Summary
Social cues contribute to the circadian entrainment of physiological and behavioral rhythms. These cues supplement the influence of daily and seasonal cycles in light and temperature. In Drosophila, the social environment modulates circadian mechanisms that regulate sex pheromone production and mating behavior. Here we demonstrate that a neuroendocrine pathway, defined by the neuropeptide Pigment-Dispersing Factor (PDF), couples the central nervous system (CNS) to the physiological output of peripheral clock cells that produce pheromones, the oenocytes. PDF signaling from the CNS modulates the phase of the oenocyte clock. Despite its requirement for sustaining free-running locomoter activity rhythms, PDF is not necessary to sustain molecular rhythms in the oenocytes. Interestingly, disruption of the PDF signaling pathway reduces male sex pheromones and results in sex-specific differences in mating behavior. Our findings highlight the role of neuropeptide signaling and the circadian system in synchronizing the physiological and behavioral processes which govern social interactions.
Introduction
The circadian timing system generates oscillations in physiology and behavior synchronized to daily cycles in environmental conditions (e.g., photoperiod and temperature). By synchronizing to the environment the internal time-keeping mechanism provides the organism with an adaptive advantage by enabling it to anticipate daily reoccurring events. This synchronization facilitates reproduction, and coordinates feeding and metabolic rhythms, among other processes (Dunlap et al., 2004). The social environment also influences clock time. Studies performed in various animals, ranging from insects to mammals, have demonstrated social influences on circadian rhythmicity (Bloch and Grozinger, 2011; Davidson and Menaker, 2003; Mistlberger and Skene, 2004; Mrosovsky, 1996). Several recent studies in the fruit fly Drosophila melanogaster have shown that social experience affects the circadian regulation of various behaviors including locomotor activity (Levine et al., 2002a), reproductive behavior (Fujii and Amrein, 2010; Fujii et al., 2007), sleep (Donlea et al., 2009), and learning and memory (Donlea et al., 2009). Our own work in Drosophila demonstrated that social context, defined by the genotypic composition of the social group, affects the circadian entrainment of a peripheral clock housed within the oenocytes, the cells responsible for the synthesis and expression of cuticular hydrocarbon pheromones (Krupp et al., 2008). Corresponding with the temporal changes to the oenocyte clock, the social environment also affected the expression of male sex pheromones and the frequency of mating. Because pheromones mediate social responses, the modulation of these signals may be important for relaying information between members of the social group. Although the underlying sensory mechanisms responsible for the social influences on the circadian clock are unknown, it is possible that the modulation of pheromonal signaling may reflect a feedback mechanism that facilitates the social synchrony necessary for effective social encounters.
The circadian system of Drosophila is composed of multiple cellular clocks located in many of the tissues and organs of the body. Because individual cells are circadian clocks, these individual oscillators must be synchronized within a tissue; likewise individual tissues must be kept in a stable phase-relationship with each other in order to build a coherent circadian system. For example, a defined network of approximately 150 clock neurons in the CNS governs behavioral rhythms in Drosophila (Allada and Chung, 2010). Communication between clock neurons via the neuropeptide Pigment Dispersing Factor (PDF) is required for free-running locomotor activity rhythms (Renn et al., 1999). PDF is expressed and rhythmically released by a small group of clock neurons, the ventral lateral neurons (vLNs) (Helfrich-Förster, 1997; Park et al., 2000), where it acts locally through its receptor, PDFR, to synchronize the molecular rhythms of other neurons within the circadian circuit (Hyun et al., 2005; Lear et al., 2005; Lin et al., 2004; Mertens et al., 2005; Park et al., 2000; Shafer et al., 2008; Yoshii et al., 2009). Although it is generally accepted that intercellular signaling temporally structures the circadian circuit in the brain and is necessary for generating rhythms in behavior, it is not clear whether similar mechanisms might regulate the timing of peripheral clock cells residing outside of the CNS.
Circadian oscillators have been identified in numerous peripheral tissues in Drosophila, including the olfactory and gustatory sensilla (Chatterjee et al., 2010; Krishnan et al., 1999; Tanoue et al., 2004), oenocytes (Krupp et al., 2008), prothoracic gland (Myers et al., 2003), epidermis (Ito et al., 2008), fat body (Xu et al., 2008), malpighian tubules (Giebultowicz and Hege, 1997), and male reproductive system (Beaver et al., 2002). Consistent with a hierarchical model of circadian regulation with the brain directing the phase of ‘slave’ peripheral oscillators, as exists in mammals, an early study in Drosophila suggested that a diffusible factor emanating from the brain coordinates behavioral rhythms in arrhythmic mutant individuals (Handler and Konopka, 1979). More recently, however, this view has been replaced by the idea that peripheral clocks are cell-autonomous in the fly. Coordinated timing between individual oscillators is thought to occur via light and temperature sensitive intracellular molecular pathways that respond to ambient conditions (Allada and Chung, 2010). Elegant transplantation experiments using malpighian tubules, the renal organ of the fly, best demonstrate the cell-autonomous, self-sustaining nature of peripheral clock cells in Drosophila. It was shown that the molecular rhythm of transplanted malpighian tubules maintains phase coherence with the donor fly after being transferred to a host entrained to a reverse light/dark cycle (Giebultowicz and Hege, 1997). Malpighian tubules express the blue-light circadian photoreceptor, Cryptochrome (CRY), and can entrain directly to light in vitro (Ivanchenko et al., 2001). Thus, it is generally accepted that peripheral clock cells in Drosophila sustain temporal coherence with each other and with behavioral rhythms by responding directly to the same entrainment cues that set the phase of the central pacemaker neurons in the brain. In this way, peripheral clocks maintain synchrony with external environmental cues independent of input from the central clock in the brain; the prothoracic gland is the only known exception (Myers et al., 2003). However, whether that the central clock a phase influence on the timing mechanism of peripheral oscillators, however, has not been rigorously tested.
Here, we propose that a neuropeptidergic pathway originating in the CNS regulates the peripheral oenocyte clock. We analyzed the contribution of the PDF signaling pathway to the temporal regulation of the peripheral oenocyte clock and its physiological output. We found that the PDF signaling pathway sets the phase of the oenocyte clock under free-running conditions, a consequence of the modulation of the period of the circadian cycle. Corresponding changes in the expression of the clock-controlled gene, desat1, the production of male sex pheromones, and the temporal pattern of mating suggest that the modulation of the oenocyte clock by PDF signaling is required for reproductive behavior. Direct stimulation of the oenocytes by PDF in vivo altered pheromone expression, indicating that PDF may act as a neuroendocrine signal with the ability to remotely regulate the circadian physiology of peripheral clock cells. Together, our results demonstrate that the CNS exerts an influence on peripheral clock function in Drosophila melanogaster, and provides insight into how a distributed circadian timing system coordinates physiological and behavioral rhythms important for social behavior.
Results
PDF Signaling Modulates the Timing of the Oenocyte Clock
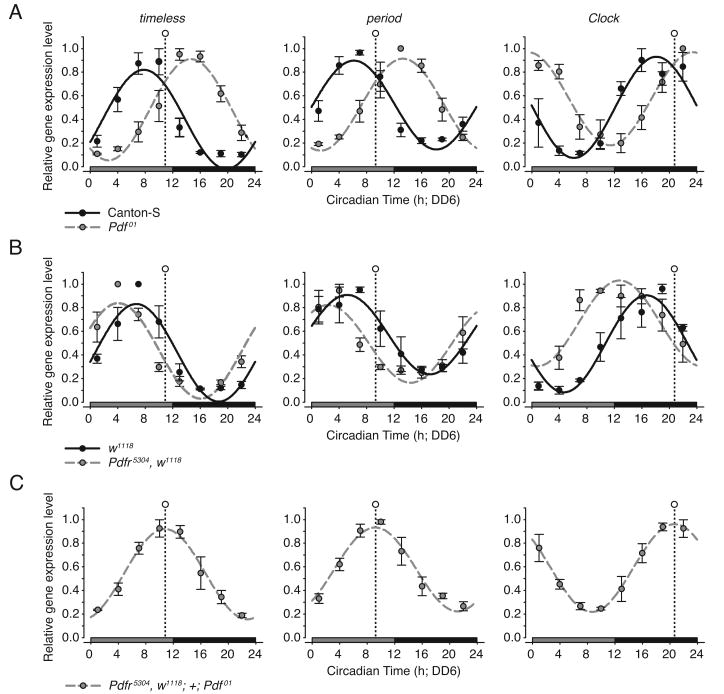
To determine whether PDF signaling plays a role in the entrainment of the peripheral oenocyte clock, we examined the temporal expression patterns of the core clock genes period (per), timeless (tim) and Clock (Clk) – three genes previously used as readouts for the temporal precision of the molecular clock mechanism (Krupp et al., 2008). The expression profiles of the clock genes were assayed in the oenocytes of both Pdf (Pdf01) and Pdf receptor (Pdfr5304) mutant flies, and compared to wild-type control strains, Canton-S and w1118. In Pdf01 and Pdfr5304 mutant flies, pacemaker neurons maintain molecular rhythms for several days under free-running conditions before becoming desynchronized (Lin et al., 2004; Peng et al., 2003; Yoshii et al., 2009); likewise, locomotor activity remains rhythmic after the onset of constant darkness before gradually damping (Hyun et al., 2005; Renn et al., 1999). To avoid possible masking effects attributable to these persistent rhythms, the experiments described below were performed on sixth day of constant darkness (DD6).
