of being the most highly discordant for IgE levels. The EBV cell lines provided a source of both DNA and RNA from these individuals. Ethical approval was given by the United Kingdom Multicentre Research Ethics Committee. All subjects or their parents gave written informed consent to the study.
Genotyping
Genotyping was performed by PCR and restriction fragment length polymorphism analysis as detailed previously13 for 3 PHF11 SNPs: rs3765526, rs9526569 and rs1046295. Details of primers are available on request.
Transcription factor binding search programs
Putative transcription factor binding sites in the sequences spanning rs3765526, rs9526569, and rs1046295 were identified by using the programs TFSEARCH,40 TFSCAN,41,42 and MatInspector (Genomatix Software GmbH, Munich, Germany).43 Programs were run using default settings.
Cell culture and nuclear protein extraction
Daudi (male B lymphoblast), CCL-114 (male B lymphoblast), CCL-159 (female B lymphoblast), Calu-3 (airway epithelial), and BEAS-2B (bronchial epithelial) cell lines were obtained from the American Type Culture Collection. Both B lymphoblasts and the airway epithelium are of importance in asthma pathogenesis, making these relevant cell types for EMSA investigation. In addition, PHF11 has been shown to be expressed in lymphocytes and lung tissue.13 Daudi, CCL-114, and CCL-159 cells were cultured in RPMI 1640 media (Sigma, Gillingham, United Kingdom [UK]) supplemented to contain final concentrations of 10% FBS (Sigma), 10 mmol/L HEPES (Sigma), 1 mmol/L sodium pyruvate (Sigma), 4.5 g/L glucose (Sigma), 2 mmol/L L-glutamine (Sigma), 20 U/mL penicillin, and 0.1 mg/mL streptomycin (Sigma). Calu-3 cells were grown in modified Eagle medium (Sigma) supplemented with 10% FBS, 1 mmol/L sodium pyruvate, 2 mmol/L L-glutamine, 20 U/mL penicillin, and 0.1 mg/mL streptomycin. BEAS-2B cells were cultured in Keratinocyte-SFM (Gibco-BRL, Paisley, UK) media supplemented with 2 mmol/L L-glutamine, 5 ng/mL epidermal growth factor (Gibco-BRL), 50 μg/mL bovine pituitary extract (Gibco-BRL), 20 U/mL penicillin, and 0.1 mg/mL streptomycin. All cell lines were cultured at 37°C and 5% CO2. Nuclear extracts were prepared by using a modified Schreiber protocol44 and quantified by using the Bradford assay.45
Electrophoretic mobility shift assays
For rs3765526, rs9526569, and rs1046295, sense and antisense single-stranded oligonucleotides for both alleles were designed and synthesized (Eurofins MWG Operon, Ebersberg, Germany). Each oligonucleotide consisted of the SNP allele and 15 bases of 5′ and 3′ flanking sequence. In addition, an AGCT tag was added to the 5′ end of each oligonucleotide to facilitate subsequent radiolabeling (rs3765526 AGCTGAAAGGTAAACATTT[A/G]TGTAACATAAGGAAT; rs9526569 AGCTAATGAAACCAAAAGC[C/T]TTTATGTAACATTAA; rs1046295 AGCTTTATTAGGGATTACC[A/G]TTTCCTAAGCCAAGA). Consensus oligonucleotides for Oct-1 (AGCTAGTGCTATATGCAAATTTTATTAGCATGCTA) and sex determining region Y (SRY) (AGCTAGTGCTTAAAACAAAAGCATGCTAGCTA) were obtained from Invitrogen (Paisley, UK). The oligonucleotides were annealed to form probes for each allele of each SNP and labeled with α-32P CTP (Amersham Biosciences, Little Chalfont, UK) by using the Klenow fragment46 (New England Biolabs, Hitchin, UK). Typically, EMSA binding reactions contained 3 to 5 μg nuclear extract, 2 μL radiolabeled probe (40 counts per second/μL), 1× binding buffer (Promega, Hampshire, UK), 0.05 to 0.1 mg/mL poly(dI-dC).poly(dI-dC) (Sigma) and 4% glycerol (Sigma) in a total reaction volume of 15 μL. For competition assays, reactions included unlabeled competitor probe at a concentration of ×10, ×50, ×75, or ×100 that of the labeled probe. For supershift assays, 2 μg antibody was added to the reaction before the first incubation step. The antibodies used for supershift assays were C/EBPβ (Santa Cruz Biotechnology, Santa Cruz, Calif), Oct-1 (Santa Cruz Biotechnology), YY1 (Santa Cruz Biotechnology), SRY (Santa Cruz Biotechnology), c-Myb (Santa Cruz Biotechnology), CDX (Chemicon, Billerica, Mass), and CDX2 (Abcam, Cambridge, UK).
Reactions were incubated without labeled probe for 10 minutes at room temperature. On addition of the labeled probe, reactions were incubated for a further 20 minutes at room temperature. Products were run on a 6% nondenaturing polyacrylamide gel at 4°C and 100 V for approximately 2 hours using 0.5× TBE run buffer before visualizing by exposure to Kodak X-Omat AR film (Sigma).
Purified recombinant Oct-1 (POU2F1) and SRY proteins were obtained from Abnova (Taipei City, Taiwan). EMSAs performed by using recombinant proteins included 1 μg BSA per reaction.
DNA and cDNA preparation
EBV transformed cells were cultured in 500 mL roller cultures. Once log phase was reached, cells were pelleted, media discarded, and a mixture of RLT buffer (Qiagen, Crawley, UK) and β-mercaptoethanol (Sigma) added. Pellets were vortexed to ensure thorough resuspension, after which they were frozen at −70°C. After defrosting at 37°C in a water bath, RNA was extracted in batches by using a homogenizer and the RNeasy Maxi Kit (Qiagen), following the manufacturers’ protocol. Quality was assessed by using a bioanalyzer (Agilent Technologies, Wokingham, UK) and quantified on a spectrophotometer.
