Abstract
Recombinant antibody fragments like single chain variable fragments (scFvs) represent an attractive yet powerful alternative to immunoglobulins and hold great potential in the development of clinical diagnostic/therapeutic reagents. Structurally, scFvs are the smallest antibody fragments capable of retaining the antigen-binding capacity of whole antibodies and are composed of an immunoglobulin (Ig) variable light (VL) and variable heavy (VH) chain joined by a flexible polypeptide linker. In the present study, we constructed a scFv against bovine IgA from a hybridoma cell line IL-A71 that secretes a monoclonal antibody against bovine IgA using recombinant DNA technology. The scFv was expressed in Escherichia coli and purified using immobilized metal affinity chromatography (IMAC). The binding activity and specificity of the scFv was established by its non-reactivity toward other classes of immunoglobulins as determined by enzyme-linked immunosorbent assay (ELISA) and immunoblot analysis. Kinetic measurement of the scFv indicated that the recombinant antibody fragment had an affinity in picomolar range toward purified IgA. Furthermore, the scFv was used to develop a sensitive ELISA for the detection of foot and mouth disease virus (FMDV) carrier animals.
Keywords: ELISA, FMDV, IgA, scFv
Introduction
Mucosal infections continue to represent a challenge for the development of vaccines that can either prevent the pathogen from colonizing the surface epithelium (non-invasive bacteria), penetrate the surface barrier and replicate within the body (invasive bacteria and viruses), and/or block the binding of microbial toxins and neutralize them. In most cases it would seem desirable to induce specific secretory IgA (sIgA) antibodies associated with immunological memory, in addition to systemic immunity, as seen in a number of gastrointestinal, respiratory, urinary, and genital tract infections caused by bacteria [1–5].
IgA being monomeric or dimeric shows very high affinity in binding or neutralizing viruses. It binds very poorly to the complement and therefore is less likely to initiate inflammatory reactions [6]. Engineered IgA antibody provides external passive immunotherapy against viral, bacterial, and eukaryotic pathogens, and the detection of IgA involves the use of monoclonal antibodies. Monoclonal antibody (Mab) generation has limitations such as cost, inconsistency in production of mAbs, and extensive screening procedures for adventitious agents. To overcome these problems, generation of scFvs by recombinant DNA technology has been widely used. These antibody fragments can be derived either from hybridomas or from bacteriophages displaying antibody fragments [7, 8] and consist of variable light (VL) and variable heavy (VH) chains of whole antibody linked by a small polypeptide linker [9]. Hence, these fragments are smaller than the whole immunoglobulins. They retain the antigen-binding site in a single linear molecule and their design, construction, and expression in Escherichia coli has demonstrated their structure–function relationship and antigen–antibody interactions. These properties make scFv useful in both clinical and medical applications [10–14]. Furthermore, the inherent properties of these antibodies can be engineered to increase their affinity, specificity and stability by affinity maturation and various mutagenesis techniques [15–18].
Mucosal IgA plays a potential role in detection of infection in oesophago-pharyngeal fluids of foot and mouth disease virus (FMDV), an important veterinary pathogen which is a highly contagious viral disease of cloven-hoofed animals and an economically devastating disease of livestock as it also affects a variety of wild animals [19, 20, 21]. FMDV is transmitted between animals naturally when they come in contact with the contaminated feed and aerosols from infected animals.
In the present study, we describe the construction of a scFv from a mouse hybridoma against bovine IgA, its expression in E. coli and its utility as a reagent for the detection of FMDV-specific IgA in salivary samples of FMDV carrier animals.
Materials and methods
Materials
Cells and hybridoma
The mouse hybridoma cell line IL-A71 (procured from European Collection of Cell Cultures (ECACC) secreting an anti-bovine Mab of IgG1 isotype was maintained in the hybridoma laboratory, Indian Immunologicals Limited (IIL), Hyderabad, India, and was used for the amplification of VH and VL.
Bacterial strains, vectors, and chemicals
All molecular biology reagents and the bacterial strain, E. coli BL21 (DE3) cells, used for the propagation of plasmids and overexpression of protein were obtained from Invitrogen (Carlsbad, USA). The bacterial expression vector, pET28a, used for cloning and expression of the scFv, was procured from Novagen (Madison, USA). Nickel–nitriloacetic acid (Ni-NTA) agarose used for the purification of 6X His-tagged proteins was purchased from Qiagen (Hilden, Germany). Bovine immunoglobulins, IgG1, IgG2, and IgM, and the commercial kit used for the detection of bovine immunoglobulin IgA were purchased from Bethyl Laboratories (Texas, USA). All other fine chemicals used were purchased from Sigma (Missouri, USA).
Methods
Isolation of total RNA and cDNA synthesis
Total RNA isolated from the hybridoma cell line (1 × 106 cells) using TRIzol reagent (Invitrogen, USA) was resuspended in diethyl pyrocarbonate (DEPC) treated water and quantified using a Biophotometer (Eppendorf, Germany). The cDNA was synthesized using random hexamers and a Thermoscript reverse transcriptase (RT) PCR kit (Invitrogen, USA) according to the manufacturer’s instructions. The cDNA was stored at –20 °C until further use.
Amplification of variable domains, assembly and cloning of scFv
cDNA encoding for the antibody variable domains (VH and VL) were PCR amplified using universal primers [22]. The variable regions were assembled using splicing by overlap extension (SOE) PCR (Table 1) to form single chain variable fragment (scFv). The resultant scFv product was subjected to PCR with the VL forward and VH reverse primers to incorporate the EcoRI and NotI restriction sites at the 5’ and 3’ end of PCR product (Table 1). The amplified PCR product and pET28a vector were restriction digested with EcoRI and NotI enzymes for 2 h, gel purified and subjected to ligation using T4 DNA ligase to obtain pET28a-scFv. The resultant product secretes scFv tagged with 6X Histidine which allows as purifying and detecting scFv.
Table 1.