In wild-type oenocytes, the temporal expression profiles of per and tim were previously shown to peak during the early night, whereas Clk peaked during the early day in a 24 h light:dark cycle (LD12:12; Krupp et al., 2008). Here the circadian expression of per, tim, and Clk in wild-type oenocytes continued to be rhythmic on DD6 (Figures 1A and 1B and Tables S1-S3). Although the phase relationships between the clock genes remained stable, the peak time of expression on DD6 was shifted by roughly 9-12 h relative to that previously observed in LD conditions. The shift in peak expression reflected a long (∼25.5-26.5 h) free-running period and a corresponding delay in phase of roughly 1.5-2.5 h for each day in DD. Consistent with a long free-running period, the oenocyte clock exhibited a corresponding phase delay on DD1 (Krupp et al., 2008) and DD3 (Figure S1 and Table S1).
Figure 1. PDF Signaling Modulates the Phase of the Peripheral Oenocyte Clock.
(A-C) Temporal expression patterns of the clock genes tim, per, and Clk in the oenocytes of (A) Pdf01, (B) Pdfr5304, and (C) Pdfr5304; +; Pdf01 flies as determined by quantitative RT-PCR. Canton-S and w1118 strains served as wild-type controls for Pdf01 and Pdfr5304, respectively. Gene expression was assayed on DD6. Relative gene expression values represent the mean of n=3 independent replicate time series (control, black circles; mutant genotype, gray circles). Error bars indicate ±SEM. Best-fit cosine curves overlie the relative gene expression values for each genotype (control, solid black line; mutant genotype, dashed gray line). The time of peak clock gene expression for the Pdfr5304; +; Pdf01 is indicated in panels A-C (open circle with vertical dashed line). See Tables S1-S4 for fit parameter values and statistics.
In Pdf01 and Pdfr5304 mutant flies, clock gene expression in the oenocytes continued to be rhythmic under free-running conditions and the phase relationship between the genes remained fixed (Figures 1A and 1B and Table S4), indicating that synchrony between individual oenocyte cells does not require PDF signaling. Although rhythmic, the temporal pattern of clock gene expression of both Pdf01 and Pdfr5304 was significantly different than wild-type controls. In Pdf01 flies, the time of peak expression of per, tim, and Clk was delayed an average of 6.4 h relative to Canton-S (Figure 1A and Tables S1-S3), whereas in Pdfr5304 peak expression was advanced by 3.1 h relative to w1118 (Figure 1B and Tables S1-S3). The direction of the drift in peak phase over successive days, as projected by comparing DD3 and DD6 (compare Figures 1A-1B and S1A-S1B) indicates that (1) the oenocyte clock of Pdf01 and Pdfr5304 run with long free-running periods (>24 h), and (2) differences in the time of peak phase on DD6 reflect differences in the period length, with the free-running period for Pdf01 being longer than CantonS and that for Pdfr5304 being shorter than w1118. The reason for the disagreement in period length between Pdf01 and Pdfr5304 is not yet known. These findings indicate that the PDF signaling pathway acts to modulate the timing of the peripheral oenocyte clock
To further explore the interaction between Pdf and Pdfr, we examined the circadian profile of clock gene expression in the oenocytes of flies mutant for both genes (Pdfr5304; +; Pdf01). Comparing the clock gene expression profile of Pdfr5304; +; Pdf01 to Pdfr5304 and Pdf01 mutants showed that the temporal profile of clock gene expression of the double mutant was significantly different from flies mutant for either gene alone and from the wild-type control strains (Figures 1A-1C and Tables S1-S4). In the double mutant, the peak phase for per, tim, and Clk expression occurred roughly mid-way between Pdf01 and Pdfr5304, and is delayed compared to Canton-S and w1118 controls. Together, these results indicate that competing signaling events involving PDF and PDFR may act in an opposing manner to either speed up or slow down the molecular rhythm of the oenocyte clock. Accordingly, when Pdf and Pdfr-associated input is absent, the oenocyte clock displays a unique molecular rhythm not observed in wild-type flies.
PDF Signaling Modulates Oenocyte Physiology
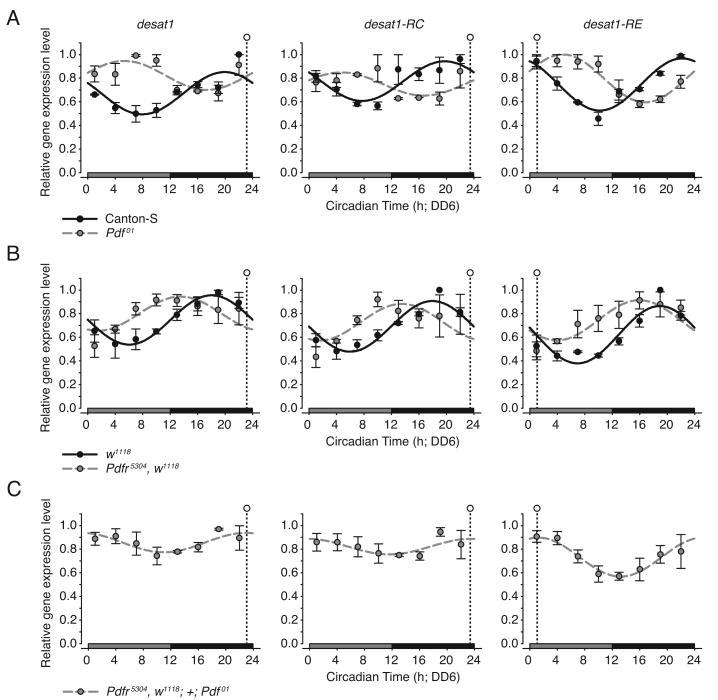
Next we examined two physiological outputs of oenocyte activity: (1) the expression of the clock-controlled gene, desaturase1 (desat1; Dallerac et al., 2000; Krupp et al., 2008; Marcillac et al., 2005), and (2) the production of cuticular hydrocarbon compounds (CHCs) (Billeter et al., 2009), several of which function as sex pheromones and influence mating behavior (Ferveur, 2005; Jallon, 1984). The desat1 gene encodes a key enzyme involved in the metabolic pathway regulating the biosynthesis of the male Drosophila sex pheromones including (z)-7-Tricosene (7-T), (z)-5-Tricosene (5-T) and (z)-7-Pentacosene (7-P; Dallerac et al., 2000; Marcillac et al., 2005). It has been suggested that the circadian regulation of desat1 expression within the oenocytes is responsible for daily fluctuations in the expression level of sex pheromones on the surface of the male cuticle (Krupp et al., 2008).
To determine if the phenotypic effects on the oenocyte clock resulting from the loss of PDF signaling correlated with a change in oenocyte physiology, we monitored the circadian expression of desat1 in Pdf01 and Pdfr5304 mutant flies under constant dark conditions (DD6; Figure 2). The desat1 locus encodes five transcriptional isoforms (annotated desat1-RA to –RE); all isoforms are expressed in the oenocytes (Billeter et al., 2009) but are differentially regulated by the clock (Figure S2 and Table S5). We focused our analysis on the expression patterns of the clock-controlled transcripts desat1-RC and -RE. RC is the most abundant transcript in the oenocytes and is expressed in most, if not all tissues, in contrast RE is expressed at a low level, but is restricted to only the oenocytes and male reproductive organs (Billeter et al., 2009). In wild-type control flies, the expression of desat1-RC and -RE remained rhythmic (Figures 2A and 2B and Tables S1 and S2) and mimicked the expression of Clk under free-running conditions. The time of peak expression for RC and RE overlaps with that of Clk early in the subjective night of DD6, representing an advance in phase of roughly 9-12 h relative to the expression profile observed under LD conditions. When PDF signaling was disrupted the expression of both RC and RE remained rhythmic (Figures 2A and 2B) and, as with the control flies, maintained a fixed phase relationship to that of Clk. Similar to expression patterns previously described for the clock genes in response to disruptions of the PDF pathway, both RC and RE showed a phase delay and a phase advancement in Pdf01 and Pdfr5304 mutant flies, respectively, relative to wild-type controls under free-running conditions (Figures 2A and 2B and Tables S1 and S2). Moreover, the profile of desat1 transcript expression of the Pdfr5304; +; Pdf01 double mutant displayed a relationship (Figures 2A-2C and Table S1) identical to that previously described for the clock genes (compare to Figure 1).
Figure 2. PDF Signaling Modulates the Phase of the Clock-Controlled Gene, desat1.