Ten micrograms of RNA was used to synthesize double-stranded cDNA by using the One-cycle cDNA synthesis kit (Affymetrix, Santa Clara, Calif).
Allelotyping
Allele peak areas in Sequenom hME assay (Sequenom, San Diego, Calif) mass spectra are directly proportional to the amount of starting material in the assay. Therefore, the peak areas provide a method of comparing the relative amounts of 2 alleles for a heterozygote in the genotyping reaction.47-49 In cDNA from a heterozygous individual equally expressing both alleles, the peak areas should have a ratio of 1:1, whereas in an individual preferentially expressing 1 allele, the ratio should differ from this. However, in genomic DNA, the ratio of alleles in a heterozygote should be 1:1, but it is known that peak areas also decrease in size with increasing mass. Therefore, to determine whether allele specific expression is occurring, the ratio of the peak areas in the genomic DNA should be compared with that in the cDNA to account for any natural bias in peak areas. If the 2 ratios differ significantly, preferential allele expression is indicated (see this article’s Fig E1).
Primers were designed by using the standard Sequenom software (sequences available on request) and obtained from Metabion (Metabion, Munich, Germany). Amplification was performed in a 5 μL reaction. Reactions contained final concentrations of reagents of 0.8 mmol/L deoxynucleotide triphosphates, 1 mmol/L MgCl2 (Bioline, London, UK), 1× PCR buffer (Bioline), 0.125 U BioTaq (Bioline), 0.2 μmol/L forward and reverse primers, and 5 ng template genomic DNA, or cDNA equivalent to 5 ng total RNA. The PCR program was as follows: 96°C for 60 seconds; 6 cycles of 94°C for 45 seconds, 60°C for 45 seconds, and 72°C for 30 seconds; 30 cycles of 94°C for 45 seconds, 65°C for 45 seconds, and 72°C for 30 seconds; and 72°C for 10 minutes. Eight replicate PCRs for each type of template DNA were performed for each subject. PCR products were prepared for mass spectrometry following the standard Sequenom method, which is a single base extension procedure in which the alleles of the variant are determined by the mass of the product via mass spectrometry.47 Allele peak areas were used for subsequent statistical analyses.
Statistical analysis of allelotyping data
Statistical analyses of the allele ratios for allelotyping were performed by using the software package Statistical Package for the Social Sciences 12.0 for Windows (SPSS Inc, Chicago, Ill). For paired sample t tests, the ratio of the peak areas from the mass spectrometry results of allele A relative to allele G in the cDNA was calculated and compared with the mean ratio in the genomic DNA for all samples to minimize the effect of any potential variation in genomic DNA ratio. Graphs describing the allelotyping results were created by using the R environment.
RESULTS
Bioinformatic analysis of transcription factor binding of 3 PHF11 SNPs
The sequences immediately flanking rs3765526, rs9526569 and rs1046295 were interrogated in silico for potential transcription factor binding sequences by using the programs TFSearch, TFScan, and MatInspector (Genomatix Software GmbH). A number of transcription factor binding sites were predicted to span each SNP with several instances of potential allele-specific binding (Table I).
TABLE I. Transcription factors predicted to have greater affinity to 1 allele of each of the rs3765526, rs9526569, and rs1046295 PHF11 polymorphisms.
| SNP | Allele | Program | Transcription factor | Strand | Binding sequence | Score |
|---|---|---|---|---|---|---|
| rs3765526 | A | TFSearch | CdxA | + | ATTTATG | 100 |
| CdxA | − | CATTTAT | 86.4 | |||
| VBP | − | TTTATGTAAC | 94.1 | |||
| E4BP4 | + | ATTTATGTAACA | 92.6 | |||
| HLF | − | TTTATGTAAC | 89.7 | |||
| CRE-BP | + | TTATGTAA | 89.0 | |||
| MatInspector | Fork head RElated ACtivator-2 | + | GAAAGGTAAACATTTAT | 0.968 | ||
| PAR-type chicken vitellogenin promoter-binding protein (avian) | − | GTTACATAAAT | 0.975 | |||
| E4BP4, bZIP domain, transcriptional repressor | + | AAACATTTATGTAACATAAGG | 0.970 | |||
| G | TFSearch | CdxA | + | ATTTGTG | 91.0 | |
| HLF | − | TTTGTGTAAC | 87.5 | |||
| C/EBPb | − | CATTTGTGTAACAT | 87.2 | |||
| TFScan | MOUSE$AAMY_01 | + | TTGTGTAAC | — | ||
| MatInspector | Fork head RElated ACtivator-2 | + | GAAAGGTAAACATTTGT | 0.968 | ||
| Hepatic leukemia factor | − | CTTATGTTACACAAATGTTTA | 0.917 | |||
| PAR-type chicken vitellogenin promoter-binding protein (avian) | + | GTTACACAAAT | 0.892 | |||
| rs9526569 | C | TFSearch | CdxA | + | CTTTATG | 96.2 |
| E4BP4 | + | CTTTA | 94.1 | |||
| MatInspector | Albumin D-box binding protein | + | AGCCTTTATGTAACA | 0.841 | ||
| Hepatic leukemia factor | − | TTAATGTTACATAAAGGCTTT | 0.932 | |||
| PAR-type chicken vitellogenin promoter-binding protein (avian) | − | GTTACATAAAG | 0.975 | |||
| T | TFSearch | E4BP4 | + | TTTTATGTAACA | 92.2 | |
| CdxA | + | TTTTATG | 92.1 | |||
| MatInspector | Albumin D-box binding protein | + | AGCTTTTATGTAACA | 0.858 | ||
| Hepatic leukemia factor | − | TTAATGTTACATAAAAGCTTT | 0.932 | |||
| PAR-type chicken vitellogenin promoter-binding protein (avian) | − | GTTACATAAAA | 0.975 | |||
| rs1046295 | A | TFSearch | C/EBPb | − | CCATTTCCTAAGCC | 89.9 |
| YY1 | + | GATTACCATTTCCTAAG | 87.3 | |||
| TFScan | HS$GMCSF_04 | + | CATTT | — | ||
| MatInspector | CCAAT/enhancer binding protein beta | − | TGGCTTAGGAAATGGTAAT | 0.96 | ||
| G | TFSearch | v_Myb | − | ACCGTTTCC | 85.6 | |
| MatInspector | v-Myb (viral?) | − | GGAAACGGTAA | 0.923 |
Searches performed by using the programs MatInspector, TFSearch, and TFScan. The polymorphism is shown in boldface in the transcription factor binding sequences. The score is as reported by the search program. No scores are reported by TFScan (indicated by a dash). For MatInspector, the matrix similarity score is given; a value of 1 indicates a perfect match. For TFSearch, a score of 100 indicates a perfect match.