Primers used for PCR of VH and VL regions and SOE PCR for construction of scFv
| Variable Light Chain Forward Primers | |
| LB1 | GCCATGGCGGA(CT)ATCCAGCTGACTCAGCC |
| LB2 | GCCATGGCGGA(CT)ATTGTTCTC(AT)CCCAGTC |
| LB3 | GCCATGGCGGA(CT)ATTGTG(AC)T(AC)ACTCAGTC |
| LB4 | GCCATGGCGGA(CT)ATTGTG(CT)T(AG)ACACAGTC |
| LB5 | GCCATGGCGGA(CT)ATTGT(AG)ATGAC(AC)CAGTC |
| LB6 | GCCATGGCGGA(CT)ATT(AC)AGAT(AG)A(AC)CCAGTC |
| LB7 | GCCATGGCGGA(CT)ATTCAGATGA(CT)(AGT)CAGTC |
| LB8 | GCCATGGCGGA(CT)AT(CT)CAGATGACACAGAC |
| LB9 | GCCATGGCGGA(CT)ATTGTTCTCA(AT)CCAGTC |
| LB10 | GCCATGGCGGA(CT)ATTG(AT)GCT(GC)ACCCAATC |
| LB11 | GCCATGGCGGA(CT)ATT(GC)T(AG)ATGACCCA(AG)TC |
| LB12 | GCCATGGCGGA(CT)(AG)TT(GT)TGATGACCCA(AG)AC |
| LB13 | GCCATGGCGGA(CT)ATTGTGATGAC(GCT)CAG(GT)C |
| LB14 | GCCATGGCGGA(CT)ATTGTGATAAC(CT)CAGGA |
| LB15 | GCCATGGCGGA(CT)ATTGTGATGACCCAG(AT)T |
| LB16 | GCCATGGCGGA(CT)ATTGTGATGACACAACC |
| LB17 | GCCATGGCGGA(CT)ATTTTGCTGACTCAGTC |
| Variable Light Chain Reverse Primers | |
| LF1 | GGAGCCGCCGCCGCCAGAACCACCACCACCAGAACCACC ACCACCACGTTTGATTTCCAGCTTGG |
| LF2 | GGAGCCGCCGCCGCCAGAACCACCACCACCAGAACCACC ACCACCACGTTTTATTTCCAGCTTGG |
| LF4 | GGAGCCGCCGCCGCCAGAACCACCACCACCAGAACCACC ACCACCACGTTTTATTTCCAACTTTG |
| LF5 | GGAGCCGCCGCCGCCAGAACCACCACCACCAGAACCACC ACCACCACGTTTCAGCTCCAGCTTGG |
| Variable Heavy Chain Forward Primers | |
| HB1 | GGCGGCGGCGGCTCCGGTGGTGGTGA(GT)GT(AG)(AC)AGCTTCAGGAGTC |
| HB2 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGT(GCT)CAGCT(GCT)CAGCAGTC |
| HB3 | GGCGGCGGCGGCTCCGGTGGTGGTCAGGTGCAGCTGAAG(GC)A(GC)TC |
| HB4 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGTCCA(AG)CTGCAACA(AG)TC |
| HB5 | GGCGGCGGCGGCTCCGGTGGTGGTCAGGT(CT)CAGCT(GCT)CAGCA(AG)TC |
| HB6 | GGCGGCGGCGGCTCCGGTGGTGGTCAGGT(CT)CA(AG)CTGCAGCAGTC |
| HB7 | GGCGGCGGCGGCTCCGGTGGTGGTCAGGTCCACGTGAAGCAGTC |
| HB8 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGTGAA(GC)(GC)TGGTGGAATC |
| HB9 | GGCGGCGGCGGCTCCGGTGGTGGTGA(AGC)GTGA(AT)G(CT)TGGTGGAGTC |
| HB10 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGTGCAG(GC)(GT)GGTGGAGTC |
| HB11 | GGCGGCGGCGGCTCCGGTGGTGGTGA(GT)GTGCA(AC)CTGGTGGAGTC |
| HB12 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGTGAAGCTGATGGA(AG)TC |
| HB13 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGTGCA(AG)CTTGTTGAGTC |
| HB14 | GGCGGCGGCGGCTCCGGTGGTGGTGA(AG)GT(AG)AAGCTTCTCGAGTC |
| HB15 | GGCGGCGGCGGCTCCGGTGGTGGTGAAGTGAA(AG)(GC)TTGAGGAGTC |
| HB16 | GGCGGCGGCGGCTCCGGTGGTGGTCAGGTTACTCT(AG)AAAG(AT)GT(GC)TG |
| HB17 | GGCGGCGGCGGCTCCGGTGGTGGTCAGGTCCAACT(AGC)CAGCA(AG)CC |
| HB18 | GGCGGCGGCGGCTCCGGTGGTGGTGATGTGAACTTGGAAGTGTC |
| HB19 | GGCGGCGGCGGCTCCGGTGGTGGTGAGGTGAAGGTCATCGAGTC |
| Variable Heavy Chain Reverse Primers | |
| HF1 | ATGCGCGGCCGCCGAGGAAACGGTGACCGTGGT |
| HF2 | ATGCGCGGCCGCCGAGGAGACTGTGAGAGTGGT |
| HF3 | ATGCGCGGCCGCCGCAGAGACAGTGACCAGAGT |
| HF4 | ATGCGCGGCCGCCGAGGAGACGGTGACTGAGGT |
Expression of scFv in E. coli
pET28a-scFv was transformed into XL-blue E. coli-competent cells and plated onto Luria-Bertani (LB) agar supplemented with 50 μg/ml kanamycin (LB-Kan). The plates were incubated overnight at 37 °C. The resultant clones were grown overnight in LB-Kan medium, and DNA was isolated using the Qiagen Miniprep kit (Germany) according to the manufacturer’s instructions. The sequence of the selected plasmid DNA was verified by automated cycle sequencing.
The selected plasmid was transformed into E. coli BL21(DE3), plated onto LB-Kan and incubated overnight at 37 °C. A single colony of E. coli BL21 (DE3) containing pET28a-scFv was inoculated in LB-Kan medium and grown overnight in an orbital shaker at 30 °C. The overnight culture was diluted 40-fold in fresh LB-Kan medium and grown at 37 °C at 200 rpm till the culture reached an A600 of 0.8–0.9. The culture was induced with 1-mM Isopropyl-β-D-1-thiogalactopyranoside (IPTG) by incubating at 28 °C for 4 h. Following completion of induction, the bacterial pellet was collected by centrifugation at 5000 × g for 20 min at 4 °C.