(A-C) Temporal expression patterns of desat1 (sum of all isoforms), and specific transcriptional isoforms desat1-RC and -RE in the oenocytes of (A) Pdf01, (B) Pdfr5304, and (C) Pdfr5304; +; Pdf01 flies as determined by quantitative RT-PCR. Canton-S and w1118 strains served as wild-type controls for Pdf01 and Pdfr5304, respectively. Gene expression was assayed on DD6. Relative gene expression values represent the mean of n=3 independent replicate time series (control, black circles; mutant, gray circles). Error bars indicate ±SEM. Best-fit cosine curves overlie the relative gene expression values for each genotype (control, solid black line; mutant, dashed gray line). The time of peak desat1 expression for the Pdfr5304; +; Pdf01 is indicated in the panels A-C (open circle with vertical dashed line). See Tables S1 and S2 for fit parameter values and statistics.
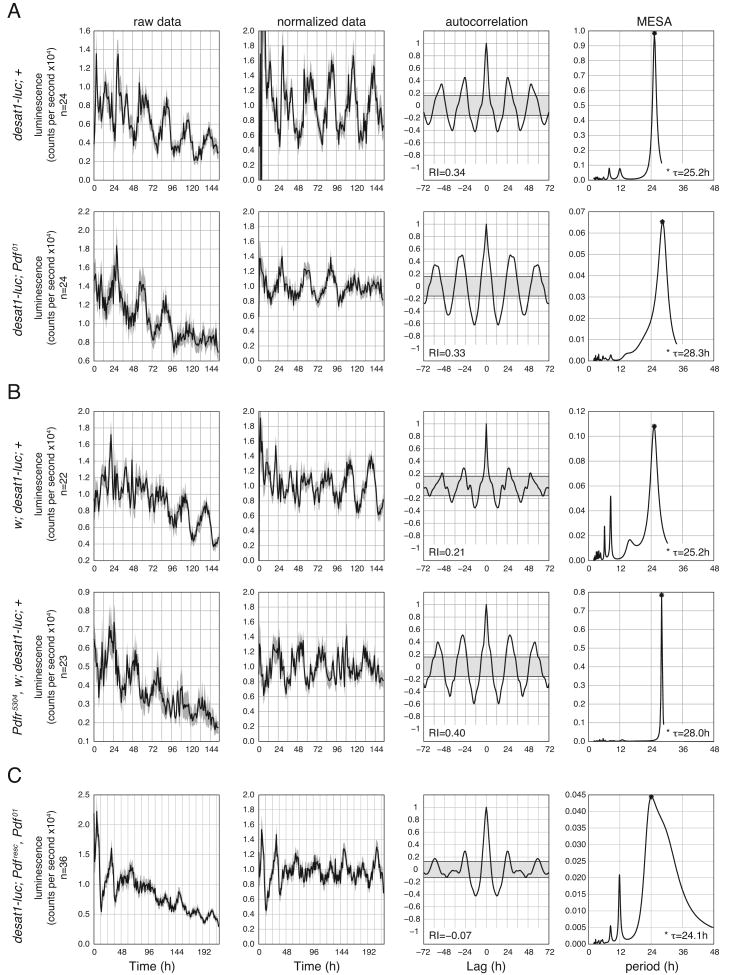
To confirm the role of PDF signaling in influencing the free-running period of the oenocyte clock, we generated a clock-controlled luciferase reporter derived from the regulatory sequence of the desat1-RE promoter. The RE promoter targets transgene expression specifically to the adult male oenocytes and reproductive organs (Billeter et al., 2009). With the desat1-luciferase reporter (desat1-luc), it was possible to continuously monitor the molecular rhythm of the oenocyte clock in living flies over many days in constant conditions. In wild-type control flies, desat1-luc expression was significantly rhythmic with an estimated periodicity of approximately 25 h (Figure 3A and 3B, top row), reproducing the circadian expression of the endogenous desat1-RE transcript. When placed in the mutant genetic background of either Pdf01 or Pdfr5304, the desat1-luc reporter ran with a long period of >28 h (Figures 3A and 3B, bottom row). Importantly, the introduction of a single transgenic copy of the wild-type Pdf gene (Pdfresc) rescued the long period phenotype of Pdf01, restoring the period to near wild-type length (Figure 3C). Thus, Pdf and Pdfr are necessary for maintaining the periodicity of the oenocyte clock and desat1 expression.
Figure 3. Pdf is Necessary for Setting the Period of the Oenocyte Clock.
(A-C) Analysis of the desat1-driven luciferase reporter (desat1-luc) expressed in (A) Pdf01, (B) Pdfr5304, and (C) Pdfrsc, Pdf01 flies. Wild-type controls are shown in the top row of (A) and (B). Luminescence was monitored in living adult male flies continuously under constant dark conditions from DD2-DD8 for (A and B), and DD1-DD9 for (C). Left most column of panels shows mean luminescence values plotted vs. time for n number of individuals. The second column shows detrended, normalized data. Gray shadings surrounding the plotted lines denote ±SEM. The third column represents the results of applying an autocorrelation function to the normalized data luminescence data. The Rhythmicity Index (RI) measures rhythm strength. The shape of the correlograms and associated RI values indicate that each data set are rhythmic; the RI for (C) is not significant, but the shape of the correleogram suggests rhythmicity. The right-most column shows the results of Maximum Entropy Spectral Analyses (MESA), a method applied to estimate periodicity. The abscissa position and height of the peak in the MESA plots indicate the principal periodicities by which desat-luc exhibited systematically fluctuating luciferase activity. Free-running period, τ
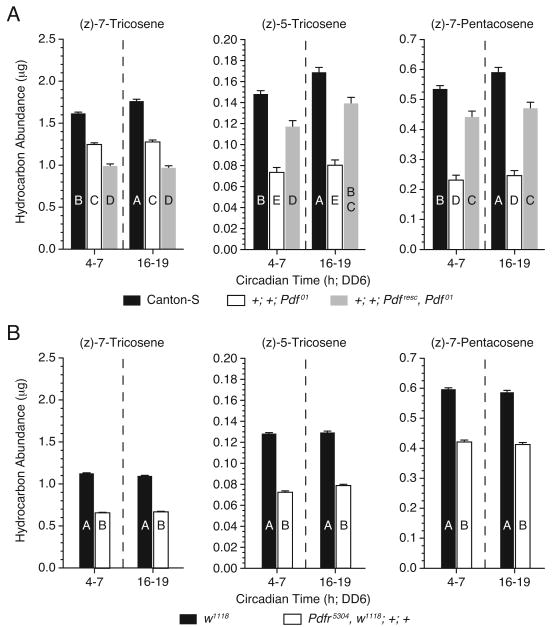
The level of desat1 expression in the oenocytes directly correlates with the amount of the sex pheromones 7-T, 5-T and 7-P expressed on the cuticular surface of male D. melanogaster (Krupp et al., 2008). Therefore, we predicted that the effects on the circadian expression pattern of desat1 in response to disruptions in PDF signaling would produce corollary changes in sex pheromone expression. We compared the sex pheromone expression profiles of wild-type controls to that of Pdf01 and Pdfr5304 mutant flies, during the subjective day and night on DD6. Canton-S control flies expressed 7-T, 5-T, and 7-P at all times of the day with significantly higher levels occurring during the subjective night (Figure 4A), a time roughly corresponding with the observed peak in desat1 expression. In comparison, Pdf01 mutants showed a dramatic reduction in the amount of 7-T, 5-T, and 7-P at all time points, and a loss of the subjective day/night difference (Figure 4A). Similarly, Pdfr5304 mutant flies showed a reduction in the absolute levels of sex pheromone expression relative to w1118 controls (Figure 4B). Conclusions, however, could not be drawn regarding the absence of temporal changes in the level of expression in Pdfr5304, since w1118 failed to show the expected subjective day/night differences. The decrease in sex pheromone levels in Pdf01 and Pdfr5304 were part of an overall reduction in the total amount of CHCs, encompassing all chemical classes of hydrocarbon compounds (Table S6). Thus, Pdf and Pdfr are necessary for regulating the expression of sex pheromones and CHCs in general.
Figure 4. PDF Signaling Influences the Expression of Male Sex Pheromones.
(A and B) Mean amounts of sex pheromones 7-T, 5-T, and 7-P expressed by (A) Pdf01 and Pdfesc, Pdf01, and (B) Pdfr5304 males at times during the subjective day (CT4-7) and night (CT16-19). Canton-S and w1118 served as wild-type control males. n>15 for each data point. Error bars indicate ±SEM. Uppercase letters signify significant differences (ANOVA, p<0.01). All cuticular hydrocarbon extracts were collected on DD6. See also Table S6.
Next we asked whether rescuing Pdf would restore sex pheromone expression. We found that the Pdfresc transgene rescued the expression of 5-T and 7-P of Pdf01 mutant males to near wild-type levels, but it failed to recover 7-T expression, or subjective day/night differences (Figure 4A and Table S6). The rescue of sex pheromone expression, although only partial, further demonstrates the requirement for PDF in regulating oenocyte physiology.
