Significance
Many studies of wild populations reveal links between heterozygosity and fitness, with relatively heterozygous individuals carrying fewer parasites, living longer and being more attractive to mates. These patterns appear ubiquitous and are often highly significant, but heterozygosity usually accounts for very little of the total variation in fitness. However, most studies analyze only around 10 loci, representing a tiny fraction of the genome. We therefore used high-throughput DNA sequencing to estimate genome-wide heterozygosity based on over 10,000 loci and found it to accurately reflect inbreeding. Applied to wild harbor seals, genome-wide heterozygosity explained almost half of the variation in parasite infection. By implication, a greater proportion of fitness variation could be linked to genotype than previously thought.
Keywords: inbreeding, genetic variability, heterozygosity fitness correlation, single nucleotide polymorphism, RAD sequencing
Abstract
Proxy measures of genome-wide heterozygosity based on approximately 10 microsatellites have been used to uncover heterozygosity fitness correlations (HFCs) for a wealth of important fitness traits in natural populations. However, effect sizes are typically very small and the underlying mechanisms remain contentious, as a handful of markers usually provides little power to detect inbreeding. We therefore used restriction site associated DNA (RAD) sequencing to accurately estimate genome-wide heterozygosity, an approach transferrable to any organism. As a proof of concept, we first RAD sequenced oldfield mice (Peromyscus polionotus) from a known pedigree, finding strong concordance between the inbreeding coefficient and heterozygosity measured at 13,198 single-nucleotide polymorphisms (SNPs). When applied to a natural population of harbor seals (Phoca vitulina), a weak HFC for parasite infection based on 27 microsatellites strengthened considerably with 14,585 SNPs, the deviance explained by heterozygosity increasing almost fivefold to a remarkable 49%. These findings arguably provide the strongest evidence to date of an HFC being due to inbreeding depression in a natural population lacking a pedigree. They also suggest that under some circumstances heterozygosity may explain far more variation in fitness than previously envisaged.
It has long been known that inbreeding can reduce fitness, mainly through the unmasking of recessive or partially recessive deleterious alleles (1). Such effects are well documented in the laboratory (2, 3) but until recently have remained largely unstudied in natural populations (4, 5). This is because an individual’s inbreeding coefficient (f) can be calculated directly only by using a deep pedigree, and these pedigrees are seldom available outside the laboratory (6). However, because inbreeding increases an individual’s homozygosity, the heterozygosity of a panel of neutral genetic markers can in theory be used as a surrogate for f.
Initial searches for heterozygosity fitness correlations (HFCs) (7, 8) used allozymes, but the results obtained are difficult to interpret because the proteins themselves may be under selection. With the discovery of microsatellites, an abundant class of putatively neutral genetic marker, it became possible to look for HFCs without concerns about selection on the markers themselves. A burgeoning literature now shows that HFCs based on small numbers of microsatellite loci are found in many bird and mammal species for a remarkable range of fitness traits, including neonatal survival (9), parasite susceptibility (10) and lifetime reproductive success (11), and even behavioral qualities such as territory-holding ability (12), song complexity (13) and attractiveness (14). This weight of evidence suggests that HFCs are an important and widespread phenomenon in the natural world. It is therefore important to understand their basis.
Theory predicts that HFCs arise as a result of inbreeding depression, which will reduce the fitness of individuals in proportion to their inbreeding coefficient f (7). Therefore, variance in inbreeding coefficients within a population is necessary to generate HFCs. However, simulation and empirical studies indicate that the variance in f in natural populations is usually low, and estimates of f based on the small numbers of markers typically deployed are often very poor (15). As a result, HFCs will usually be weak or nonsignificant even when inbreeding actually explains a large proportion of fitness. Szulkin et al. (16) summarized this by saying that HFCs allow one to see only the “tip of the iceberg” and provided two examples in which heterozygosity explains 3% and 6% of trait variation even though inbreeding is expected to account for 24% and 30% of the variance, respectively. The most favorable situations to observe HFCs are in populations where inbred individuals are not rare due to factors such as small population size, extreme reproductive skew, and natal philopatry (15), but even in these cases using only a few markers severely curtails the power to observe the actual impact of inbreeding.