Purification of scFv by immobilized metal affinity chromatography (IMAC)
The bacterial pellet was resuspended in lysis buffer (50 mM Tris–HCl, 155 mM NaCl, pH 7.6) to prepare a 10% (w/v) suspension. Lysozyme was added to a final concentration of 50 mg/10 ml of lysate and incubated overnight at –20 °C. The sample was subjected to sonication, centrifuged at 9200 × g for 30 min at 4 °C, and the supernatant was subjected to IMAC.
The supernatant was loaded onto an IMAC column (5 ml volume) equilibrated with 10 column volumes of 50 mM Tris–HCl, 155 mM NaCl, pH 7.6 (equilibration buffer) at a flow rate of 1ml/min and washed with 20 column volumes of washing buffer (equilibration buffer with 30 mM Imidazole, pH 7.6). Bound scFv was eluted with 5 column volumes of elution buffer containing equilibration buffer with 300 mM Imidazole, pH 7.6, as 1 ml fractions. All the eluted fractions were analyzed by SDS-PAGE and immunoblotting. Fractions containing the recombinant scFv were pooled and dialyzed against phosphate-buffered saline (PBS). Protein concentration was determined by the BCA method before storing it at –20 °C until further use.
Detection of scFv by SDS-PAGE and immunoblot analysis
The purified scFv was electrophoresed on SDS-PAGE (12% w/v acryl-amide gel) [23] and electroblotted onto a PVDF membrane (Hybond-C, GE Healthcare, USA) using a transblot apparatus (Bio-Rad, USA) following the manufacturer’s instructions. The blot was probed with His-probe (Pierce, Thermoscientific, Rockford, USA) and developed using 0.05% 3,3´-diaminobenzidine tetrahydrochloride (DAB) (Sigma, Saint Louis, USA) and 0.3% hydrogen peroxide in PBS.
Antigen-binding activity of scFv against bovine IgA by sandwich ELISA
Sandwich ELISA was performed using a commercial kit from Bethyl laboratories to evaluate the antigen binding activity of purified scFv. Titration of scFv (200 µg/ml) was performed against varying concentrations of bovine IgA (1 µg/ml to 1.95 ng/ml) in a precoated mAb strips as described by the manufacturer’s instructions. The binding of the scFv to bovine IgA was detected by addition of His-probe followed by 3,3´,5,5´-tetramethylbenzidine (TMB) substrate. The plate was incubated at 37 °C for 10 min and the reaction was stopped by addition of 1.25 M H2SO4. The absorbance was measured at 450 nm using a microplate reader (BIO-TEK, US).
Competitive ELISA
A competitive ELISA was performed to determine the competition between scFv and its parent Mab IL-A71 over the binding site on bovine IgA. Briefly, a microtiter plate was coated with 200 ng/well of bovine IgA in 50 mM carbonate-bicarbonate buffer (pH 9.6) and incubated overnight at 4 °C. The plate was washed thrice with PBS-T and blocked with 1% bovine gelatin in phosphate-buffered saline (PBS) containing 0.05% (v/v) Tween 20 (PBS-T) followed by washing with PBS-T to remove the excess gelatin. ScFv (1000 ng/100 µl) was added by serial two-fold dilution and incubated at 37 °C for 1 h. E. coli lysate was used as a negative control. A Mab IL-A71 specific for bovine IgA was added to each well containing scFv and E. coli lysate, incubated at 37 °C for 1 h. The plate was washed with PBS-T and dried by flicking. Goat anti-mouse IgG HRP conjugate (1:5000) was added to each well and the plate was incubated at 37 °C for 1 h. The plate was washed five times with PBS-T and 100 µl of H2O2-activated TMB (Sigma, USA) was added. The reaction was stopped after 10 min by addition of 100 µl of 1.25 M H2SO4 to each well, and absorbance was read at 450 nm using a microplate reader (BIO-TEK, USA).
Determination of specificity of the scFv against different classes of bovine Igs and IgA of different species
The binding specificity of scFv toward bovine IgA was evaluated by testing its reactivity with bovine IgG1, IgG2, IgM, and IgA of cattle, buffalo, sheep, goat, and canine by indirect ELISA. Briefly, serially diluted bovines IgA, IgG1, IgG2, IgM (100, 80, 60, 40, 20 ng/well) were coated onto microtiter wells. The wells were washed with PBS-T and blocked with 1% (w/v) bovine gelatin by incubating at 37 °C for 1 h. The wells were washed as described above and scFv was added and incubated for 1 h at 37 °C. The wells were washed with PBS-T, and scFv were detected by adding His-probe (1: 5000 dilutions) followed by TMB substrate. The plate was incubated at 37 °C for 10 minutes and the reaction was stopped by addition of 1.25 M H2SO4. The absorbance was measured at 450 nm using a microplate reader (BIO-TEK, US).
Subsequently, in another set of experiments, saliva samples of cattle, buffalo, sheep, goat, and canine was coated onto a micro titer wells at a dilution of 1:5. The wells were washed and blocked as mentioned above, and IgA present in the saliva samples was detected by both scFv and Kit Mab, respectively.
Affinity measurement
Affinity of scFv was determined using BIACore X-100 following ligand capture protocol. NTA sensor chip (GE, USA) was used to capture 6X His-tagged ScFv on to the chip surface. Ligand capture and analyte binding conditions were optimized prior to performing the kinetic analysis, using BIACore Applications wizard (GE, USA). For affinity measurement, 50 ng/ml (2 nM) scFv was used. The standard IgA was diluted to concentrations ranging from 20 to 125 ng/ml (12–78 nM) in 1×HBS-EP buffer (GE, USA). Affinity measurement was performed using multiple injection workflows with contact time of 180 s for binding and dissociation time of 400 s for each concentration of IgA. Sensor chip regeneration and kinetic analysis workflow was setup as suggested by the BIAcore Wizard template (GE, USA). Surface plasmon resonance (SPR) data were evaluated, assuming 1:1 interaction, using BIAEvaluation software (GE, USA). The kinetic measurements were verified with internal consistency tests.