Oenocytes Respond to Direct Activation by Membrane-tethered PDF
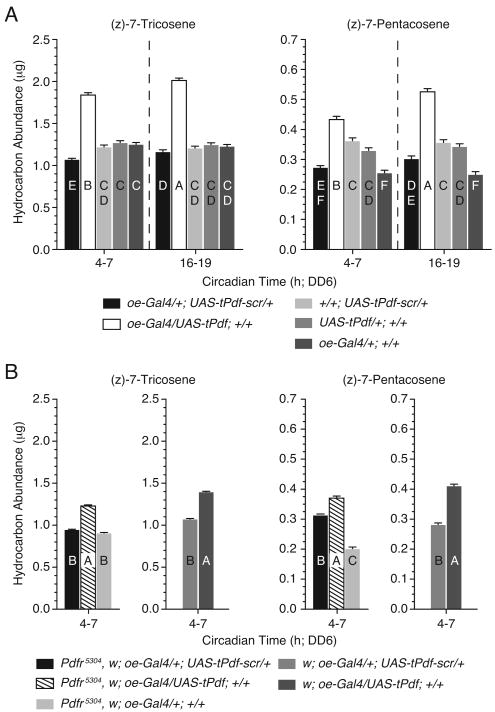
The gene encoding the PDF receptor, Pdfr, is expressed by the oenocytes (Figure S3), an indication that the oenocytes are primed to respond to direct stimulation by PDF. To determine whether the oenocytes can respond directly to the PDF peptide, we used the Gal4/UAS system to specifically target the expression of a cell membrane-tethered form of PDF (tPDF) (Choi et al., 2009) to the oenocytes. The oenocyte Gal4 driver, oe-Gal4 (Billeter et al., 2009) was used to drive tPDF expression. Flies expressing tPDF in the oenocytes were compared to those expressing a scrambled PDF peptide (tPDF-scr); a peptide containing the disarranged component amino acids present in PDF. Flies heterozygous for oe-Gal4, UAS-tPDF and UAS-tPDF-scr transgenes served as negative controls.
The misexpression of tPDF in the oenocytes resulted in a significant increase in the levels of 7-T and 7-P relative to tPDF-scr and heterozygous controls at all time points sampled on DD6 (Figure 5A). Opposite to the decreased expression found with the Pdf01 and Pdfr5304 mutations, tPDF induced an overall increase in the total amount of CHCs, affecting all chemical classes of cuticular hydrocarbon compounds analyzed, except CVA (Table S7). Thus, the oenocytes have the ability to directly respond to the PDF peptide. Moreover, the relationship between the gain-of-function expression of tPDF and the loss-of-function Pdf01 and Pdfr5304 mutants with regard to sex pheromone expression suggests that a common regulatory pathway may be alternately activated and repressed by these genetic manipulations.
Figure 5. Targeted Expression of Membrane Tethered-PDF in the Oenocytes Increases Sex Pheromone Biosynthesis.
(A and B) Mean amounts of 7-T and 7-P expressed by (A) oe-gal4/UAS-tPDF, and (B) Pdfr5304; oe-gal4/UAS-tPDF at times during the subjective day (CT4-7) and night (CT16-19). Genotypes of control strains are as stated. n=30 (A) and n=18 (B) for each data point. Error bars indicate ±SEM. Uppercase letters signify significant differences (ANOVA, p<0.01). All cuticular hydrocarbon extracts were collected on DD6. See also Table S7.
The PDF ligand is thought to signal exclusively through PDFR to affect the circadian rhythm in locomotor activity. To test the requirement of PDFR as it relates to oeonocyte physiology, we expressed tPDF within the oenocytes of Pdfr5304 flies and assayed the expression of 7-T and 7-P. If PDFR is required for tPDF activity in the oenocytes, then loss of PDFR function would be expected to block the phenotypic increase in sex pheromone expression. Surprisingly, the loss of PDFR did not mitigate the phenotypic effects resulting from the expression of tPDF (Figure 5B). The expression of 7-T and 7-P remained significantly elevated in w, Pdfr5304; oe-Gal4/UAS-tPDF relative to negative control flies w, Pdfr5304; oe-Gal4/+; UAS-tPDF-scr/+. Although there remain unresolved questions regarding the activities of PDF and PDFR, these results suggest that PDF may signal through a second, unidentified PDF-responsive receptor.
Neuronal PDF Expression is Necessary for Oenocyte Physiology
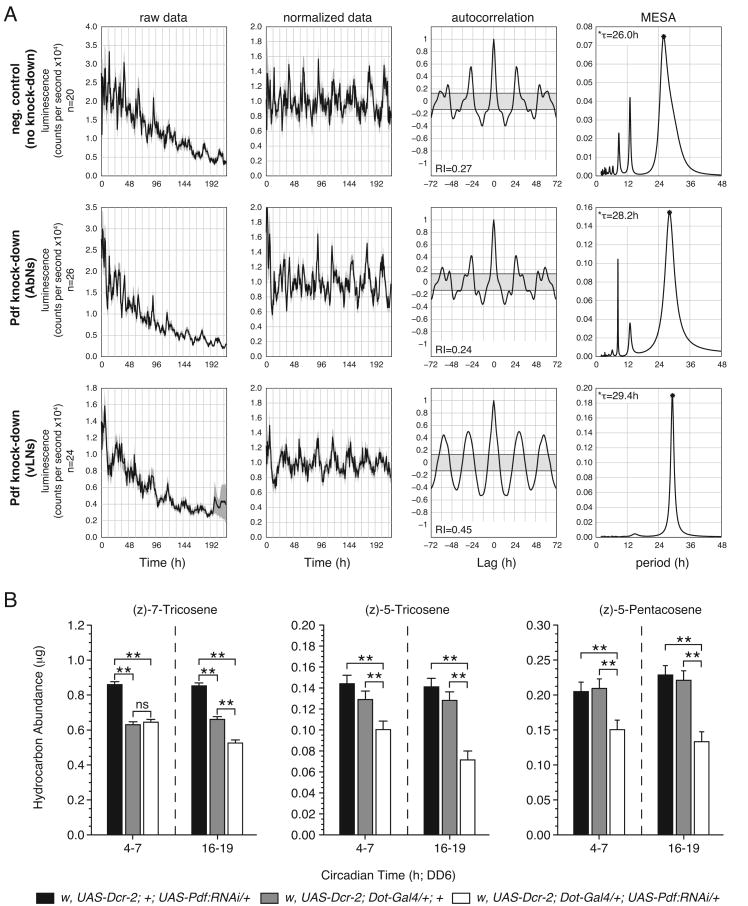
Several populations of neurons express PDF in the adult fly. These include the 16 ventral lateral clock neurons (vLNs) in the brain and a cluster of approximately 8 abdominal ganglia neurons (AbNs) in the ventral nerve cord. To determine which population of PDF-expressing neurons is responsible for influencing oenocyte physiology, we utilized the Gal4/UAS system to knockdown Pdf expression by RNAi (Shafer and Taghert, 2009). The Dorothy(Dot)-Gal4 and tim-Gal4 drivers were used to target RNAi to the AbNs and vLNs, respectively (Figure S4) . Using the desat1-luc reporter, we asked which population of PDF-expressing neurons is involved in regulating the free running rhythm of the oenocyte clock. Surprisingly, both the AbNs and the vLNs appear to play a role in modulating the period of the oenocyte clock. Knockdown of PDF in either population of neurons resulted in a long period (∼29h) relative to negative controls (∼25-26 h; Figure 6A and Figure S5), consistent with the phenotypes of Pdf01 and Pdfr5304 (Figure 3).
Figure 6. Neuronal PDF Expression is Necessary for Oenocyte Physiology.
(A) Analysis of the desat1-driven luciferase reporter (desat1-luc) expressed in control (w, UAS-Dcr-2; desat1-luc/+ ; UAS-Pdf-RNAi/+), AbN knockdown (w, UAS-Dcr-2; desat1-luc/Dot-Gal4; UAS-Pdf-RNAi/+), and vLN knockdown (w, UAS-Dcr-2; desat1-luc/tim-Gal4; UAS-Pdf-RNAi/+) flies. Luminescence was monitored in living adult male flies continuously under constant dark conditions from DD1-DD9. Left most column of panels shows mean luminescence values plotted vs. time for n number of individuals. The second column shows detrended, normalized data. Gray shadings surrounding the plotted lines denote ±SEM. The third column represents the results of applying an autocorrelation function to the normalized data luminescence data. The shape of the correlogram and associated RI values indicate that each data set is rhythmic. The right-most column shows the results of MESA, a method applied to estimate periodicity in these time series. The abscissa position and height of the peak in the MESA plots indicate the principal periodicities by which desat-luc exhibited systematically fluctuating luciferase activity. Free-running period, τ See Figure S4 for additional experimental controls.
(B) Mean amounts of 7-T, 5-T and 7-P expressed by control and AbN knockdown flies at times during the subjective day (CT4-7) and night (CT16-19). Genotypes of control strains are as stated. n=12 for each data point. Error bars indicate ±SEM. ANOVA, ** p<0.001; ns, not significant. All cuticular hydrocarbon extracts were collected on DD6. See also Table S8.