To explain HFCs, authors also frequently invoke “local effects” where one or a small number of the microsatellites used as markers are by chance linked to a gene experiencing heterozygote advantage (8). Local effects are widely discussed in the literature, but their importance is unclear given that balanced polymorphisms are thought to be rare (17) and strong linkage between random pairs of loci is infrequent (16). It has also been argued that the contribution of local effects to HFCs may be overstated due to many studies having used an inappropriate statistical framework (16).
One means of unambiguously testing whether inbreeding alone can explain HFCs is to deploy a larger number of genetic markers (18). If an HFC is due to inbreeding depression, the use of more markers will reduce the error variance in the estimation of genome-wide heterozygosity and thereby strengthen the correlation. In contrast, if the HFC is highly dependent on one or a few marker loci being by chance linked to a fitness locus with strong effects, as in the local effect model, adding more markers located throughout the genome will reduce the strength of the HFC as the contribution of any one marker becomes progressively diluted. Until recently, however, this approach was not available to most studies because of the prohibitive costs of developing and screening large numbers of additional microsatellites.
Restriction site associated DNA (RAD) sequencing (19) has recently emerged as a rapid and economic means of genotyping thousands of single nucleotide polymorphisms (SNPs) in virtually any organism. This approach concentrates high-throughput sequencing effort around restriction enzyme cut sites that are distributed across the genome, thereby generating sufficiently deep local sequence coverage to reliably call SNPs as being either heterozygous or homozygous. Although SNPs are individually less informative than microsatellites due to their lower allelic diversity, this should be more than compensated for by the large numbers of markers screened, in principle allowing genome-wide heterozygosity to be estimated with far greater precision than previously possible.
Here, we evaluated the ability of RAD sequencing to accurately estimate genome-wide heterozygosity using an experimental population of oldfield mice (Peromyscus polionotus subgriseus) with known pedigree-based inbreeding coefficients (20). We then applied RAD sequencing to a natural population of harbor seals (Phoca vitulina) to determine whether inbreeding explains previously reported HFCs for survivorship and parasite infection (21). We hypothesized that these HFCs should strengthen with the deployment of many thousands of SNPs if they are due to inbreeding.
Results
Relationship Between RAD Heterozygosity and f.
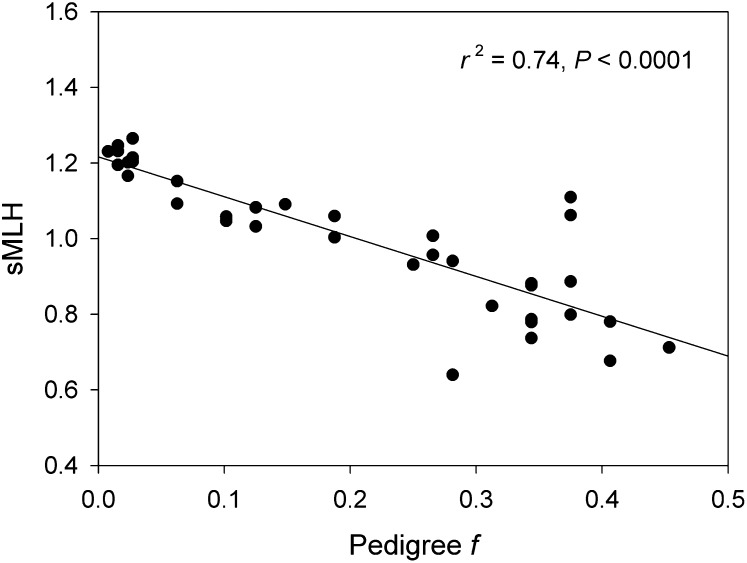
Illumina RAD sequencing of 36 oldfield mice yielded 265 million paired-end reads (Fig. S1) that were assembled into 63,129 RAD tags (Fig. S2A) as described in SI Methods and SI Results. After SNP calling and filtering (Figs. S3 and S4), 13,198 tags were retained for further analysis, each of which contained a single biallelic SNP (Fig. S5). Individual heterozygosity was expressed using the measure sMLH, which corrects for the fact that not all individuals are genotyped for the same loci by standardizing average multilocus heterozygosity in an individual by the average observed heterozygosities in the population of the subset of loci for which the focal individual was genotyped (10). RAD sMLH was strongly correlated with pedigree-based f (Fig. 1, r2 = 0.74, P < 0.0001), far more so than sMLH at 12 microsatellites (r2 = 0.28, P = 0.0003). A highly significant relationship was also obtained between RAD allele sharing and pedigree-based pairwise relatedness (r = 0.94, P < 0.0001), which was stronger than the equivalent relationship based on microsatellites (r = 0.68, P < 0.0001).