For rs3765526, 5 transcription factors were predicted to bind specifically only to 1 allele (allele A: VBP, E4BP4, CRE-BP; allele G: C/EBPb, Mouse$AAMY_01), with a further 3 predicted to bind with greater affinity to 1 allele than the other (allele A: CdxA, HLF, PAR-type chicken vitellogenin promoter-binding protein). No transcription factors were predicted to bind 1 allele of rs9526569 alone, although 3 were reported as binding with higher affinity to a particular allele (allele C: CdxA, E4BP4; allele T: albumin D-box binding protein). Finally, for rs1046295, 5 transcription factors were reported as binding specifically to 1 allele (allele A: C/EBPb, YY1, HS$GMCSF_04, CCAAT/enhancer binding protein β; allele G: v_Myb). We consequently tested whether the alleles affected binding in vitro by using EMSAs.
Transcription factor binding modulated in vitro by rs1046295
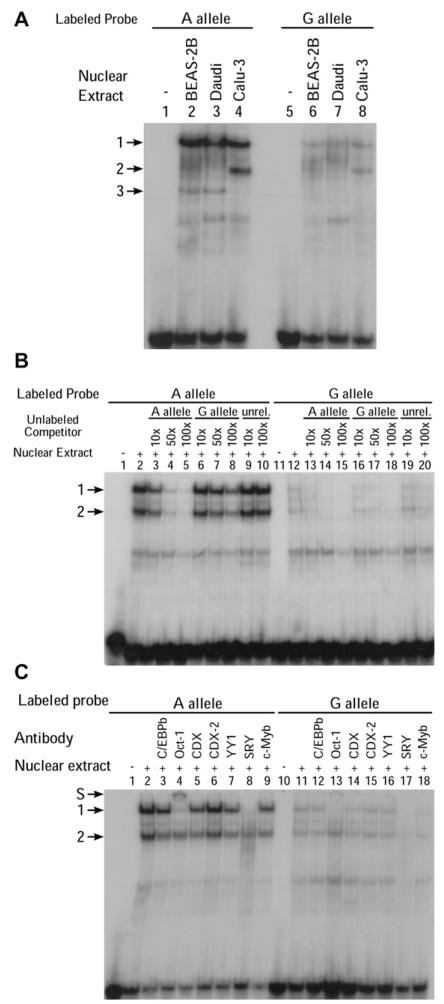
EMSAs performed for rs3765526 and rs9526569 revealed no significant allele-specific band patterns (data not shown). However, EMSAs using probes spanning rs1046295 incubated with nuclear extract from a human B-cell line (Daudi) identified 2 major complexes (denoted 1 and 2) with significantly greater affinity for allele A than allele G (Fig 2, A). The same pair of complexes was also identified in CCL-114 (male B lymphoblast) and CCL-159 (female B lymphoblast) cells (see this article’s Fig E2). This band pattern was consistent across separate experiments and independent extractions of nuclear proteins. Using nuclear extracts from human bronchial epithelial cells (BEAS-2B) or human lung adenocarcinoma cells (Calu-3), the intensity of the faster migrating complex 2 was greatly reduced; however, an additional complex was seen (denoted complex 3; Fig 2, A). We showed that complexes 1 and 2 were specific by using competition assays with molar excess of unlabeled probes corresponding to each of the 2 alleles and an unrelated probe. Significantly higher affinity for complex 1 was seen with the A versus the G allele by using this in vitro approach (Fig 2, B). Complex 3 was shown to be a result of nonspecific binding in subsequent competition assays by using Calu-3 nuclear extract (data not shown). Therefore, because the 2 specific bands of interest were strongest using Daudi nuclear extract, subsequent experiments were performed by using this cell line. Results were consistent using probes labeled and Daudi nuclear extracts from different dates.
FIG 2. Allele-specific protein-DNA interactions at PHF11 involving rs1046295.
A, EMSAs corresponding to the A or G alleles of rs1046295 with BEAS-2B, Calu-3, and Daudi nuclear extracts. B, Competition EMSAs using Daudi nuclear extract. Rs1046295 allele A (lanes 1-10) and G (lanes 11-20). Molar excesses of unlabeled competitor oligonucleotides for allele A, allele G, and an unrelated probe, rs71307668 (unrel.), indicated. C, Supershift EMSA experiments for rs1046295 using Daudi nuclear extract. Supershift indicated by S.