Using the same means to knockdown PDF expression, we also asked which population of neurons was necessary to support wild-type expression levels of male sex pheromones. Here, only PDF derived from the AbNs played a role in regulating oenocyte physiology. The PDF knockdown in the AbNs resulted in a significant decrease in the amount of 7-T, 5-T, and 7-P during the both subjective day and night on DD6 (Figure 6B and Table S8), whereas the vLN knockdown had no affect on pheromone levels (data not shown and Table S8). The extent of the decrease in the expression of these pheromones in response to the AbN PDF knockdown is consistent with that shown for both Pdf01 and Pdfr304 (Figure 4). Thus, it appears that while both the vLNs and the AbNs contribute to the regulation of the oenocyte clock, only the AbNs influence the physiological output of the oenocytes.
Circadian Regulation of desat1, Sex Pheromone Expression, and Mating Behavior Requires a Functional Oenocyte Clock
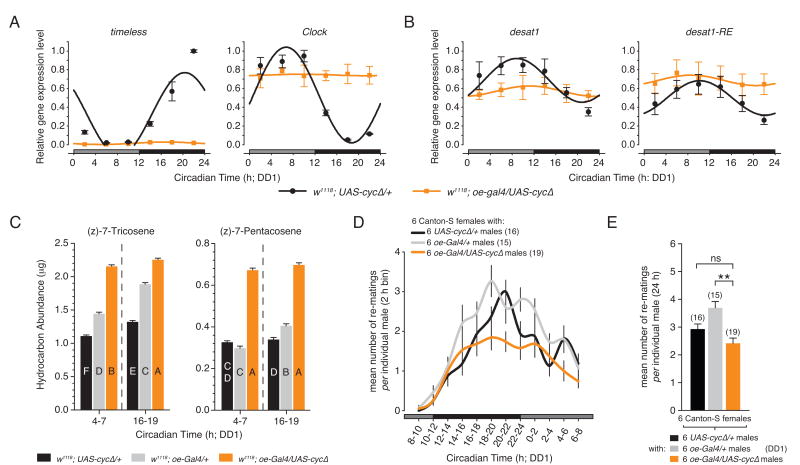
The results above demonstrate that PDF signaling is involved in the regulation of the oenocyte clock, desat1 expression, and cuticular hydrocarbon production. Although desat1 has been demonstrated to be under circadian control and integral to the regulation of cuticular hydrocarbon synthesis, it is unclear whether the rhythm of desat1 expression is dependent upon the cell-autonomous clock mechanism of the oenocytes. To determine the role of the oenocyte clock on the regulation of desat1 expression, we used genetic means to disrupt the molecular clock mechanism specifically in the oenocytes, while leaving the central clock and other peripheral oscillators intact. To do so, we used the oe-Gal4 to drive the expression of a dominant negative form of the core clock gene, cycle (cyc; UAS-cycΔ ; Tanoue et al., 2004). CYCΔ acts by sequestering the endogenous CLK protein, thereby reducing the efficiency of CLK to bind regulatory DNA sequences and blunting its ability to activate the transcription of per and tim. Flies expressing CYCΔ in the oenocytes (referred to as oeclock- flies) were compared to those heterozygous for the oe-Gal4 or the UAS-cycΔ transgenes.
In oeclock- flies maintained under constant conditions (DD1), tim expression was dramatically reduced relative to controls, but maintained a weak, low amplitude rhythm, whereas Clk exhibited a constant high level of expression but with no discernible circadian pattern (Figure 7A and Table S9). The altered expression profiles of tim and Clk indicate that both limbs (i.e. the PER/TIM and CLK/CYC limbs) of the interconnected transcriptional/translational molecular feedback mechanism of the oenocyte clock are disrupted by the targeted expression of CYCΔ.
Figure 7. Targeted Disruption of the Oenocyte Clock Impacts the Circadian Expression of desat1, Sex Pheromone Biosynthesis, and Mating Behavior.
(A and B) Temporal expression patterns of (A) the core clock genes, tim and Clk, and (B) desat1 in the oenocytes of control (w1118; UAS-cycΔ/+) and oenocyte clock disrupted (w1118; oe-gal4/UAS-cycΔ) males as determined by quantitative RT-PCR. Gene expression was assayed on DD1. Relative gene expression values represent the mean of n=3 independent replicate time series. Within each replicate experiment, the relative expression of w1118; oe-gal4/UAS-cycΔ was calibrated to the peak expression value of the control w1118; UAS-cycΔ/+ (peak expression is equal to 1). Best-fit cosine curves overlie the relative gene expression values for each genotype. See Table S9 for fit parameter values and statistics.
(C) Mean amounts of 7-T and 7-P expressed by control (w1118; UAS-cycΔ/+ and w1118; oe-gal4/+) and oenocyte clock disrupted (w1118; oe-gal4/UAS-cycΔ) males at times during the subjective day (CT4-7) and subjective (CT16-19). n=18 for each data point. Uppercase letters signify significant differences (ANOVA, p<0.01). All cuticular hydrocarbon extracts were collected on DD1. Error bars indicate ±SEM. See also Table S10.
(D and E) The temporal distribution (D) and mean number of re-matings (E) occurring over a 24-hour observation period for control (w1118; UAS-cycΔ/+ and w1118; oe-gal4/+) and oenocyte clock disrupted (w1118; oe-gal4/UAS-cycΔ) flies on DD1. The reduced level of mating for for w1118; UAS-cycΔ/+ males may indicate a low level of leaky CYCΔ expression in the brain possibly affecting behavior. n for each genotype is shown in parentheses. ANOVA: ** p<0.01. Error bars indicate ±SEM in all panels.
Targeted expression of CYCΔ also altered the profile of desat1 expression in the oenocytes. In contrast to controls, oeclock- males exhibited a flat but stable level of desat1 expression (i.e., the sum of all desat1 transcripts; Figure 7B and Table S9). The oenocyte-specific transcript, desat1-RE, showed a similar disruption in its circadian expression profile. However, RE displayed an elevated steady-state level of expression (Figure 7B and Table S9). Thus, the circadian expression of desat1 requires the activity of CLK in a way that is likely dependent upon the molecular clock mechanism.
The oenocyte clock may directly contribute to the regulation of pheromone production by regulating desat1 expression. Indeed, in response to the targeted expression of CYCΔ we observed significant changes in the absolute levels of 7-T and 7-P. Correlating with the elevated steady-state expression level of desat1-RE, flies with a disrupted oenocyte clock showed a significant increase in the level of both 7-T and 7-P relative to controls (Figure 7C and Table S10). Even with apparent disruptions to the oenocyte clock and desat1 transcription, oeclock- males continued to show a significant difference in the level of 7-T between the subjective day and night, with peak levels occurring during the night (Figure 7C). The amplitude change between day and night was lower relative to controls, possibly an indication of some residual clock activity. Together these results indicate that the endogenous circadian clock mechanism within the oenocytes regulates desat1 expression, which in turn affects the level of pheromone production and expression on the surface of the cuticle.
The effects on male sex pheromones suggested that the oenocyte clock may play a role in regulating the reproductive behavior of Drosophila. To investigate this possibility, we utilized a group-mating assay in which 6 virgin males were housed with 6 virgin females and allowed to interact continuously over a 24-hour testing period. Under these conditions, individual wild-type females will re-mate multiple times over a single 24 h reproductive episode. The temporal distribution and overall number of re-matings of oeclock- males was compared to UAS-cycΔ/+ and oe-Gal4/+ heterozygous controls when separately grouped with wild-type females. Mating assays were performed under constant conditions on DD1.
The temporal distribution showed that oeclock- males and controls re-mated at roughly the same frequency for the first 6-8 h of the 24 h testing cycle (Figure 7D). Thereafter, the re-mating frequency for oeclock- males flattened, remaining constant for the rest of the subjective night and continuing into the next day. In contrast, the re-mating frequency of the UAS-cycΔ/+ and oe-Gal4/+ control males continued to increase before peaking sharply during the middle-to-late portion of the subjective night (CT16-22). The mean number of re-matings per male for oeclock- was significantly different then that for oe-Gal4/+, but not UAS-cycΔ/+ controls (Figure 7E), suggesting that differences in the temporal pattern of re-mating behavior are not dependent on the total number matings per individual. Thus, the loss of a functioning oenocyte clock resulted in a temporal difference in re-mating behavior, without affecting the total number of matings.
Mating Behavior of Drosophila is Modulated by PDF
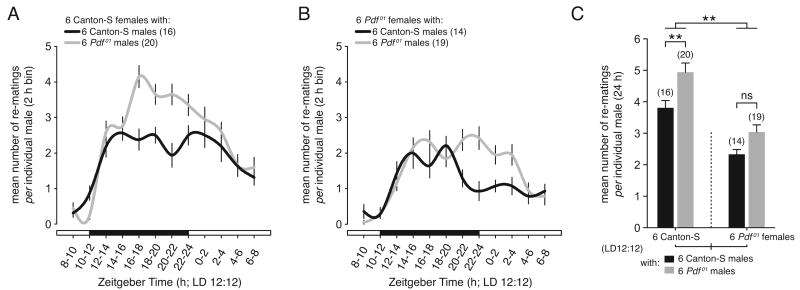
The oenocyte clock, as shown above, is necessary for normal sex pheromone expression and mating behavior. This raised the question: what role does the modulation of the oenocyte clock and its physiological outputs by PDF signaling play in the regulation of mating behavior? To address this question, we again used the group-mating assay. Here, the temporal distribution and overall number of re-matings of control Canton-S males were compared to that of Pdf01 males. Mating assays were performed in a light/dark cycle (LD 12:12) to more closely simulate the light conditions flies might typically experience in nature.