Fig. 1.
Correlation between pedigree-based inbreeding coefficient (f) and RAD standardized multilocus heterozygosity (sMLH) for 36 oldfield mouse individuals. The r2 for the equivalent relationship based on 12 microsatellites is substantially lower at 0.276.
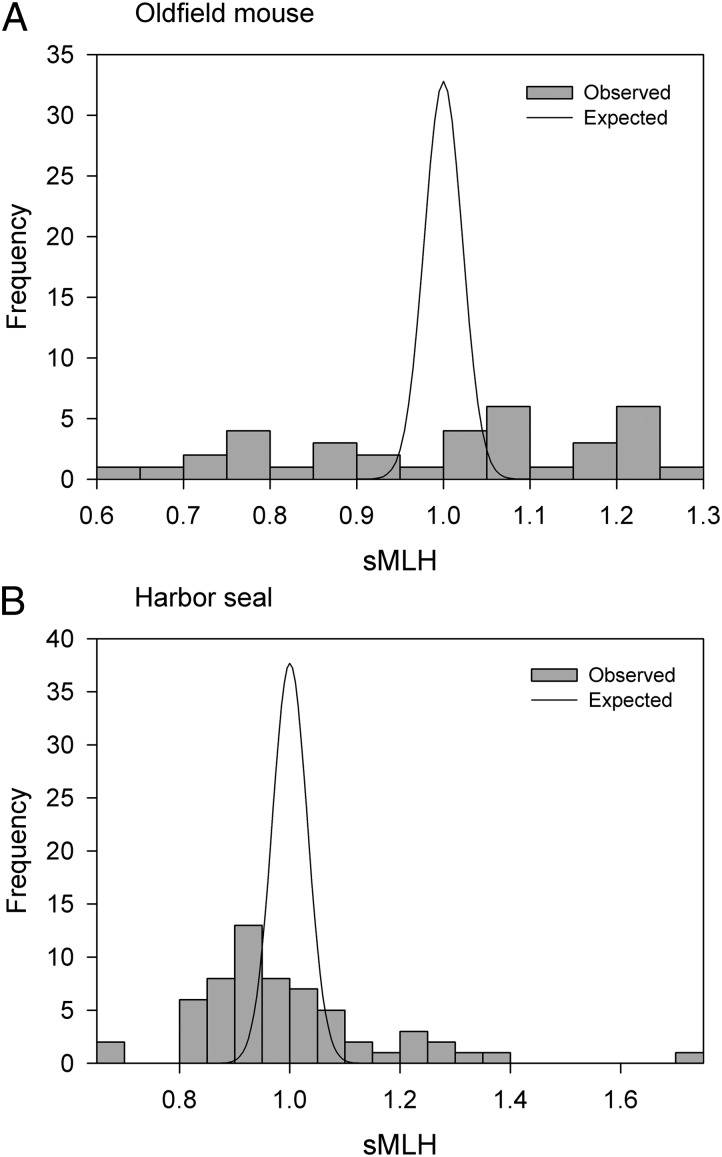
Variation in inbreeding within a population increases the variance in sMLH beyond that expected if heterozygosities at different loci were statistically independent. As expected, this genetic signature of inbreeding is conspicuous in a plot of the distribution of RAD sMLH in our sample of oldfield mice (Fig. 2A). The excess in variance is quantified by a parameter called g2, which measures the extent to which heterozygosities are correlated across loci and can be computed from genotype data using the method of David et al. (22). In our sample of oldfield mice, g2 calculated from 13,198 SNPs was 0.035 ± 0.007 SD, which is significantly different from zero (P < 0.001). With so many markers, the sampling of loci does not result in any appreciable estimation error, and even with as few as 2,000 SNPs this error is still very small (Fig. S6).
Fig. 2.