We proceeded to identify the transcription factors responsible for these allele-specific complexes by performing supershift assays for a number of candidate proteins including C/EBPb, YY1, Cdx, Cdx2, Oct-1, SRY, and c-Myb. The transcription factor c-Myb is the human homolog of v-Myb and was predicted to bind the G allele only. The human homolog of CdxA, Cdx1, has been proposed to be of importance in the functioning of another asthma susceptibility gene, dipeptidyl-peptidase 10 (DPP10).14 No antibody was available for Cdx1, hence the inclusion of both Cdx and Cdx2 antibodies. Anti–Oct-1 (Transfac matrix ID V$OCT1_03, consensus binding sequence RTAATNA), predicted to bind both alleles, and anti-SRY (Transfac matrix IDV$SRY_01, consensus binding sequence AAACWAM), predicted to bind neither allele, were included as positive and negative controls, respectively.
The supershift assays revealed that complex 1, which binds with much greater affinity to allele A, was abolished in the presence of antibodies for Oct-1 and SRY (Fig 2, C), indicating that these transcription factors might be part of the allele-specific DNA binding complex seen with these B-cell nuclear extracts. Because of the surprising supershift occurring with the negative control anti-SRY, the experiment was repeated, with the same result. The experiment was then repeated with 2 SRY antibodies on a separate Daudi extract and probes labeled on a separate date, and a supershift obtained with only 1 antibody (data not shown). Complex 2 was unaffected by any of the antibodies tested.
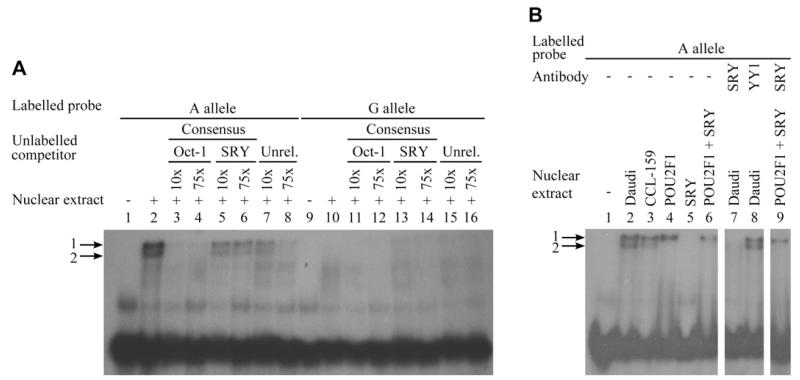
To confirm further the identity of the transcription factors resulting in band 1, 2 additional experiments were performed. A competition assay using Daudi nuclear extract and Oct-1 and SRY cold consensus sequence probes as competitors was performed. This revealed that the Oct-1 consensus sequence was able to compete efficiently against the rs1046295 probe for protein binding, reducing the intensity of bands 1 and 2, whereas the SRY consensus probe had no effect (Fig 3, A). Second, to confirm whether Oct-1 and/or SRY proteins bound the rs1046295 A allele probe, EMSAs were performed with purified recombinant proteins. This revealed that recombinant Oct-1 protein produced a band identical to the complex 1 with Daudi protein, whereas SRY produced no band shift. A reaction combining the probe, recombinant Oct-1, and recombinant SRY produced a band identical to the reaction with Oct-1 alone. Assays performed including anti-SRY confirmed the supershift with Daudi nuclear extract. However, they produced no supershift with recombinant Oct-1 and Oct-1 + SRY proteins (Fig 3, B), indicating the previous results using Daudi nuclear extract were not a result of the SRY antibody binding Oct-1.
FIG 3. Allele-specific protein-DNA interactions at PHF11 confirming the ability of only Oct-1 to bind at rs1046295.
A, Competition EMSAs using Daudi nuclear extract. Rs1046295 allele A (lanes 1-8) and G (lanes 9-16). Molar excesses of unlabeled competitor oligonucleotides for the Oct-1 and SRY consensus sequences, and an unrelated probe (Unrel.), are indicated. B, EMSAs performed for rs1046295 allele A using Daudi and CCL-159 nuclear extracts and Oct-1 (POU2F1) and SRY recombinant proteins. Addition of antibodies for SRY or YY1 (negative control) is indicated (lanes 7-9).
Allelotyping
To investigate whether the effect on transcription factor binding was associated with allele-specific differences in gene expression, we analyzed the relative expression of transcripts arising from the 2 alleles. Because rs1046295 is located in the 3′ UTR of PHF11, and therefore transcribed, we were able to compare directly the relative transcription of the 2 alleles in heterozygous individuals. To do so, we performed allelotyping50 with quantification by matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometry (Sequenom).
To demonstrate the robustness of the allelotyping method, experiments were performed for a series of control template genomic DNA samples, consisting of ratios of known homozygous individuals. Linear regression showed that the ratio of the allele peak areas was directly proportional to the ratio of the alleles in the template DNA (rs1046295 r2 = 0.972; data not shown), thus underlining the reliability of this method to detect preferential allele expression.
We genotyped rs1046295 in the MRC-A/RNA population and identified 36 heterozygotes. Subsequently, 8 replicate genotyping reactions were performed in both the genomic DNA and cDNA of each of these individuals. Because the alleles should be present in equal amounts in the genomic DNA, the mean ratio of the peak areas (A:G) in the genomic DNAwas calculated to determine any bias toward 1 allele resulting from the genotyping. cDNA ratios were then compared with the mean ratio for the genomic DNA (Table II).