The temporal distribution showed that Canton-S and Pdf01 males when grouped with Canton-S females mated at roughly the same rate for the first 6-8 hours of the 24 h testing cycle (Figure 8A). Thereafter, Pdf01 males sustained a higher frequency of re-mating than Canton-S during the late night, and continued to re-mate for several hours past dawn (ZT 2-4). Corresponding to this temporal difference, Pdf01 males mated more on average than Canton-S, amounting to >1 additional re-mating per Pdf01 male (p=0.0085) relative to Canton-S controls when paired with Canton-S females (Figure 8C, left). This relationship also was present in mating experiments performed under constant conditions on DD6 (Figure S6).
Figure 8. Pdf Affects the Amount and Temporal Distribution of Mating Behavior.
(A-C) Temporal distribution (A and B) and mean number of re-matings (C) occurring over a 24-hour observation period for Canton-S control and Pdf01 male flies in LD12:12. Male flies were mated with either Canton-S (A) or Pdf01 (B) females. (C) The interaction between genotype and sex is shown in the mean number of re-matings per male. Pdf01 males mate more frequently than Canton-S males regardless of the genotype of the females, but less often with Pdf01 females than with Canton-S females. n for each genotype is shown in parentheses. ANOVA: ** p<0.01. Error bars indicate ±SEM in all panels.
When males were grouped with Pdf01 females, Pdf01 males continued to mate more frequently than Canton-S males (Figure 8C, right), however this difference failed to reach significance. The distribution of matings showed that Pdf01 males mated at a higher frequency relative to Canton-S during the late night and continuing past dawn (Figure 8B). Overall, the genotype of the female members of the group played a significant role in the total number of re-matings, regardless of the male genetype, with Pdf01 females showing a stark reduction in re-matings relative to their Canton-S counterparts (Figure 8C). Thus, Pdf01 males mate more, while Pdf01 females appear to be more selective and mate less than Canton-S. The role of Pdf in regulating oenocyte physiology and sex pheromone expression may account for the effects on mating behavior.
Discussion
The circadian system contributes to the temporal regulation of social behavior. However, it is unclear how the circadian rhythms of central and peripheral oscillators are integrated to temporally organize social interactions. Here, we demonstrate that in D. melanogaster the CNS conveys temporal information to peripheral clock cells via a neuroendocrine signaling pathway. Specifically, we found that the neuropeptide PDF, a factor required for circadian behavior, modulates the timing and physiological output of the peripheral oenocyte clock. We propose that the PDF signaling pathway may act to temporally couple the circadian mechanism in the oenocytes mediating sex pheromone biosynthesis with mating behavior.
Temporal Regulation of Peripheral Oscillators by the PDF Signaling Pathway
The PDF signaling pathway serves to coordinate the circadian oscillations of clock neurons in the brain of Drosophila (Lin et al., 2004; Park et al., 2000; Yoshii et al., 2009), a precondition generally thought to be necessary for the generation of free-running rhythms in circadian behavior. Here we show that PDF also plays an ancillary role in directing the physiological rhythms of peripheral oscillators. Our results demonstrate that the PDF signaling pathway, although not required for entrainment or sustained rhythmicity, conveys phase information to the peripheral oenocyte clock. The free-running molecular rhythm of the oenocyte clock of Pdf01 flies showed a lengthened period and a subsequent phase delay under constant conditions, while that of Pdfr5304 flies showed a shortened period and a phase advance. The relationship between Pdf and Pdfr confirmed that both ligand and receptor are involved in setting the phase of the oenocyte clock. Interestingly, this relationship also indicated that an unidentified feature of the PDF signaling pathway (which may include a second ligand or PDF-responsive receptor) retains activity in the absence of either PDF or PDFR, actively delaying or advancing the phase of the clock, respectively. Only in the absence of both ligand and receptor did temporal input to the oenocyte clock appear to be lost.
In contrast to the rhythms in clock gene or desat1 transcription, the desat1-luc reporter in Pdfr5304 flies showed a long period phenotype equivalent to that of Pdf01 flies. The reason for the temporal difference between the transcriptional rhythms and the desat1-luc reporter in Pdfr5304 flies is not understood. However, given that the desat1-luc transgene contains the promotor and the 5' untranslated region (5'UTR) of the desat1-RE transcript, it is plausible that the expression of the luciferase protein is subject to additional regulatory influences which are not observable when measuring clock gene and desat1 transcription alone. Mechanisms of post-transcriptional regulation mediated by the 5'UTR, such as transcript stability and translation, are involved in the circadian regulation of clock-controlled genes in plants and mammals (Kim et al., 2011; Kim et al. 2007; Ovadia et al., 2010). Similarly, post-transcriptional regulation via micro-RNAs plays a role in the circadian biology of Drosophila (Kadener et al., 2009), and although there are no published examples of such regulation through interactions with the 5'UTR such a mechanism is possible. As we have shown here, there are five desat1 isoforms expressed in the oenocytes; each is identical in the protein coding sequence and only distingusishable by the 5'UTR. The differential regulation of these transcripts by the oenocytes likely occurs at the level of promoter mediated transcription, but the diversity of 5'UTRs indicates a post-transcriptional mechanism directing a higher level of regulation of desat1 expression. How PDF signaling events link to the clock and the regulation of clock-controlled genes is not known. As we have discussed, our results indicate that the PDF signaling pathway may involve complex regulatory interactions occurring at multiple levels during the process of gene expression.
The ability of the oenocytes to maintain a molecular rhythm, albeit shifted, in the absence of a coordinated central clock and behavioral rhythms indicates that the oenocytes, like other peripheral clocks, maintain a high degree of autonomy. As with other identified peripheral clocks in Drosophila, the oenocytes express the gene encoding for the blue-light photoreceptor Cryptochrome (CRY; J.J.K.and J.D.L,unpublished data) suggesting that the oenocytes may directly entrain to the light/dark cycle. Therefore, proper phasing between physiological and behavioral rhythms may involve a mechanism whereby semi-autonomous, photosensitive peripheral clocks independently tune to the solar day, yet remain responsive to temporal input from the CNS. It is conceivable that such a circadian system may allow independently entrained oscillators to maintain close phase coherence under varying environmental conditions.
How might the PDF neuropeptide reach the peripherally located oenocytes? In flies, PDF-expressing neurons located in the ventral lateral protocerebrum (vLNs) and abdominal ganglia (AbNs) display many of the molecular and anatomical characteristics of neurosecretory (neuroendocrine) cells (Kula-Eversole et al., 2010; Park et al., 2011; Park et al., 2008). The vLNs are clock neurons and rhythmically release PDF from their axon terminals, where as the AbNs, not considered to be clock cells, do not show a circadian change in PDF immunoreactivity (Park et al., 2000). Our results suggest that both the vLNs and AbNs contribute to the regulation of the oenocyte clock. Recently, PDF released by the AbNs terminals on the gut has been shown to affect the motor activity of non-innervated regions of the renal system (Talsma et al., 2012). Thus, it appears that PDF released by the AbNs is able to remotely control the activity of distant tissues. Since the oenocytes do not appear to be innervated (J.-C.B. and J.D.L, unpublished data), there is no reason to expect that the oenocytes receive direct synaptic input from PDF expressing neurons. Instead, we suggest that PDF released into the hemolymph, possibly by both the vLNs and AbNs, may function as a circulating neurohormone to be received by the oenocytes and possibly other tissues expressing PDFR. Although not shown in flies, PDF has been demonstrated to be present within the hemolymph of locusts (Persson et al., 2001), thus supporting the possibility that the PDF peptide may act as a neuroendocrine factor.
The role of PDF in synchronizing the circadian oscillations of clock neurons has been hypothesized to reside in its ability to adjust the intrinsic speed (and, subsequently, the period and phase) of the molecular timekeeping mechanism (Yoshii et al., 2009). The network of circadian clock neurons shows widespread receptivity to PDF (Shafer et al., 2008). Depending on the subgroup of clock neurons, PDF either lengthens or shortens the period of the molecular rhythm, while in other neurons, PDF is required to maintain rhythmicity (Yoshii et al., 2009). How the same signaling pathway differentially affects the rhythms of different groups of clock neurons is not known. Due to the fact that we observed analogous phase effects on the molecular rhythm of the oenocytes (even though both effects were observed in a single cell type) indicates that the synchronizing role of PDF signaling may generally apply to both central and peripheral oscillators. Moreover, the phase-regulatory function of PDF (whether the period is shortened or lengthened) may be dependent on cell-autonomous factors expressed by the responding cell. It will be important to determine if other peripheral clocks are likewise regulated by the PDF signaling pathway, and if so, whether there are cell-type specific differences in the intracellular signaling events linking PDFR to the molecular clock mechanism.