Observed distributions of RAD sMLH (shaded bars) together with expected distributions under the null hypothesis of g2 = 0 (solid curves), meaning that there is no variance in inbreeding. See Methods for details. Results are shown separately for (A) oldfield mice genotyped at 13,198 SNPs and (B) harbor seals genotyped at 14,585 SNPs.
The large number of SNPs deployed also suggests that RAD heterozygosity should provide a very precise estimate of individual f: From table 2 in Szulkin et al. (16) the expected correlation between sMLH and f, denoted rsMLH,f, is −1.03 (±0.024 SD). Pedigree-based estimates of f can be used to provide direct estimates of g2 and of rsMLH,f, which turn out to be slightly lower (g2 = 0.028 ± 0.009 SD; rsMLH,f = −0.86) than those based on the RAD data. As discussed later, this is to be expected because pedigree f does not capture all of the variance in inbreeding due to unknown founder relatedness, possible pedigree errors, and the random deviation of actual heterozygosity from the statistical expectation.
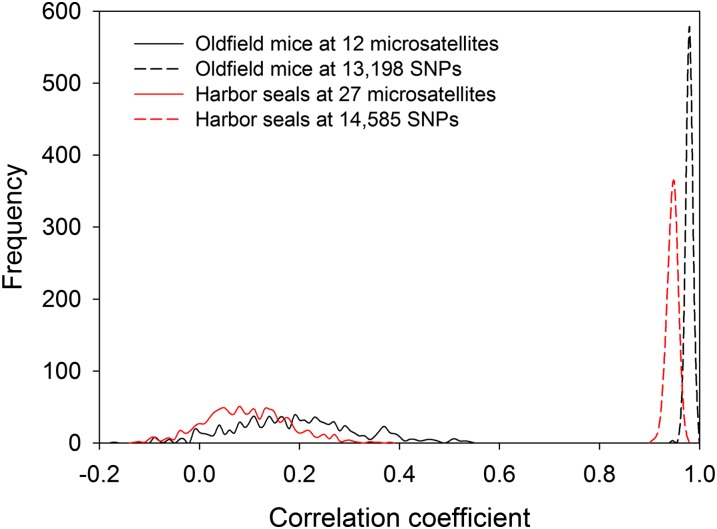
The accuracy of RAD-based heterozygosity estimates was further confirmed by dividing the markers into two random subsets, computing the correlation in sMLH between the two, and repeating this 1,000 times to obtain a distribution of heterozygosity–heterozygosity (het-het) correlation coefficients (15). The resulting values are tightly centered on a mean of 0.975 (±0.006 SD; Fig. 3) for the RAD data, whereas the equivalent value based on 12 microsatellites is only 0.187 (±0.125 SD). Again, as few as 2,000 SNPs are sufficient to obtain a strong positive correlation (Fig. S7A). This suggests that genome-wide heterozygosity can be reliably estimated from the many thousands of markers generated by RAD sequencing.
Fig. 3.
The strength of correlation in heterozygosity among loci. The markers for which animals were genotyped were randomly divided into two equal groups and the correlation coefficient (r) between the resulting sMLH values was calculated. This procedure was repeated 1,000 times to generate distributions of r values. Values centering around zero suggest that inbred individuals are rare or absent, whereas increasingly positive values indicate that inbred individuals are present.
Data on body mass at weaning were also available for the oldfield mice, allowing us to quantify the strength of association with f and marker heterozygosity. Separate generalized linear models (GLMs) of mass were constructed in which f, microsatellite sMLH, and RAD sMLH were fitted respectively as predictor variables, together with sex and the two-way interactions. Only the genetic terms were retained in the reduced models, in which f was significant at P = 0.027 (F1,35 = 5.35), microsatellite sMLH at P = 0.037 (F1,35 = 4.69), and RAD sMLH at P = 0.025 (F1,35 = 5.51). Thus, RAD sMLH is at least as good a predictor of body mass as pedigree f.
Application to a Natural Vertebrate Population.