TABLE II. Rs1046295 paired-sample t tests.
| Sample | N | Mean difference (genomic DNA – cDNA) | 95% CI of the difference | Sig. (2-tailed) (2 S.F.) | Overexpressed allele | Fold change | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| 8016-4 | 4 | −0.64 | −0.88 | −0.4 | 0.0035 | A | 1.49 |
| 8033-3 | 8 | 1.30 | 1.29 | 1.32 | 2.5 × 10−14 | G | 0.00764 |
| 8036-3 | 5 | −0.57 | −0.67 | −0.47 | 8.2 × 10−5 | A | 1.44 |
| 8036-4 | 1 | −1.02 | — | — | — | — | 1.78 |
| 8046-3 | 7 | −0.16 | −0.30 | −0.027 | 0.026 | A | 1.12 |
| 8046-4 | 8 | −0.26 | −0.53 | 0.0056 | 0.054 | — | 1.20 |
| 8052-3 | 7 | −0.41 | −0.7 | −0.12 | 0.013 | A | 1.31 |
| 8060-5 | 7 | −0.44 | −0.59 | −0.29 | 0.00032 | A | 1.34 |
| 8062-4 | 8 | −0.22 | −0.34 | −0.11 | 0.0030 | A | 1.17 |
| 8067-3 | 8 | −0.25 | −0.43 | −0.059 | 0.017 | A | 1.19 |
| 8081-4 | 8 | −0.14 | −0.26 | −0.022 | 0.027 | A | 1.11 |
| 8099-4 | 7 | −0.32 | −0.58 | −0.061 | 0.023 | A | 1.24 |
| 8100-3 | 8 | −0.13 | −0.35 | 0.094 | 0.21 | — | 1.10 |
| 8103-4 | 8 | −4.19 | −4.93 | −3.45 | 2.9 × 10−6 | A | 4.20 |
| 8110-4 | 8 | −0.037 | −0.17 | 0.095 | 0.53 | — | 1.03 |
| 8111-5 | 8 | 0.080 | 0.0041 | 0.16 | 0.042 | G | 0.939 |
| 8122-4 | 6 | −0.48 | −1.08 | 0.12 | 0.095 | — | 1.37 |
| 8127-3 | 8 | −0.25 | −0.34 | −0.17 | 0.00023 | A | 1.19 |
| 8135-3 | 8 | −0.035 | −0.16 | 0.086 | 0.52 | — | 1.03 |
| 8136-3 | 6 | −0.044 | −0.14 | 0.048 | 0.27 | — | 1.03 |
| 8146-3 | 8 | 0.050 | −0.11 | 0.21 | 0.48 | — | 0.962 |
| 8146-4 | 8 | −0.16 | −0.30 | −0.031 | 0.023 | A | 1.13 |
| 8148-4 | 7 | −0.23 | −0.33 | −0.13 | 0.0015 | A | 1.17 |
| 8148-5 | 8 | −0.061 | −0.23 | 0.11 | 0.42 | — | 1.05 |
| 8154-3 | 8 | 0.075 | −0.11 | 0.26 | 0.38 | — | 0.943 |
| 8158-3 | 7 | −0.098 | −0.30 | 0.11 | 0.29 | — | 1.07 |
| 8169-3 | 6 | −0.43 | −0.57 | −0.30 | 0.00043 | A | 1.33 |
| 8169-4 | 5 | −0.25 | −0.46 | −0.045 | 0.028 | A | 1.19 |
| 8176-5 | 8 | 0.18 | −0.51 | 0.87 | 0.56 | — | 0.863 |
| 8179-4 | 8 | −0.54 | −0.71 | −0.36 | 0.00019 | A | 1.41 |
| 8187-3 | 8 | −0.016 | −0.12 | 0.092 | 0.74 | — | 1.01 |
| 8189-3 | 8 | −0.072 | −0.23 | 0.083 | 0.31 | — | 1.05 |
| 8198-4 | 8 | — | — | — | — | A | — |
| 8199-3 | 7 | −0.16 | −0.50 | 0.18 | 0.29 | — | 1.12 |
| 8199-4 | 7 | −0.022 | −0.25 | 0.20 | 0.82 | — | 1.02 |
| 8199-5 | 6 | −0.14 | −0.20 | −0.078 | 0.0020 | A | 1.11 |
| Population | 231 | 0.18 | 0.14 | 0.22 | 6.5 × 10−16 | A | 1.14 |
| Population (edited) | 136 | 0.11 | 0.054 | 0.16 | 8.7 × 10−5 | A | 1.08 |
S.F., Significant figures; Sig., significance.
Population, Results for all individuals used, excluding 8033-3, 8103-4, and 8198-4. Population (edited), In addition, only results for individuals with 8 replicates for cDNA were used, and in the case of samples with more than 8 replicates, only 8 were used. t Test significance is for a 2-tailed test. 8036-4 was not analyzed individually because of a lack of cDNA replicate results. Fold changes were calculated by dividing the mean cDNA peak area allele A:G ratio by the mean genomic DNA ratio. For sample 8198-4, the peak area of allele G was 0; hence, no meaningful value could be obtained for the ratio of A:G. Dashes for mean difference, 95% CI of the difference and significance indicate data could not be analyzed due to lack of replicates (8036-4) or due to a peak area for allele G of 0 (8198-4). Dashes for overexpressed allele indicate that there was no significant overexpression of either allele.
Three individuals with extreme preferential expression of 1 allele were identified by visual inspection of the data (subject ID numbers 8033-3, 8198-4 and 8103-4). These 3 outliers were therefore excluded from further analyses. Outlying cDNA replicate results were also removed from further analysis (Fig 4).
FIG 4. Boxplots showing the allelotyping results.
Combined genomic DNA results (blue), combined means of cDNA ratios of all individuals (red), with and without sample outliers, and replicate cDNA results for each individual (brown). Peak area ratio of allele A:allele G used for 8 PCR replicates. Moderate outliers (outside 1.5*interquartile range) are displayed as a circle, extreme outliers (outside 2*interquartile range) as a circle in the box-whiskers. Samples 8198-4 (ratio is infinite), 8033-3, and 8103-4 (outlying results) not shown. Significance of any difference between cDNA and genomic DNA peak ratios indicated (*P < .05; **P < .01; ***P < .001).