PDF Signaling Affects Sex Pheromone Biosynthesis and Mating Behavior
The involvement of the PDF signaling pathway in the regulation of the oenocyte clock is indicative of a hierarchically structured circadian system, with timing information provided by the CNS serving to modulate the output of autonomous peripheral oscillators. Consistent with the effects exhibited by the oenocyte clock, the disruption of the PDF signaling pathway also phase-shifted the circadian expression of the clock-controlled gene, desat1. Notably in gene expression experiments, the expression of desat1 closely tracked Clk, indicating that desat1 may be regulated directly by an output mechanism of the cell-autonomous oenocyte clock possibly via the transcriptional regulators of the Clk gene, VRILLE and PDP1ɛ (Allada and Chung, 2010), or possibly by CLK itself. Consistent with the possibility of direct regulation, consensus binding sites or VRI, PDF1ɛ, and CLK are present within the desat1 locus (Figure S2A).
Genetic manipulations affecting PDF expression also affected the display of cuticular hydrocarbon compounds; compounds including the male sex pheromones, 7-T ,5-T, and 7-P. Loss of Pdf or Pdfr expression reduced sex pheromone expression, while misexpression of Pdf increased these compounds. We suggest that these effects on pheromone expression reflect asynchrony between components of the circadian system, those being primarily the central pacemaker neurons and the oenocyte clock. In the absence of phase information provided by the CNS via PDF, the oenocyte clock and by extension the circadian expression of desat1 may become uncoupled from rhythms in other physiological and behavioral processes necessary for proper pheromonal output. In this way, seemingly subtle changes in phase may lead to a misalignment between rhythms and an amplified response in physiological output.
Several studies have demonstrated daily rhythmicity in courtship and mating (Hardeland, 1972; Sakai and Ishida, 2001; Tauber et al., 2003), thus implicating the circadian system in the regulation of sexual behavior in Drosophila. Recently, others have shown that the PDF-expressing vLNs are involved in mediating a male sex drive rhythm (MSDR), a novel activity rhythm displayed by males when individually paired with a female and allowed to interact continuously for 24 h (Fujii and Amrein, 2010; Fujii et al., 2007). Our results extend these findings by demonstrating the circadian system not only influences courtship and mating, but also regulates the physiology mediating the production and display of chemical signals critically important to sexual behavior. We propose that the PDF signaling pathway and its ability to synchronize the activity of peripheral and central oscillators may couple reproductive physiology with behavior. In this regard, we suggest that the PDF signaling pathway may act at two levels: within the individual (i.e., the male fly), PDF signaling may influence both sexual characteristics (pheromone expression) and sex drive, while between individuals of the group PDF-dependent effects on male pheromone expression may alter female mating behavior.
Studies in several organisms have demonstrated that fitness benefits of the circadian system are evident in a light/dark cycle, but not in constant conditions or when out of phase with environment cues (Dodd et al., 2005; Ouyang et al., 1998). These studies offer the best evidence that the synchronization of physiology and behavior with environmental cycles is important for reproductive success. Here, we have shown that under normal light conditions the re-mating frequency of Pdf01 males is increased but reduced in females. Thus, it seems that PDF signaling affects male attractiveness or sex appeal, while also influencing female receptivity and the choice of potential mates, possibly acting to balance these sexually dimorphic features of reproductive behavior. How this sexual balancing act, as it relates to PDF signaling in males and females, affects the reproductive success of flies under natural environmental conditions offers an interesting avenue for future studies.
The circadian system of Drosophila is affected by input from the social environment. Social experience can reset the daily activity rhythms of the fly (Levine et al., 2002a), and alter the molecular rhythm of oscillators present in the head, as well as those residing in the oenocytes (Krupp et al., 2008). Given our results here, it is conceivable that the modulation of the PDF signaling pathway may account for the broad effects on the circadian system in response to the social experience. Consistent with this possibility, Immonen and Ritchie recently showed that Drosophila females exhibit elevated expression of Pdf RNA after exposure to male courtship song (Immonen and Ritchie, 2012), suggesting that the modulation of Pdf expression at the level of transcription may play a significant role biologically in response to socio-sexual interactions. Indeed, the level of Pdf transcription does seem to play a regulatory role within the circadian system, as an increase in the relative expression of Pdf within the vLNs has been shown to affect behavioral rhythms – producing a slightly shortened period and a slightly advanced evening peak in activity – without causing arrhythmicity (Helfrich-Förster et al., 2000). Determining if and how endogenous Pdf expression is modulated by social cues, and how this relates to the release of the PDF peptide will provide further insight into the PDF-dependent signaling mechanism that regulates the timing of the oenocyte clock as well as other circadian oscillators that influence social behavior.
Experimental Procedures
Fly Strains and Rearing
All fly strains were reared on standard medium containing agar, glucose, sucrose, yeast, cornmeal, wheat germ, soya flour, molasses, propionic acid, and Tegosept in a 12 h light/dark cycle (LD 12:12) at 25°C in 40 to 50% humidity. Previously described mutant and transgenic strains applied to this study include Pdf01 and Pdfresc (Renn et al., 1999), Pdfr5304 (Hyun et al., 2005), oe-gal4 (Billeter et al., 2009), UAS-cycΔ (Tanoue et al., 2004), UAS-t-Pdf-ML (M6a) and UAS-t-Pdf-Scr (B3) (Choi et al., 2009), UAS-Pdf:RNAi, UAS-DCR-2, tim-Gal4, and Dot-Gal4 (Shafer and Taghert, 2009). Oe-Gal4 drives expression in the oenocytes and male reproductive organs. To control for genetic background strains were outcrossed to either Canton-S or the w1118. Canton-S and w1118 were used as wild-type control strains for Pdf01 and Pdfr5304, respectively.
For quantitative PCR and cuticular hydrocarbon analyses, adult males were collected within 24 h post-eclosion, and maintained in mixed-gender groups for 24 h prior to being separated using CO2 anaesthesia. Male pairs were subsequently raised in vials (10 × 75 mm) containing 1 ml of food medium and entrained for 3-4 days in LD 12:12 conditions prior to testing under the indicated environmental conditions (LD, light/dark; DD1 or DD6, first or sixth full day constant dark, respectively). For mating experiments, virgin adult males and females were collected shortly after eclosion using CO2 anaesthesia, kept in same-sex groups of 20 in food vials (12 × 95 mm), and aged for 5-6 days in LD 12:12 conditions prior to testing. For DD mating experiments flies were aged according to the LD treatment prior to being placed in constant conditions and tested on DD6.
Oenocyte Dissection and Quantitative RT-PCR
Oenocyte dissections were performed as previously described (Krupp and Levine, 2010; Krupp et al., 2008). Oenocytes were isolated from the dorsal abdominal segments 2-5 of filleted adult male abdomens and immediately placed into cell lysis buffer for RNA isolation. Individual samples consisted of the oenocytes pooled from 8 male flies collected over a 2-3 h period. Full time series experiments consisted of oenocyte samples collected at eight successive time points (six for CYCΔ experiments) spanning a 24 h period. Control and test oenocyte samples were collected and processed in tandem at all stages of analysis.
RNA was isolated from dissected oenocyte preparations using the RNeasy Micro kit (QIAGEN), and total RNA was reverse transcribed with the qScript cDNA Supermix (Quanta Biosciences). Quantitative PCR reactions were performed with the Perfecta SYBR Green Supermix (Quanta Biosciences), on an Mx3005P Real-Time PCR System (Stratagene). The relative level of gene transcript expression was determined separately for each gene analyzed from cDNA prepared from a common pool of dissected oenocytes. qPCR reactions were performed in triplicate, and the specificity of each reaction was evaluated by dissociation curve analysis. Each experiment was replicated 3-4 times. Relative expression amounts were calculated with the REST relative expression method (Pfaffl, 2001) with Rp49 serving as an internal reference gene. Within each replicate time series, all time point values were calibrated to the peak level of expression, with the peak value set equal to 1. Expression values for each genotype were calibrated independently except where indicated. See Supplemental Information for the list of gene specific primer sets were used in quantitative PCR reactions.
Luciferase Assay
Luminometric monitoring was performed under DD condtions as described by Plautz et al. (1997). Molecular timecourse data was evaluated using analytical tools in Matlab (see Krishnan et al., 2001; Levine et al., 2002b). Data showing a regular rise and fall in the correlogram plot were deemed rhythmic. The Rhythmicity Index assessed the strength of the rhythms with higher values representing stronger periodic fluctuations. Maximum Entropy Spectral Analysis (MESA) was used to estimate period. When multiple peaks were present the highest one was taken to estimate the primary periodicity. Mean period values were computed for a given genotype from n individuals. See Supplemental Information for details related to the desat1-luc construct.
Cuticular Hydrocarbon Analysis
For hydrocarbon analysis, flies were anesthetized with ether and placed into individual glass microvials containing 50 μl of hexane containing 10 ng/μl of octadecane (C18) and 10 ng/μl of hexacosane (C26) as injection standards. To achieve efficient extraction, the microvials were gently agitated for 5 min. Hydrocarbons were analysed using a Varian CP3800 gas chromatograph with a flame ionization detector (GC/FID) as described previously (Krupp et al., 2008). Varian Star Integrator software (Varian Inc.) was used to quantify compounds based on peak areas.