Sixty harbor seals were RAD sequenced, generating 374 million paired-end reads (Fig. S1). These were assembled into 83,148 RAD tags (Fig. S2B), of which 14,585 were retained for further analysis (details in SI Methods and SI Results). Similarly to the oldfield mouse, a strong signal of inbreeding was present in the RAD data, with g2 estimated at 0.028 ± 0.009 SD (P < 0.001), which in our experience is a high value for a natural population (16). This is also illustrated by the empirical sMLH distribution (Fig. 2B), which has a much larger variance than expected in the absence of variation in inbreeding. Consequently, RAD heterozygosity is a very precise estimate of individual inbreeding (rsMLH,f = −0.97) in this sample, whereas the equivalent number based on 27 microsatellites is only −0.21. Accordingly, the mean het-het correlation coefficient increases from 0.090 ± 0.084 SD based on 27 microsatellites to 0.941 ± 0.011 SD (Fig. 3) for the RAD data. This signal is slightly weaker than in the oldfield mouse both overall and when equivalent-sized random subsets of SNPs are considered (Fig. S7A). However, this is expected, as g2 is lower than in the experimental oldfield mouse population where inbreeding coefficients range from 0 to 0.45. Moreover, the SNPs are on average less heterozygous in the harbor seals than in the mice (mean unstandardized MLH = 0.062 ± 0.011 SD and 0.164 ± 0.029 SD, respectively) and are therefore relatively less informative (Fig. S5).
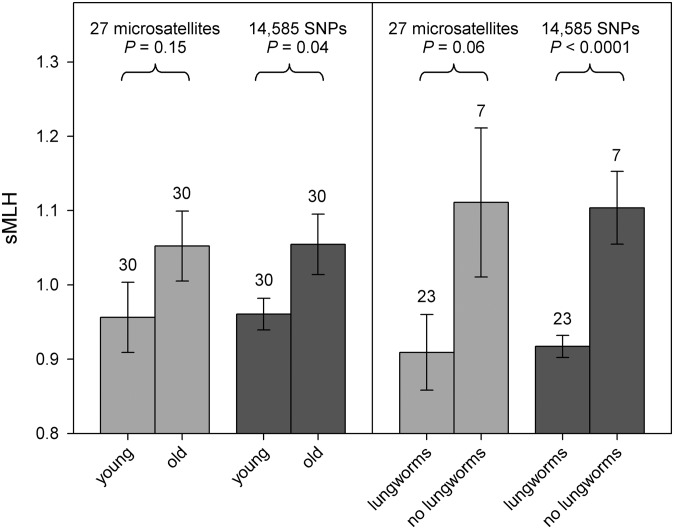
If an HFC is due to inbreeding depression, adding more markers should improve the estimate of f, and the relationship between fitness and average marker heterozygosity should strengthen (18). To test this prediction empirically, we fitted RAD sMLH as a predictor variable in GLMs of two fitness-related traits, longevity and parasite infection, both of which were coded as binary response variables (details in SI Methods). Young harbor seals, defined as being less than 1 y of age, showed a nonsignificant tendency to be less heterozygous at microsatellites than older seals (Fig. 4, F1,59 = 2.09, P = 0.15). This difference strengthened to become marginally significant with RAD heterozygosity (F1,59 = 4.39, P = 0.04). However, when a single highly heterozygous outlier (RAD sMLH = 1.73, two-tailed Grubbs’ test, α = 0.05) was removed from the analysis, RAD heterozygosity no longer remained significant (F1,58 = 3.14, P = 0.08).
Fig. 4.
Relationship between sMLH, estimated from microsatellites and RAD genotypes, and two fitness-related traits in harbor seals. Bars with light shading represent mean (± SEM) microsatellite sMLH and bars with dark shading represent mean (± SEM) RAD sMLH. Sample sizes are given above the bars. (Left) The mean (± SEM) sMLH of young and older seals, the former being classified as those that died before reaching 1 y of age (details in Methods). A nonsignificant tendency based on 27 microsatellites for younger seals to have lower heterozygosity than older seals strengthens to P = 0.04 when 14,585 SNPs are deployed. (Right) The mean (± SEM) sMLH of young seals with and without lungworms. A nonsignificant trend for infected seals to have lower heterozygosity than uninfected seals based on 27 microsatellites becomes significant at P < 0.0001 with 14,585 SNPs.
As infection and mortality due to lungworm burden occur mainly during the first year of life, we restricted our analysis of parasite infection to the young seals, 23 of which were infected with lungworms. We found that a close to significant trend for lungworm-infected seals to have lower microsatellite heterozygosity than uninfected seals (Fig. 4, F1,29 = 3.51, P = 0.06) became highly significant with the RAD data (F1,29 = 16.03, P = 6.23 × 10−5). Moreover, the deviance in lungworm infection explained by heterozygosity increased almost fivefold from 10.8% to 49.2%. By implication, genome-wide heterozygosity strongly influences susceptibility to lungworm infection.