For the remaining 33 individuals, the ratio of peak areas A:G in the cDNA was compared with the mean ratio from the genomic DNA by using a paired sample t test. This revealed a modest, but very significant, difference (cDNA mean, 1.49; genomic DNA mean, 1.31; mean difference, 0.18, 95% CI of the difference, 0.14-0.22; fold change, 1.14; P = 6.5 × 10−16). Significant differences were also obtained for both sexes independently (male-only: cDNA mean, 1.47; genomic DNA mean, 1.31; mean difference, 0.16; 95% CI of the difference, 0.10-0.22; fold change, 1.12; P = 1.5 × 10−7; female-only: cDNA mean, 1.51; genomic DNA mean, 1.31; mean difference, 0.20; 95% CI of the difference, 0.15-0.26; fold change, 1.15, P = 8.0 × 10−11).
DISCUSSION
Previous positional cloning of chromosome 13q14 implicated PHF11 as a determinant of total serum IgE levels, although because of the possibility of a joint SETDB2-PHF11 transcript and further distant alleles affecting linkage, all 3 genes in the region, SETDB2, PHF11, and RCBTB1, could influence asthma susceptibility.13 Here we have shown by EMSA experiments that the A allele of rs1046295 is capable of interacting with the transcription factor Oct-1 with greater affinity than the alternate G allele. Oct-1 is a POU domain transcription factor that is ubiquitously expressed and has been reported as being both a positive and negative regulator of transcription, with its function depending on the context in which it binds the DNA.51-54 It is also believed that Oct-1 can regulate transcription by interacting with other transcription factors/regulators.54 Oct-1 has been shown to influence transcription by binding an element in the 3′ region of the mouse IL-5 gene,55 possibly of relevance for rs1046295, which is located in the 3′ UTR of PHF11.
From the initial supershift assay, there was evidence that the transcription factor SRY might also be capable of binding rs1046295 allele dependently and be involved in the formation of complex 1, despite no predicted binding site for the protein being present in the probe sequence. This supershift was a robust result, replicated across multiple experiments. However, subsequent EMSAs indicated that this result is unlikely to be due to SRY binding the probe. In addition, no difference was observed in band patterns between male and female B lymphoblast cell lines. Because SRY is a male-specific gene found on the Y chromosome, the lack of difference in binding between the sexes again indicates that the supershift occurred for reasons other than SRY being present in the complex.
We were unable to identify the factor causing formation of complex 2, visible most strongly in the B lymphoblast cell lines. However, the presence of such a complex, specific to B cells, indicates the possibility of varying regulation of PHF11 in different cell types.
To determine whether rs1046295 had an in vivo effect on transcription, allelotyping experiments were performed. The results indicated that allele-specific expression of the SNP occurs, although individual variation was observed in this response. In general, the A allele was predominantly overexpressed with the fold change for the population being small—approximately 1.14. Although the scale of preferential allele expression appears to be subtle in these resting cells, the changes could still be biologically significant in conditions enhancing IgE production in vivo. When the results were analyzed by sex, a significant preferential expression of the A allele was observed in both males and females. This again indicates that the supershift with SRY antibody is not a result of the SRY protein itself binding at this SNP locus.
That not all individuals heterozygous for the rs1046295 polymorphism showed preferential allele expression, that its extent varied between individuals, and that both alleles for each polymorphisms were seen to be overexpressed are similar to observations made for other genes.50,56-58 Several possible explanations have been proposed,57 including allelic heterogeneity, epigenetic effects, or incomplete linkage disequilibrium between the SNP and polymorphism causing the preferential expression. In addition, incomplete penetrance has been observed in other studies.57
Previous work published has also identified a relative decrease in expression of the G allele of rs1046295. Clarke et al59 compared the ratio of rs1046295 alleles expressed by 5 heterozygotes in TH1 and TH2 cells. They found a significant difference in the ratio of the alleles between the 2 cell types, caused by a marked underexpression of the G allele in the TH1 cells only. They also observed a 2-fold increase in a PHF11 splice variant lacking exon 2 in individuals homozygous for the G allele of rs1046295 relative to those homozygous for the A allele. Therefore, this indicates that rs1046295 affects PHF11 at the level of splicing as well as transcription. The nature of the experiments in this report are such that they are not negatively affected by alternative splicing of PHF11, but are also insufficient to draw conclusions about the role of rs1046295 in alternative splicing of the gene. Therefore, further investigation is required to reveal the full impact of rs1046295 on PHF11 function.
Studies of rs1046295 have shown the G allele to be associated with both increased IgE levels13 and asthma.39 Based on the results presented here and by Clarke et al,59 this may be a result of decreased expression and alternative splicing caused by the G allele because of differential binding of transcription factors. The chromosome 13q14 locus modulates total serum IgE levels, raised amounts of which are a main feature of atopic diseases such as asthma. Further understanding of the mechanisms by which this locus influences susceptibility to asthma may allow the development of new treatments for this common disease.
Supplementary Material
Key messages.
Rs1046295 affects Oct-1 transcription factor binding in PHF11.
The A allele of rs1046295 is significantly overexpressed compared with the G allele.
Rs1046295 may modulate PHF11 expression and susceptibility to atopic disease.
Acknowledgments
We thank the families participating in this project and the individuals involved with their clinical testing and phenotyping.
Supported by the Wellcome Trust (075491/Z/04).
Abbreviations used
- EMSA
Electrophoretic mobility shift assay
- MALDI-TOF
Matrix-assisted laser desorption/ionization-time-of-flight
- Oct-1
Octamer-binding transcription factor-1
- PHF11
PHD finger protein 11
- SNP
Single nucleotide polymorphism
- UK
United Kingdom
- UTR
Untranslated region
Footnotes
Disclosure of potential conflict of interest: The authors have declared that they have no conflict of interest.