Saliva sample collection
Saliva samples were collected from both naïve cattle that were neither vaccinated nor infected with FMDV and also from cattle that had been experimentally infected with FMDV. Saliva samples were collected by placing a 1/6th portion of a regular size cotton tampon (Tampax®, Kiev, Ukraine) pre-dampened by the addition of 0.5 ml of PBS (pH 7.3–7.5) underneath the tongue and also from the vestibule. Approximately 1–2 ml of saliva was extracted from each tampon by compression within the barrel of a syringe or by centrifugation for 10 min at 1862 × g before storage at –20 °C. Saliva samples were also collected from buffaloes, sheep, goats, and canines for cross-reactivity study.
Detection of bovine IgA in standard using both scFv and commercial anti-bovine IgA
Indirect ELISA was performed for the qualitative analysis of bovine IgA in standard sample using both scFv and commercial anti-bovine IgA. Briefly, standard IgA was coated onto microtiter wells by incubating at 4 °C for overnight. ScFv (250 ng/well) and anti-bovine IgA were added and serially diluted to the coated well and incubated further for 1 h at 37 °C. The binding of the scFv and anti-bovine IgA toward bovine IgA was detected by addition of His-Probe and anti-mouse HRPO, respectively, followed by the addition of 3,3´,5,5´-tetramethylbenzidine (TMB) substrate. The plate was incubated at 37 °C for 10 min and the reaction was stopped by addition of 1.25 M H2SO4. The absorbance was measured at 450 nm using a microplate reader (BIO-TEK, US).
Detection of FMDV specific bovine IgA in saliva samples by immunocapture ELISA
An immunocapture ELISA was performed for the detection of IgA antibodies raised against the structural proteins of FMDV. A polyclonal rabbit anti-FMDV antibody was coated to the microtiter wells at a dilution of 1:6000 in carbonate buffer pH 9.6 and blocked using blocking buffer (3% w/v skim milk containing 0.05% v/v Tween 20) by incubating at 37 °C for 1 h. The wells were washed with PBS-T and concentrated, inactivated FMDV antigen (1:10 in blocking buffer) was added to the wells and incubated for 1 h. After washing the microtiter wells, test saliva was added (1:5 in blocking buffer) and further incubated for 1 h at 37 °C. The microtiter wells were washed with PBS-T and detected using both anti-bovine IgA HRPO and anti-bovine IgA scFv (375 ng/well). The scFv was detected with His-probe. The reaction was developed by the addition of substrate (OPD: Sigma, USA and H2O2: Merck, Germany) and stopped using 1 M H2SO4, the plates were read on a multi-channel spectrophotometer (Versamax, Molecular device, USA) at 492 nm (A492). A sample was considered negative when the optical density (OD) value was less than the mean value for the negative control plus two standard deviations. The sample was considered positive when the OD value is above the mean plus two standard deviations.
Results
Assembly, cloning and expression of scFv
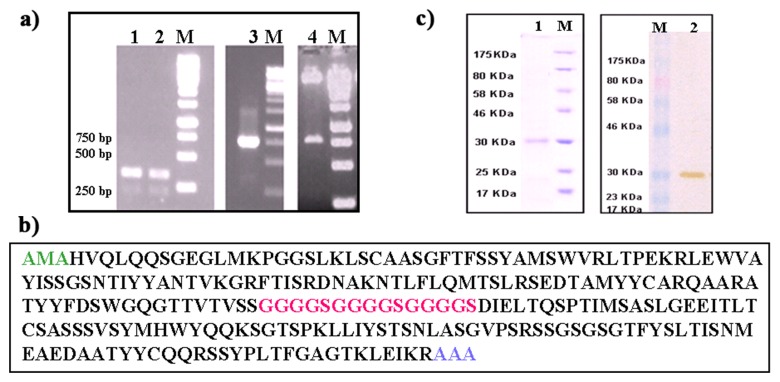
The VH and VL domains of the antibody fragment were PCR amplified from a mouse hybridoma-secreting anti-bovine IgA monoclonal antibody (Fig. 1a, lanes 1 and 2). The amplified VH and VL chains were joined together with the 45-mer-polynucleotide linker using SOE PCR and the resultant 723 bp long PCR product (Fig. 1a, lane 3) was cloned into pET28a vector (Fig. 1a, lane 4) to yield pET28a-scFv. The pET28a-scFv was sequenced and the entire sequence was submitted to the international ImmunoGeneTics information system (IMGT) for sequence verification. Sequence information revealed the presence of 120 aminoacids long VH, 106 aminoacids long VL and a 15 aminoacids long linker region (Fig. 2c). The pET28a-scFv was transformed into competent E. coli expression cells. Following 4 h of induction, the bacterial cells were harvested, lysed, and the soluble cytoplasmic solution was loaded onto Ni-NTA column, and eluted fractions were collected. The purified recombinant scFv analyzed by SDS-PAGE and immunoblotting indicated the presence of a ~30 kDa band (Fig. 1b, lanes 1 and 2).
Fig. 1.
(a) Electrophoretic analysis of PCR amplified variable heavy and light chain domains. These gene sequences were amplified from total RNA isolated from a hybridoma secreting cell line IL-A71. Lane M: Molecular weight markers. Lanes 1, 2, 3 show variable heavy and light chain genes and assembled scFv PCR products. Lane 4 shows the release of 750 bp product, recombinant expression cassette after EcoRI and NotI digestion. (b) Amino acid sequence of anti-bovine IgA scFv containing VL, linker peptide and VH. The linker peptide is marked in italics. Underlining indicates the restriction enzyme sites for cloning the scFv gene. (c) SDS-PAGE and Western blot analysis of scFv expression. Lane M: molecular mass marker. Lanes 1 and 2 are purified samples from Ni-NTA agarose column. Sample was separated with 12% SDS-PAGE, followed by Coomassie blue staining (lane 1). For Western blotting of purified sample from Ni-NTA affinity column was transferred onto a PVDF membrane and probed with anti-His probe for 1 h. The antibody was detected by using DAB substrate (lane 2)
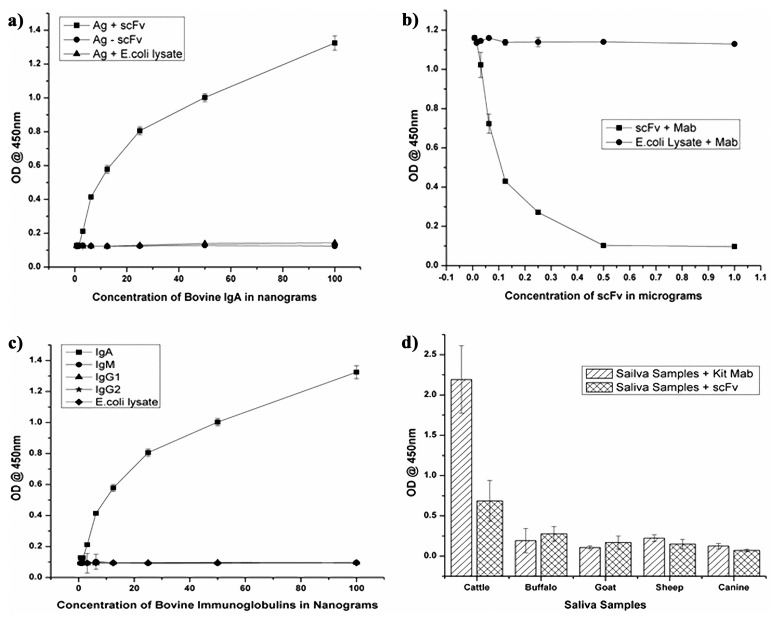
Fig. 2.