Group-mating Assay
Group-mating assays were performed in disposable 55 × 8 mm Petri dishes containing a fly food slice (22 × 5 mm). Assays were set up by sequentially introducing six virgin females followed by six virgin males of the indicated genotypes using a mouth pipette. Assays were started at zeitgeber time 8 (17.00 hr) in an incubator set at 25°C and at LD12:12. The ZoomBrowser EX software (Canon, Inc.) controlled a Canon S10 digital camera to take images of the assays at 2 min intervals for 24 h. Constant red light illumination (λ>620 nm) was used to monitor mating during the dark phase. Images were surveyed for copulating pairs and scored if a pair was observed for at least four consecutive frames. The frequency and time of re-mating events (after the first six mating) were assayed.
Statistical Analyses
Non-linear best cosine curve fitting of gene expression data was performed in SPSS (v16.0). Student's t test was used to test for differences in fit curve parameters. Two-way ANOVA followed by the post-hoc Tukey-Kramer test was used to determine whether pheromone levels differed between genotypes at the given time points; it was also used to assess significance in mating behavior. See Supplemental Information for further details related to statistical analyses.
Supplementary Material
Highlights.
Oenocytes receive regulatory input from the CNS via the neuropeptide PDF PDF modulates the phase of the oenocyte clock and affects pheromone expression Oenocytes respond directly to PDF, indicating that PDF may act as a neurohormone PDF signaling couples sex pheromone expression with mating behavior
Acknowledgments
Thanks to J. Atallah, J. Schneider, A. Rooke and R. Rooke for comments on the text, and to S. Jagadeesh for assistance with mating experiments. This work was supported by grants to J.D.L. from Canadian Institutes of Health Research, the Natural Science and Engineering Research Council, the Canada Research Chair Program, and the Child and Brain Development Program of the Canadian Institutes of AdvancedResearch. J.J.K. was supported by Sleep and Biological Rhythms Toronto, a CIHR-funded trans-disciplinary research and training program at the University of Toronto. J.-C.B. was supported by a fellowship for advanced researcher from the Swiss National Science Foundation. Work in the laboratory of M.N.N. was supported in part by the National Institute of Neurological Disorders and Stroke (NINDS) and National Institute of General Medical Sciences (NIGMS), National Institutes of Health (NIH) (R01NS055035, R01NS056443, R01GM098931).
Footnotes
SUPPLEMENTAL INFORMATION: Supplemental Information includes six figures and ten tables and can be found with this article online at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM. Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:2134–2139. doi: 10.1073/pnas.032426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. 2009;461:987–991. doi: 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- Bloch G, Grozinger CM. Social molecular pathways and the evolution of bee societies. Philos Trans R Soc Lond B Biol Sci. 2011;366:2155–2170. doi: 10.1098/rstb.2010.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Tanoue S, Houl JH, Hardin PE. Regulation of gustatory physiology and appetitive behavior by the Drosophila circadian clock. Curr Biol. 2010;20:300–309. doi: 10.1016/j.cub.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Fortin JP, McCarthy EV, Oksman L, Kopin AS, Nitabach MN. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Curr Biol. 2009;19:1167–1175. doi: 10.1016/j.cub.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallerac R, Labeur C, Jallon JM, Knipple DC, Roelofs WL, Wicker-Thomas C. A delta 9 desaturase gene with a different substrate specificity is responsible for the cuticular diene hydrocarbon polymorphism in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:9449–9454. doi: 10.1073/pnas.150243997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AJ, Menaker M. Birds of a feather clock together–sometimes: social synchronization of circadian rhythms. Curr Opin Neurobiol. 2003;13:765–769. doi: 10.1016/j.conb.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Ramanan N, Shaw PJ. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science. 2009;324:105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sinauer Associates Inc.; Sunderland, MA: 2004. [Google Scholar]
- Ferveur JF. Cuticular hydrocarbons: their evolution and roles in Drosophila pheromonal communication. Behav Genet. 2005;35:279–295. doi: 10.1007/s10519-005-3220-5. [DOI] [PubMed] [Google Scholar]
- Fujii S, Amrein H. Ventral lateral and DN1 clock neurons mediate distinct properties of male sex drive rhythm in Drosophila. Proc Natl Acad Sci USA. 2010;107:10590–10595. doi: 10.1073/pnas.0912457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–251. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebultowicz JM, Hege DM. Circadian clock in Malpighian tubules. Nature. 1997;386:664. doi: 10.1038/386664a0. [DOI] [PubMed] [Google Scholar]
- Handler AM, Konopka RJ. Transplantation of a circadian pacemaker in Drosophila. Nature. 1979;279:236–238. doi: 10.1038/279236a0. [DOI] [PubMed] [Google Scholar]
- Hardeland R. Species differences in the diurnal rhythmicity of courtship behaviour within the melanogaster group of the genus Drosophila. Anim Behav. 1972;20:170–174. doi: 10.1016/s0003-3472(72)80188-x. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C, Täuber M, Park JH, Mühlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Immonen E, Ritchie MG. The genomic response to courtship song stimulation in female Drosophila melanogaster. Proc Biol Sci. 2012;279:1359–1365. doi: 10.1098/rspb.2011.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito C, Goto SG, Shiga S, Tomioka K, Numata H. Peripheral circadian clock for the cuticle deposition rhythm in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:8446–8451. doi: 10.1073/pnas.0800145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- Jallon JM. A few chemical words exchanged by Drosophila during courtship and mating. Behav Genet. 1984;14:441–478. doi: 10.1007/BF01065444. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Kwak E, Kim SH, Lee KH, Woo KC, Kim KT. hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res. 2011;39:8901–8914. doi: 10.1093/nar/gkr605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- Krishnan B, Levine JD, Lynch MK, Dowse HB, Funes P, Hall JC, Hardin PE, Dryer SE. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- Krupp JJ, Levine JD. Dissection of oenocytes from adult Drosophila melanogaster. J Vis Exp. 2010;41 doi: 10.3791/2242. doi:10.3791–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Kent C, Billeter JC, Azanchi R, So AKC, Schonfeld JA, Smith BP, Lucas C, Levine JD. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr Biol. 2008;18:1373–1383. doi: 10.1016/j.cub.2008.07.089. [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci USA. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G proteincoupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Resetting the circadian clock by social experience in Drosophila melanogaster. Science. 2002a;298:2010–2012. doi: 10.1126/science.1076008. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002b;3:1–25. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcillac F, Bousquet F, Alabouvette J, Savarit F, Ferveur JF. A mutation with major effects on Drosophila melanogaster sex pheromones. Genetics. 2005;171:1617–1628. doi: 10.1534/genetics.104.033159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Mistlberger RE, Skene DJ. Social influences on mammalian circadian rhythms: animal and human studies. Biol Rev Camb Philos Soc. 2004;79:533–556. doi: 10.1017/s1464793103006353. [DOI] [PubMed] [Google Scholar]
- Mrosovsky N. Locomotor activity and non-photic influences on circadian clocks. Biol Rev Camb Philos Soc. 1996;71:343–372. doi: 10.1111/j.1469-185x.1996.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadia A, Tabibian-Keissar H, Cohen Y, Kenigsbuch D. The 5'UTR of CCA1 includes an autoregulatory cis element that segregates between light and circadian regulation of CCA1 and LHY. Plant Mol Biol. 2010;72:659–671. doi: 10.1007/s11103-010-9605-8. [DOI] [PubMed] [Google Scholar]
- Park D, Hadžić T, Yin P, Rusch J, Abruzzi K, Rosbash M, Skeath JB, Panda S, Sweedler JV, Taghert PH. Molecular Organization of Drosophila Neuroendocrine Cells by Dimmed. Curr Biol. 2011;21:1515–1524. doi: 10.1016/j.cub.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Veenstra JA, Park JH, Taghert PH. Mapping Peptidergic Cells in Drosophila: Where DIMM Fits In. PLoS One. 2008;3:e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:e13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson MG, Eklund MB, Dircksen H, Muren JE, Nässel DR. Pigment-dispersing factor in the locust abdominal ganglia may have roles as circulating neurohormone and central neuromodulator. J Neurobiol. 2001;48:19–41. doi: 10.1002/neu.1040. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Renn S, Park JH, Rosbash M, Hall JC, Levine JD. A pdf Neuropeptide Gene Mutation and Ablation of PDF Neurons Each Cause Severe Abnormalities of Behavioral Circadian Rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci USA. 2001;98:9221–9225. doi: 10.1073/pnas.151443298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Taghert PH. RNA-Interference Knockdown of Drosophila Pigment Dispersing Factor in Neuronal Subsets: The Anatomical Basis of a Neuropeptide's Circadian Functions. PLoS One. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talsma AD, Christov CP, Terriente-Felix A, Linneweber GA, Perea D, Wayland M, Shafer OT, Miguel-Aliaga I. Remote control of renal physiology by the intestinal neuropeptide pigment-dispersing factor in Drosophila. Proc Natl Acad Sci USA. 2012;109:12177–12182. doi: 10.1073/pnas.1200247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Tauber E, Roe H, Costa R, Hennessy JM, Kyriacou CP. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr Biol. 2003;13:140–145. doi: 10.1016/s0960-9822(03)00004-6. [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.