REFERENCES
- 1.Fleming DM, Sunderland R, Cross KW, Ross AM. Declining incidence of episodes of asthma: a study of trends in new episodes presenting to general practitioners in the period 1989-98. Thorax. 2000;55:657–61. doi: 10.1136/thorax.55.8.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skrepnek GH, Skrepnek SV. Epidemiology, clinical and economic burden, and natural history of chronic obstructive pulmonary disease and asthma. Am J Manag Care. 2004;10:S129–38. [PubMed] [Google Scholar]
- 3.Gupta R, Sheikh A, Strachan DP, Anderson HR. Burden of allergic disease in the UK: secondary analyses of national databases. Clin Exp Allergy. 2004;34:520–6. doi: 10.1111/j.1365-2222.2004.1935.x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson HR, Ruggles R, Strachan DP, Austin JB, Burr M, Jeffs D, et al. Trends in prevalence of symptoms of asthma, hay fever, and eczema in 12-14 year olds in the British Isles. BMJ. 2004;328:1052–3. doi: 10.1136/bmj.38057.583727.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson HR. Prevalence of asthma. BMJ. 2005;330:1037–8. doi: 10.1136/bmj.330.7499.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousquet J, Bousquet PJ, Godard P, Daures JP. The public health implications of asthma. Bull World Health Organ. 2005;83:548–54. [PMC free article] [PubMed] [Google Scholar]
- 7.Daniels SE, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, et al. A genome-wide search for quantitative trait loci underlying asthma. Nature. 1996;383:247–50. doi: 10.1038/383247a0. [DOI] [PubMed] [Google Scholar]
- 8.Cookson W. A new gene for asthma: would you ADAM and Eve it? Trends Genet. 2003;19:169–72. doi: 10.1016/S0168-9525(03)00025-8. [DOI] [PubMed] [Google Scholar]
- 9.Cookson W. The immunogenetics of asthma and eczema: a new focus on the epithelium. Nat Rev Immunol. 2004;4:978–88. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- 10.Los H, Postmus PE, Boomsma DI. Asthma genetics and intermediate phenotypes: a review from twin studies. Twin Res. 2001;4:81–93. doi: 10.1375/1369052012191. [DOI] [PubMed] [Google Scholar]
- 11.Weiss ST, Raby BA. Asthma genetics 2003. Hum Mol Genet. 2004;13(review issue 1):R83–9. doi: 10.1093/hmg/ddh080. [DOI] [PubMed] [Google Scholar]
- 12.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, et al. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003;34:181–6. doi: 10.1038/ng1166. [DOI] [PubMed] [Google Scholar]
- 14.Allen M, Heinzmann A, Noguchi E, Abecasis G, Broxholme J, Ponting CP, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet. 2003;35:258–63. doi: 10.1038/ng1256. [DOI] [PubMed] [Google Scholar]
- 15.Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004;304:300–4. doi: 10.1126/science.1090010. [DOI] [PubMed] [Google Scholar]
- 16.Noguchi E, Yokouchi Y, Zhang J, Shibuya K, Shibuya A, Bannai M, et al. Positional identification of an asthma susceptibility gene on human chromosome 5q33. Am J Respir Crit Care Med. 2005;172:183–8. doi: 10.1164/rccm.200409-1223OC. [DOI] [PubMed] [Google Scholar]
- 17.Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76:349–57. doi: 10.1086/427763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–4. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 19.Himes EH, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84:581–93. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–91. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S-H, Cho B-Y, Park C-S, Shin E-S, Cho E-Y, Yang E-M, et al. Alpha-T-catenin (CTNNA3) gene was identified as a risk variant for toluene diisocyanate-induced asthma by genome-wide association analysis. Clin Exp Allergy. 2009;39:203–12. doi: 10.1111/j.1365-2222.2008.03117.x. [DOI] [PubMed] [Google Scholar]
- 22.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhry S, Taub M, Mei R, Rodriguez-Santana J, Rodriguez-Cintron W, Shriver MD, et al. Genome-wide screen for asthma in Puerto Ricans: evidence for association with 5q23 region. Hum Genet. 2008;123:455–68. doi: 10.1007/s00439-008-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui J, Oka A, James A, Palmer LJ, Musk AW, Beilby J, et al. A genome-wide association scan for asthma in a general Australian population. Hum Genet. 2008;123:297–306. doi: 10.1007/s00439-008-0477-9. [DOI] [PubMed] [Google Scholar]
- 25.Wiltshire S, Bhattacharyya S, Faux JA, Leaves NI, Daniels SE, Moffatt MF, et al. A genome scan for loci influencing total serum immunoglobulin levels: possible linkage of IgA to the chromosome 13 atopy locus. Hum Mol Genet. 1998;7:27–31. doi: 10.1093/hmg/7.1.27. [DOI] [PubMed] [Google Scholar]
- 26.The Collaborative Study on the Genetics of Asthma (CSGA) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet. 1997;15:389–92. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 27.Hizawa N, Freidhoff LR, Chiu YF, Ehrlich E, Luehr CA, Anderson JL, et al. Genetic regulation of Dermatophagoides pteronyssinus – specific IgE responsiveness: a genome-wide multipoint linkage analysis in families recruited through 2 asthmatic sibs. J Allergy Clin Immunol. 1998;102:436–42. doi: 10.1016/s0091-6749(98)70132-0. [DOI] [PubMed] [Google Scholar]
- 28.Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES, Changyaleket B, et al. Genome-wide search for asthma susceptibility loci in a founder population. Hum Mol Genet. 1998;7:1393–8. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- 29.Kimura K, Noguchi E, Shibasak M, Arinami T, Yokouchi Y, Takeda K, et al. Linkage and association of atopic asthma to markers on chromosome 13 in the Japanese population. Hum Mol Genet. 1999;8:1487–90. doi: 10.1093/hmg/8.8.1487. [DOI] [PubMed] [Google Scholar]