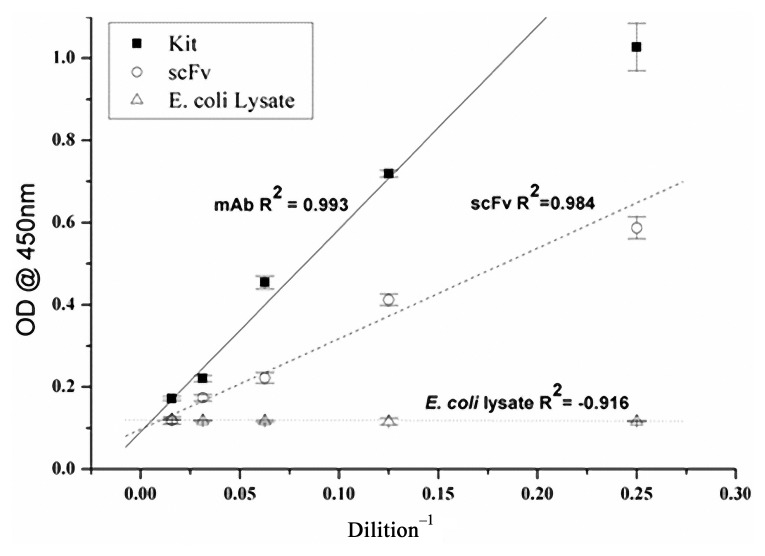
(a) Analysis of antigen-binding affinity of scFv by ELISA to evaluate the activity of scFv towards IgA. (b) Competitive ELISA using the scFv and the anti-bovine IgA Mab. X-axis values are the concentrations of scFv and Y axis shows the optical density values. (c) Reactivity of scFv toward different immunoglobulins of bovine (IgA, IgG1, IgG2, and IgM) by indirect ELISA. (d) Reactivity of scFv toward saliva samples of cattle, buffalo, sheep, goat, and canine by indirect ELISA
Determination of scFv binding to IgA by sandwich ELISA
The antigen-binding activity of the purified scFv was measured by sandwich ELISA. Titration of the scFv against bovine IgA revealed a concentration-dependent reduction of the optical density values as shown in Fig. 2a. No cross-reactivity was observed with the lysate of E. coli, indicating the binding specificity of scFv.
Competitive ELISA
Competitive ELISA was performed to determine the competition between scFv and parent Mab IL-A71 for the binding site on bovine IgA. Competition was observed when the constant amount of Mab IL-A71 was allowed to compete with varying amount of the scFv. Gradual increase in optical density values following the dilution of the scFv indicated that the scFv competed with Mab IL-A71 for the same antigenic site on bovine IgA (Fig. 2b).
Cross-reactivity by ELISA of scFv with different classes of bovine immunoglobulins and different species salivary IgA
The specificity of the scFv was assayed by performing indirect ELISA which showed reactivity with bovine IgA and no cross-reactivity was observed with other classes of bovine immunoglobulins (IgG1, IgG2, and IgM) (Fig. 2c) and it is also observed that scFv is reactive to saliva of cattle and not to saliva samples of other species (buffalo, sheep, goat, and canine) (Fig. 2d).
Affinity of scFv toward purified IgA
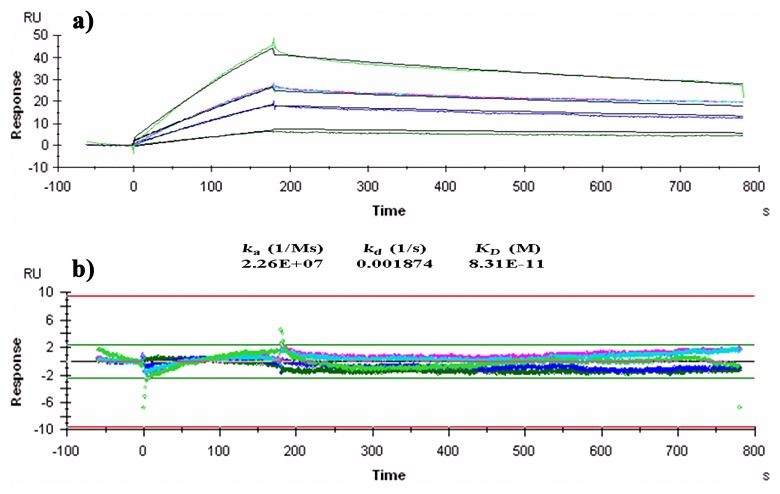
Since the parent monoclonal antibody, IL-A71, reacts to the Fc portion of IgA, immobilization of IgA or scFv could potentially interfere with the three-dimensional structure of the molecule. Therefore, ligand capture method was employed for measuring affinity of scFv. The interaction between scFv and purified IgA standards was studied at 25 °C over a concentration range of 12–78 nM of purified IgA. The association and dissociation rate constants for the interaction was found to be 2.26 × 107 M–1s–1 and 1.87 × 103 s–1, respectively. The affinity constant of the scFv to IgA was found to be 8.1 × 1011 M–1 (Fig. 3a). The residual distribution plot was linear with ±2 RU deviation (Fig. 3b) and no significant bulk contribution (RI) to the sensogram was found, these quality parameters validate affinity measurement.
