Abstract
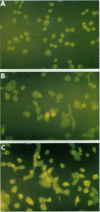
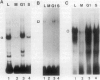
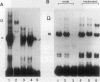
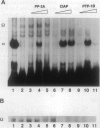
The histone gene family in mammals consists of 15-20 genes for each class of nucleosomal histone protein. These genes are classified as either replication-dependent or -independent in regard to their expression in the cell cycle. The expression of the replication-dependent histone genes increases dramatically as the cell prepares to enter S phase. Using mouse histone genes, we previously identified a coding region activating sequence (CRAS) involved in the upregulation of at least two (H2a and H3) and possibly all nucleosomal replication-dependent histone genes. Mutation of two seven-nucleotide elements, alpha and omega, within the H3 CRAS causes a decrease in expression in stably transfected Chinese hamster ovary cells comparable with the effect seen upon deletion of the entire CRAS. Further, nuclear proteins interact in a highly specific manner with nucleotides within these sequences. Mutation of these elements abolishes DNA/protein interactions in vitro. Here we report that the interactions of nuclear factors with these elements are differentially regulated in the cell cycle and that protein interactions with these elements are dependent on the phosphorylation/dephosphorylation state of the nuclear factors.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artishevsky A., Wooden S., Sharma A., Resendez E., Jr, Lee A. S. Cell-cycle regulatory sequences in a hamster histone promoter and their interactions with cellular factors. 1987 Aug 27-Sep 2Nature. 328(6133):823–827. doi: 10.1038/328823a0. [DOI] [PubMed] [Google Scholar]
- Baldin V., Lukas J., Marcote M. J., Pagano M., Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993 May;7(5):812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- Bowman T. L., Hurt M. M. The coding sequences of mouse H2A and H3 histone genes contains a conserved seven nucleotide element that interacts with nuclear factors and is necessary for normal expression. Nucleic Acids Res. 1995 Aug 25;23(16):3083–3092. doi: 10.1093/nar/23.16.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Roberts S. B., Heintz N. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 1988 Dec;2(12B):1700–1712. doi: 10.1101/gad.2.12b.1700. [DOI] [PubMed] [Google Scholar]
- Fletcher C., Heintz N., Roeder R. G. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987 Dec 4;51(5):773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Gallinari P., La Bella F., Heintz N. Characterization and purification of H1TF2, a novel CCAAT-binding protein that interacts with a histone H1 subtype-specific consensus element. Mol Cell Biol. 1989 Apr;9(4):1566–1575. doi: 10.1128/mcb.9.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppi V. E., Jr, Coffino P. G1 and S phase mammalian cells synthesize histones at equivalent rates. Cell. 1980 Aug;21(1):195–204. doi: 10.1016/0092-8674(80)90127-0. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Böhni R., Schneiderman M. H., Ramamurthy L., Schümperli D., Marzluff W. F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991 May;11(5):2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991 Mar 26;1088(3):327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- Hurt M. M., Bowman T. L., Marzluff W. F. A common transcriptional activator is located in the coding region of two replication-dependent mouse histone genes. Mol Cell Biol. 1991 Jun;11(6):2929–2936. doi: 10.1128/mcb.11.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt M. M., Pandey N. B., Marzluff W. F. A region in the coding sequence is required for high-level expression of murine histone H3 gene. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4450–4454. doi: 10.1073/pnas.86.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaludov N. K., Pabón-Peña L., Hurt M. M. Identification of a second conserved element within the coding sequence of a mouse H3 histone gene that interacts with nuclear factors and is necessary for normal expression. Nucleic Acids Res. 1996 Feb 1;24(3):523–531. doi: 10.1093/nar/24.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Bella F., Gallinari P., McKinney J., Heintz N. Histone H1 subtype-specific consensus elements mediate cell cycle-regulated transcription in vitro. Genes Dev. 1989 Dec;3(12A):1982–1990. doi: 10.1101/gad.3.12a.1982. [DOI] [PubMed] [Google Scholar]
- La Bella F., Heintz N. Histone gene transcription factor binding in extracts of normal human cells. Mol Cell Biol. 1991 Dec;11(12):5825–5831. doi: 10.1128/mcb.11.12.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBella F., Sive H. L., Roeder R. G., Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988 Jan;2(1):32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- Larner A. C., David M., Feldman G. M., Igarashi K., Hackett R. H., Webb D. S., Sweitzer S. M., Petricoin E. F., 3rd, Finbloom D. S. Tyrosine phosphorylation of DNA binding proteins by multiple cytokines. Science. 1993 Sep 24;261(5129):1730–1733. doi: 10.1126/science.8378773. [DOI] [PubMed] [Google Scholar]
- Osley M. A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Quelle D. E., Ashmun R. A., Shurtleff S. A., Kato J. Y., Bar-Sagi D., Roussel M. F., Sherr C. J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993 Aug;7(8):1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Roberts S. B., Segil N., Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991 Aug 30;253(5023):1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- Schneiderman M. H., Dewey W. C., Highfield D. P. Inhibition of DNA synthesis in synchronized Chinese hamster cells treated in G1 with cycloheximide. Exp Cell Res. 1971 Jul;67(1):147–155. doi: 10.1016/0014-4827(71)90630-6. [DOI] [PubMed] [Google Scholar]
- Segil N., Roberts S. B., Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991 Dec 20;254(5039):1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- Shapiro D. J., Sharp P. A., Wahli W. W., Keller M. J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988 Jan-Feb;7(1):47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- Sherr C. J. Mammalian G1 cyclins. Cell. 1993 Jun 18;73(6):1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C., Schümperli D. 3' processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 1988 Oct 25;16(20):9399–9414. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TERASIMA T., TOLMACH L. J. Growth and nucleic acid synthesis in synchronously dividing populations of HeLa cells. Exp Cell Res. 1963 Apr;30:344–362. doi: 10.1016/0014-4827(63)90306-9. [DOI] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- van Wijnen A. J., Ramsey-Ewing A. L., Bortell R., Owen T. A., Lian J. B., Stein J. L., Stein G. S. Transcriptional element H4-site II of cell cycle regulated human H4 histone genes is a multipartite protein/DNA interaction site for factors HiNF-D, HiNF-M, and HiNF-P: involvement of phosphorylation. J Cell Biochem. 1991 Jun;46(2):174–189. doi: 10.1002/jcb.240460211. [DOI] [PubMed] [Google Scholar]