To further test the prediction that an HFC should strengthen with marker number if inbreeding depression is responsible, we reran the GLM of parasite infection after randomly sampling five subsets each of between 50 and 14,000 SNPs and recalculating RAD sMLH. The average percentage deviance explained by heterozygosity increased gradually with SNP number before leveling off at around 7,000 loci (Fig. S7B). This pattern is very similar to the observed relationship between the number of randomly subsampled SNPs and the mean correlation in sMLH among markers (Fig. S7A). We also explored sensitivity to SNP minor allele frequency (MAF) by partitioning the marker set into loci with MAFs above and below 0.1, recalculating RAD sMLH for both and fitting these together in the model. Both categories of SNP were highly significant and thereby contribute toward the overall signal (MAF < 0.1, F1,29 = 18.40, P = 1.79 × 10−5; MAF > 0.1, F1,29 = 8.27, P = 0.004). That the lower-frequency SNPs were more significant probably reflects the far larger sample size of loci (12,455 vs. 2,130, respectively).
Discussion
We evaluated the ability of RAD sequencing to estimate genome-wide heterozygosity using a pedigreed oldfield mouse population with substantial inbreeding (up to f = 0.45) over six generations. We show that RAD heterozygosity is far more strongly correlated with f than microsatellite heterozygosity and that the r2 further increases to 0.93 if three outliers are removed. Additionally, the RAD data yield high het-het correlation coefficients and RAD sMLH is a marginally better predictor of body mass than f. Taken together, these findings suggest that RAD heterozygosity is a better estimate of genome-wide heterozygosity than pedigree-based estimates of f in our study. Although the exact reasons for the three outliers are unclear, in this case they are more likely the result of pedigree errors or mislabeled samples than unknown levels of relatedness among the founders. First, any effects of founder relatedness would be propagated throughout the pedigree and should therefore affect the majority of individuals. Second, although we do not have RAD genotypes of the founders, we were able to obtain relatively crude estimates of founder relatedness based on microsatellite and amplified fragment length polymorphism data reported previously (20). Adjusting the pedigree inbreeding coefficients with these estimates of founder relatedness did not improve the correlation with RAD heterozygosity. Regardless of the exact causes, our findings suggest that typing a very large number of loci may under certain circumstances allow one to estimate inbreeding even more precisely than from a pedigree. Furthermore, RAD sequencing provides a tool for error checking and otherwise refining pedigrees.
SNPs are attractive markers for correlating genome-wide heterozygosity with fitness (23). Although they are individually less variable than microsatellites, SNPs can be rapidly and economically genotyped in large numbers. Even so, it has been argued that for a large mammalian genome, a “herculean survey of 3,000 markers” will be required to produce a correlation between heterozygosity and f of around 0.4 (24). This is supported by two recent studies that correlated sMLH with f in captive zebra finches, obtaining r values of −0.07 based on 771 SNPs (25) and −0.46 based on 1,359 SNPs (26). However, we show that RAD sequencing is capable of screening the heterozygosity of at least an order of magnitude more SNPs, bringing a commensurate improvement in the estimation of genome-wide heterozygosity. In our study, much of the inbreeding signal is captured by a couple of thousand markers (Figs. S6 and S7), but clearly the number of loci required in other systems will depend upon the extent to which genome-wide heterozygosity varies among individuals (15). This is currently unknown for most natural populations, although approaches such as ours make it increasingly amenable to study.