Fig. 3.
(a) BIAcore analysis of anti-bovine IgA scFv. Sensogram demonstrates the binding and dissociation of scFv. Affinity was calculated assuming 1:1 binding and curve fitting of the sensogram. RU = Response Unit; Time in seconds; ka = association rate constant; kd = dissociation rate constant; KD = equilibrium dissociation constant/affinity constant. (b) Residual distribution plot of response in the kinetic analysis. In both figures, ligand (anti-IgA scFv) concentration was kept constant (50 ng/ml), and analyte (bovine IgA) concentration was varied and represented in different colors, 20 ng/ml (dark green), 50 ng/ml (purple), 100 ng/ml in duplicate (pink and sky blue), and 125 ng/ml (light green)
Determination of the FMDV specific bovine IgA in salivary samples by Immunocapture ELISA (IC-ELISA)
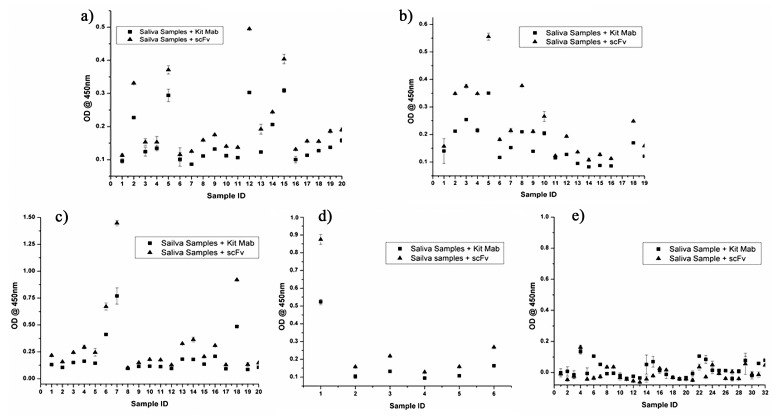
The bovine IgA was detected using anti-bovine IgA scFv and commercial anti-bovine IgA in both standard (Fig. 4) and saliva samples (Fig. 5). Detection of the bovine IgA standard using scFv and the commercial anti-bovine IgA showed a good fit with r2 values of >0.9 in both cases, indicating that detection of IgA using either of the methods remains linear over a wide concentration range. FMD antigen-specific bovine IgA was detected in 1 out of 18 vaccinated and protected animals (Fig. 5b), 3 out of 21 vaccinated and infected animals (Fig. 5c), and 1 out of 6 unvaccinated and infected animals (Fig. 5d). All the five animals which were found IgA positive were FMDV carriers and the carrier status was confirmed by isolation of virus from probang samples and detection of NSP antibodies at 35 days post-challenge. No FMDV antigen specific IgA was detected in 20 vaccinated animals (Fig. 5a), 17 vaccinated and protected animals (Fig. 5b), 18 vaccinated and infected animals (Fig. 5c), 5 unvaccinated and infected animals (Fig. 5d) and in 32 naïve animals as well (Fig. 5e). The cutoff OD for positive samples was set at 0.520 for the samples detected with anti-bovine IgA scFv and 0.372 for the samples detected with commercial anti-bovine IgA.
Fig. 4.
Detection of the IgA standard using both scFv and commercial anti-bovine IgA. X axis shows the different dilutions of scFv/commercial antibody and Y axis shows the optical density values
Fig. 5.
Detection of the FMDV-specific IgA content using scFv and commercial anti-bovine IgA in bovine salivary samples by immunocapture ELISA. (a) Salivary samples of FMDV-vaccinated animals. (b) Salivary samples of FMDV-vaccinated and protected animals. (c) Salivary samples of FMDV-vaccinated and infected animals. (d) Salivary samples of FMDV-unvaccinated and infected animals. (e) Salivary samples of naïve animals. The cut-off values of kit Mab and scFv is 0.372 and 0.520 at OD450, respectively
Discussion
Immunoglobulin A (IgA) is a predominant class of immunoglobulin present on the mucosal surfaces, which constitutes the first line of defense against various infectious diseases. It is known to be one of the primary determinants that would indicate enhanced mucosal immune response/protection [24]. Secretory IgA is transported into mucosal secretions and is resistant to proteases, prevents adhesion of bacteria/toxins to target cells, and neutralizes viruses and toxins, among other characteristics [25–28].
All current in vitro estimation tests rely on polyclonal or monoclonal hybridoma derived antibody reagents. Monoclonal antibodies may offer substantial advantages in terms of potency, reproducibility and freedom from contaminants but they are not easy to use as they require eukaryotic expression platform or the maintenance of viable hybridoma cell line, which is difficult to prepare and maintain in a quality-assured manner and in the quantities required. To overcome these problems, generation of single chain antibody fragments (scFv) through phage display technology has been utilized as the methodology of choice [29, 30] for antibodies and these antibodies have many practical applications, including immuno assay and therapy.
Recombinant DNA technology has been used to a great extent in the expression of antibodies/antibody fragments. Antibody fragments can be readily produced from the genes encoding antibody variable domains which can be derived either from hybridoma [31] or from bacteriophage displaying antibody fragments [32]. These recombinant antibodies are identical to traditional monoclonal antibodies in their basic functionality. They can be manufactured using fully in vitro processes thus offering greater flexibility during their production and greater opportunities for optimization after their creation than typical monoclonal antibodies.