- 30.Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, Annesi-Maesano I, Boussaha M, Bousquet J, et al. Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med. 2000;162:1812–8. doi: 10.1164/ajrccm.162.5.2002113. [DOI] [PubMed] [Google Scholar]
- 31.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67:1154–62. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokouchi Y, Nukaga Y, Shibasaki M, Noguchi E, Kimura K, Ito S, et al. Significant evidence for linkage of mite-sensitive childhood asthma to chromosome 5q31-q33 near the interleukin 12 B locus by a genome-wide search in Japanese families. Genomics. 2000;66:152–60. doi: 10.1006/geno.2000.6201. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Postma DS, Howard TD, Koppelman GH, Zheng SL, Stine OC, et al. Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet. 2000;67:1163–73. doi: 10.1086/321190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koppelman GH, Stine OC, Xu J, Howard TD, Zheng SL, Kauffman HF, et al. Genome-wide search for atopy susceptibility genes in Dutch families with asthma. J Allergy Clin Immunol. 2002;109:498–506. doi: 10.1067/mai.2002.122235. [DOI] [PubMed] [Google Scholar]
- 35.Blumenthal MN, Ober C, Beaty TH, Bleecker ER, Langefeld CD, King RA, et al. Genome scan for loci to mite sensitivity: the Collaborative Study on the Genetics of Asthma (CSGA) Genes Immun. 2004;5:226–31. doi: 10.1038/sj.gene.6364063. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya S, Leaves NI, Wiltshire S, Cox R, Cookson WO. A highdensity genetic map of the chromosome 13q14 atopy locus. Genomics. 2000;70:286–91. doi: 10.1006/geno.2000.6398. [DOI] [PubMed] [Google Scholar]
- 37.Anderson GG, Leaves NI, Bhattacharyya S, Zhang Y, Walshe V, Broxholme J, et al. Positive association to IgE levels and a physical map of the 13q14 atopy locus. Eur J Hum Genet. 2002;10:266–70. doi: 10.1038/sj.ejhg.5200801. [DOI] [PubMed] [Google Scholar]
- 38.Jang N, Stewart G, Jones G. Polymorphisms within the PHF11 gene at chromosome 13q14 are associated with childhood atopic dermatitis. Genes Immun. 2005;6:262–4. doi: 10.1038/sj.gene.6364169. [DOI] [PubMed] [Google Scholar]
- 39.Gao J, Li W, Willis-Owen SA, Jiang L, Ma Y, Tian X, et al. Polymorphisms of PHF11 and DPP10 are associated with asthma and related traits in a Chinese population. Respiration. 2010;79:17–24. doi: 10.1159/000235545. [DOI] [PubMed] [Google Scholar]
- 40.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, et al. Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:362–67. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faisst S, Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992;20:3–36. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wingender E. Compilation of transcription regulating proteins. Nucleic Acids Res. 1988;16:1879–902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–84. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 46.Hacking D, Knight JC, Rockett K, Brown H, Frampton J, Kwiatkowski DP, et al. Increased in vivo transcription of an IL-8 haplotype associated with respiratory syncytial virus disease-susceptibility. Genes Immun. 2004;5:274–82. doi: 10.1038/sj.gene.6364067. [DOI] [PubMed] [Google Scholar]
- 47.Ding C, Cantor CR. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc Natl Acad Sci U S A. 2003;100:3059–64. doi: 10.1073/pnas.0630494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–75. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 49.Ding C, Maier E, Roscher AA, Braun A, Cantor CR. Simultaneous quantitative and allele-specific expression analysiswithreal competitive PCR. BMC Genet. 2004;5:8–13. doi: 10.1186/1471-2156-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 51.Mordvinov VA, Schwenger GT, Fournier R, De Boer ML, Peroni SE, Singh AD, et al. Binding of YY1 and Oct1 to a novel element that downregulates expression of IL-5 in human T cells. J Allergy Clin Immunol. 1999;103:1125–35. doi: 10.1016/s0091-6749(99)70188-0. [DOI] [PubMed] [Google Scholar]
- 52.Thomas MA, Mordvinov VA, Sanderson CJ. The activity of the human interleukin-5 conserved lymphokine element 0 is regulated by octamer factors in human cells. Eur J Biochem. 1999;265:300–7. doi: 10.1046/j.1432-1327.1999.00732.x. [DOI] [PubMed] [Google Scholar]
- 53.Salerno MS, Schwenger GT, Sanderson CJ, Mordvinov VA. Binding of octamer factors to the murine IL-5 CLE0 in primary T-cells and a T-cell line. Cytokine. 2001;15:4–9. doi: 10.1006/cyto.2001.0897. [DOI] [PubMed] [Google Scholar]
- 54.Cheng CK, Yeung CM, Hoo RL, Chow BK, Leung PC. Oct-1 is involved in the transcriptional repression of the gonadotropin-releasing hormone receptor gene. Endocrinology. 2002;143:4693–701. doi: 10.1210/en.2002-220576. [DOI] [PubMed] [Google Scholar]
- 55.Salerno MS, Mordvinov VA, Sanderson CJ. Binding of octamer factors to a novel 39-positive regulatory element in the mouse interleukin-5 gene. J Biol Chem. 2000;275:4525–31. doi: 10.1074/jbc.275.6.4525. [DOI] [PubMed] [Google Scholar]
- 56.Bray NJ, Buckland PR, Owen MJ, O’Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113:149–53. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- 57.Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics. 2004;16:184–93. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 58.Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, et al. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–62. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke E, Rahman N, Page N, Rolph MS, Stewart GJ, Jones GJ. Functional characterization of the atopy-associated gene PHF11. J Allergy Clin Immunol. 2008;121:1148–54. doi: 10.1016/j.jaci.2008.02.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.