Single-chain antibody variable region fragments by virtue of their size and method of construction are potentially useful as therapeutic reagents and as tools for exploring cell surface receptor function. ScFv offer several advantages over the intact immunoglobulin molecule. For instance, they are expressed from a single transcript and can be molecularly linked with other proteins to generate bispecific scFv molecules or single chain immunotoxins. The relatively small size of scFv is an advantage in allowing for easy penetrance into the tissue spaces, and their clearance rate is exceedingly rapid. ScFv are useful for gene therapy since they can be directed to a specific cellular localization and can be fused to retroviral envelope genes to control viral host range [33]. Diagnostic assays are associated with the use of polyclonal and monoclonal antibodies. The production of these antibodies includes labour-intensive multistep processes for purification which results in poor-defined nature of the final product caused as a result of being genetically instable [34], resulting in lack of quality and quantity of diagnostic kits [35]. To overcome the above-mentioned limitations recombinant antibodies are widely used as an alternative to conventional monoclonal antibodies for the selection, screening and production of custom-produced immunological reagents while preserving the advantages of Mabs [36]. Moreover, the expression of scFv required a prokaryotic system compared to the monoclonal antibodies expression in complex eukaryotic platform which involves high cost. Added to this, the present study shows that scFv is much sensitive in detecting bovine IgA in the biological samples when compared to the full-length monoclonal antibody of the kit.
In the present paper, we describe the construction, expression, purification and immunological characterization of an anti-bovine IgA scFv and its application for the detection of FMDV-specific IgA in salivary samples by ELISA. We constructed the recombinant scFv from hybridoma and expressed in bacterial system, and the ease of purification reiterated the fact that expression of a functional recombinant antibody in bacteria offered many advantages over the maintenance of a hybridoma cell line at minimal expense.
In order to demonstrate the binding of the scFv to the IgA, ELISA was performed using commercial kit from Bethyl laboratories (USA). Cross-reactivity with IgG1, IgG2 and IgM immunoglobulins was studied which showed that the scFv reacted only with IgA but not with other immunoglobulins. In addition, scFv also did not show any reactivity with IgA derived from other species such as buffalo, sheep, goat, and canine. Affinity of the recombinant scFv was determined to be in the picomolar range, 8.31 × 1011 M–1. Affinity constant of scFv–IgA interaction was uniquely determined; confirming such high affinity values using alternate analytical techniques such as iso-thermal titration calorimetry may be required.
Further, the scFv competed with parent mAb IL-A71 for binding to IgA in a concentration-dependent manner, suggesting that scFv and parent mAb IL-A71 bound to the same epitope. Detection of the bovine IgA standard by ELISA using in-house and the commercial anti-bovine IgA Bethyl Laboratories (Texas, USA) showed a near perfect linear fit with r2 of >0.9 in both cases.
In the present study, scFv was used to detect the FMD-specific IgA from unvaccinated-infected, vaccinated-infected cattle samples at 35 days post-challenge. However, naïve and vaccinated and protected animals did not show any FMDV-specific IgA antibody at 35 days post-challenge. Specific salivary IgA levels were elevated after infection and oropharyngeal (OP) persistence of FMD virus, regardless of vaccination status, and IgA in saliva was correlated to persistence of virus or viral genome in the OP fluids. This IgA ELISA has considerable potential for the detection of subclinical infection and persistently infected cattle following vaccination and challenge exposure.
To our knowledge, this is the first report of the development of an anti-bovine IgA scFv and its potential use in a diagnostic assay for the detection of FMDV-specific IgA antibody in salivary samples. The development of a diagnostic test for FMDV could provide a novel tool to be utilized as part of an effective and viable FMDV control strategy.
Glossary
Abbreviations
- scFv:
single chain fragment variable
- VH:
variable heavy chain
- VL:
variable light chain
- IgA:
immunoglobulin A
- FMDV:
foot and mouth disease virus
- cDNA:
complementary deoxyribonucleic acid
- IMAC:
immobilized metal affinity chromatography
- IPTG:
Isopropyl-β-D-1-thiogalactopyranoside
- Ni-NTA:
nickel-nitrilotriacetic acid
- SDS-PAGE:
Sodium dodecyl sulphate-Polyacrylamide gel electrophoresis
- TMB:
3,3´,5,5´-tetramethylbenzidine
- DAB:
3-3´-diaminobenzidine
Contributor Information
N. V. Sridevi, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India
A. M. Shukra, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India
B. Neelakantam, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India
J. Anilkumar, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India
M. Madhanmohan, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India
S. Rajan, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India
Dev Chandran, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India.
V. A. Srinivasan, Research and Development Center, Indian Immunologicals Limited, Rakshapuram, Gachibowli, Hyderabad, Andhra Pradesh, 5000032, India.
References
- 1.Renegar KB, Small PA., Jr. Passive transfer of local immunity to influenza virus infection by IgA antibody. J Immunol. 1991 Mar 15;146(6):1972–1978. [PubMed] [Google Scholar]
- 2.Winner L, 3rd, Mack J, Weltzin R, Mekalanos JJ, Kraehenbuhl JP, Neutra MR. New model for analysis of mucosal immunity: intestinal secretion of specific monoclonal immunoglobulin A from hybridoma tumors protects against Vibrio cholerae infection. Infect Immun. 1991 Mar;59(3):977–982. doi: 10.1128/iai.59.3.977-982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stubbe H, Berdoz J, Kraehenbuhl JP, Corthésy B. Polymeric IgA is superior to monomeric IgA and IgG carrying the same variable domain in preventing Clostridium difficile toxin A damaging of T84 monolayers. J Immunol. 2000 Feb 15;164(4):1952–1960. doi: 10.4049/jimmunol.164.4.1952. [DOI] [PubMed] [Google Scholar]
- 4.Friman V, Nowrouzian F, Adlerberth I, Wold AE. Increased frequency of intestinal Escherichia coli carrying genes for S fimbriae and haemolysin in IgA-deficient individuals. Microb Pathog. 2002 Jan;32(1):35–42. doi: 10.1006/mpat.2001.0477. [DOI] [PubMed] [Google Scholar]
- 5.Burnett PR, VanCott TC, Polonis VR, Redfield RR, Birx DL. Serum IgA-mediated neutralization of HIV type 1. J Immunol. 1994 May 1;152(9):4642–4648. [PubMed] [Google Scholar]
- 6.Russell MW, Kilian M. Biological activities of IgA. In: Mestecky J, Bienenstock J, Lamm ME, Mayer L, Strober W, et al., editors. Mucosal Immunology. San Diego: Academic Press; 2005. pp. 267–289. [Google Scholar]
- 7.Orlandi R, Güssow DH, Jones PT, Winter G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989 May;86(10):3833–3837. doi: 10.1073/pnas.86.10.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990 Dec 6;348(6301):552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 9.Brumester J, Pluckthun A. Construction of scFv fragments from hybridoma or spleen cells by PCR assembly. In: Kontermann R, Dubel S, editors. Antibody Engineering. Berlin, Germany: Springer-Verlag; 2001. pp. 19–40. [Google Scholar]
- 10.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotný J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M. Single-chain antigen-binding proteins. Science. 1988 Oct 21;242(4877):423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- 12.Condra JH, Sardana VV, Tomassini JE, Schlabach AJ, Davies ME, Lineberger DW, Graham DJ, Gotlib L, Colonno RJ. Bacterial expression of antibody fragments that block human rhinovirus infection of cultured cells. J Biol Chem. 1990 Feb 5;265(4):2292–2295. [PubMed] [Google Scholar]
- 13.Chen LH, Huang Q, Wan L, Zeng LY, Li SF, Li YP, Lu XF, Cheng JQ. Expression, purification, and in vitro refolding of a humanized single-chain Fv antibody against human CTLA4 (CD152) Protein Expr Purif. 2006 Apr;46(2):495–502. doi: 10.1016/j.pep.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Padiolleau-Lefevre S, Alexandrenne C, Dkhissi F, Clement G, Essono S, Blache C, Couraud JY, Wijkhuisen A, Boquet D. Expression and detection strategies for an scFv fragment retaining the same high affinity than Fab and whole antibody: Implications for therapeutic use in prion diseases. Mol Immunol. 2007 Mar;44(8):1888–1896. doi: 10.1016/j.molimm.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 15.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005 Sep;23(9):1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 16.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006 May;6(5):343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 17.Filpula D. Antibody engineering and modification technologies. Biomol Eng. 2007 Jun;24(2):201–215. doi: 10.1016/j.bioeng.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Luginbühl B, Kanyo Z, Jones RM, Fletterick RJ, Prusiner SB, Cohen FE, Williamson RA, Burton DR, Plückthun A. Directed evolution of an anti-prion protein scFv fragment to an affinity of 1 pM and its structural interpretation. J Mol Biol. 2006 Oct 13;363(1):75–97. doi: 10.1016/j.jmb.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 19.Amadori M, Haas B, Moos A, Zerbini I. IgA response of cattle to FMDV infection in probang and saliva samples. EU FMD, Ras Gr, Borovets. 2000;(Appendix 9):88–106. [Google Scholar]
- 20.Salt JS, Mulcahy G, Kitching RP. Isotype-specific antibody responses to foot-and-mouth disease virus in sera and secretions of "carrier' and "non-carrier' cattle. Epidemiol Infect. 1996 Oct;117(2):349–360. doi: 10.1017/s0950268800001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Archetti IL, Amadori M, Donn A, Salt J, Lodetti E. Detection of foot-and-mouth disease virus-infected cattle by assessment of antibody response in oropharyngeal fluids. J Clin Microbiol. 1995 Jan;33(1):79–84. doi: 10.1128/jcm.33.1.79-84.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks JD, Hoogenboom HR, Griffiths AD, Winter G. Molecular evolution of proteins on filamentous phage. Mimicking the strategy of the immune system. J Biol Chem. 1992 Aug 15;267(23):16007–16010. [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Mestecky J, Russell MW, Elson CO. Intestinal IgA: novel views on its function in the defence of the largest mucosal surface. Gut. 1999 Jan;44(1):2–5. doi: 10.1136/gut.44.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazanec MB, Kaetzel CS, Lamm ME, Fletcher D, Nedrud JG. Intracellular neutralization of virus by immunoglobulin A antibodies. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6901–6905. doi: 10.1073/pnas.89.15.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazanec MB, Nedrud JG, Kaetzel CS, Lamm ME. A three-tiered view of the role of IgA in mucosal defense. Immunol Today. 1993 Sep;14(9):430–435. doi: 10.1016/0167-5699(93)90245-G. [DOI] [PubMed] [Google Scholar]
- 27.Mazanec MB, Coudret CL, Fletcher DR. Intracellular neutralization of influenza virus by immunoglobulin A anti-hemagglutinin monoclonal antibodies. J Virol. 1995 Feb;69(2):1339–1343. doi: 10.1128/jvi.69.2.1339-1343.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaetzel CS, Robinson JK, Chintalacharuvu KR, Vaerman JP, Lamm ME. The polymeric immunoglobulin receptor (secretory component) mediates transport of immune complexes across epithelial cells: a local defense function for IgA. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8796–8800. doi: 10.1073/pnas.88.19.8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003 Feb;21(2):163–170. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 30.Hanes J, Jermutus L, Weber-Bornhauser S, Bosshard HR, Plückthun A. Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc Natl Acad Sci U S A. 1998 Nov 24;95(24):14130–14135. doi: 10.1073/pnas.95.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlandi R, Güssow DH, Jones PT, Winter G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989 May;86(10):3833–3837. doi: 10.1073/pnas.86.10.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990 Dec 6;348(6301):552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 33.Gilliland LK, Norris NA, Marquardt H, Tsu TT, Hayden MS, Neubauer MG, Yelton DE, Mittler RS, Ledbetter JA. Rapid and reliable cloning of antibody variable regions and generation of recombinant single chain antibody fragments. Tissue Antigens. 1996 Jan;47(1):1–20. doi: 10.1111/j.1399-0039.1996.tb02509.x. [DOI] [PubMed] [Google Scholar]
- 34.Ali M, Hitomi K, Nakano H. Generation of monoclonal antibodies using simplified single-cell reverse transcription-polymerase chain reaction and cell-free protein synthesis. J Biosci Bioeng. 2006 Mar;101(3):284–286. doi: 10.1263/jbb.101.284. [DOI] [PubMed] [Google Scholar]
- 35.Foord AJ, Muller JD, Yu M, Wang LF, Heine HG. Production and application of recombinant antibodies to foot-and-mouth disease virus non-structural protein 3ABC. J Immunol Methods. 2007 Apr 10;321(1-2):142–151. doi: 10.1016/j.jim.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Cheung SC, Dietzschold B, Koprowski H, Notkins AL, Rando RF. A recombinant human Fab expressed in Escherichia coli neutralizes rabies virus. J Virol. 1992 Nov;66(11):6714–6720. doi: 10.1128/jvi.66.11.6714-6720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]