A number of factors could potentially introduce bias into our sMLH estimates, although these would be expected to affect most individuals within a sample to a similar extent. For example, in the absence of linkage maps we were unable to discriminate between SNPs on autosomes and sex chromosomes, meaning that our estimate of heterozygosity for males will be slightly lower than heterozygosity averaged over the autosomes. A second bias stems from the fact that RAD markers are affected by nonamplifying “null alleles,” which result mainly from mutations in the restriction enzyme recognition site. Based on the number of RAD tags without any SNP polymorphisms in our datasets, the level of nucleotide polymorphism, π, is estimated to be of the order of 0.002. Therefore, with an 8-base restriction enzyme, null alleles are expected to be found in at most only 2% of RAD markers. Null alleles are an issue for many marker types, including microsatellites, and result in a downward bias in estimated heterozygosity. Third, RAD markers paired on either side of restriction cut sites are in strong linkage and are therefore nonindependent. In our datasets, as only 10–20% of RAD tags contain a SNP (SI Results), the likelihood of tags on either side of a restriction cut site each carrying a SNP is relatively low. Consistent with this, based on calculations of pairwise linkage disequilibrium among all SNPs, ∼10% of SNPs are estimated to be in strong linkage disequilibrium with another SNP in both study systems. These factors will introduce some imprecision into estimates of heterozygosity and may be responsible for some of the unexplained variance in Fig. 1.
Despite these caveats, there are reasons to believe that RAD sequencing may produce less biased estimates of heterozygosity than many other SNP genotyping approaches. High-density genotyping arrays, for example, are usually based on SNPs identified from small “discovery panels” of individuals. This can distort the allele frequency spectrum, as SNPs with intermediate allele frequencies tend to be overrepresented and rare SNPs are often missing (27). As a result, estimates of heterozygosity based on the polymorphic SNPs tend to be inflated, whereas genome-wide heterozygosity will be underestimated because low-frequency SNPs are ignored (28). Thus, although no single approach is perfect, RAD sequencing at least eliminates ascertainment bias because the entire sample serves as the discovery panel.
Contrasting MAF distributions were obtained for the oldfield mice and the harbor seals, the latter being strongly left skewed (Fig. S5). Although we are unable to discount the possibility that at least some of the low-frequency variants in the harbor seals could be technical artifacts, we thoroughly explored the SNP calling parameter space in the oldfield mouse, using pedigree information to derive optimal parameters that were then applied equally to both species (SI Methods and SI Results). We also guarded against spurious genotypes by constructing the reference genome against which the reads were aligned using tags that were present in at least two individuals. Finally, SNPs with MAFs below 0.1 were found to contribute significantly toward the overall HFC signal, suggesting that they carry useful information about inbreeding. A plausible explanation for these contrasting distributions is therefore that they reflect species-specific differences in population size and history. Oldfield mice occur across much of the southeastern United States and can reach densities of 18–26 animals per hectare (29). Relatively high average heterozygosity and an abundance of SNPs with intermediate allele frequencies could therefore be explained by a large and stable effective population size. In contrast, many pinniped species show evidence of strong demographic responses to historical changes in the marine environment, usually through rapid population expansion during periods of increased food or habitat availability (30, 31). Little is known about the historical demography of harbor seals, although the Waddensee population has been reported to have low heterozygosity (32), which is consistent with the effective population size having at some point in the past been very small. The excess of low-frequency polymorphisms could therefore be attributable to postglacial population expansion.
Het-het correlations (15) are often used to test for the presence of inbred individuals, but with only around 10 markers commonly deployed, this approach lacks power because the marker subsets are very small. This is exactly what we found (Fig. 3): the distribution of microsatellite-based het-het correlation coefficients in the oldfield mouse was slightly positive but with a large SD, despite the sample containing highly inbred individuals (f ≤ 0.45). In contrast, the RAD data yielded het-het correlation coefficients well in excess of 0.9 for both the oldfield mice and harbor seals. This provides clear evidence for inbred individuals being present not only in the mouse pedigree but also in the natural harbor seal population.
A second line of evidence also points toward the HFCs for survival and parasite infection being due to inbreeding depression in harbor seals. With inbreeding, the deployment of additional markers should strengthen the HFC detected, whereas with a local effect adding more markers is expected to weaken the relationship between mean heterozygosity and fitness (18). We found that the microsatellite-based HFC for lungworm infection strengthened considerably with RAD heterozygosity, the statistical significance increasing from P = 0.06 to P < 0.0001. Such a dramatic strengthening of effect has not been previously reported, although one study was able to replicate an HFC for birth weight (but not one for juvenile survival) using independent panels of 10 microsatellites (33). Furthermore, the deviance in lungworm infection explained by heterozygosity steadily increases with the number of SNPs deployed (Fig. S7B), which is also strongly suggestive of genome-wide effects contributing toward inbreeding depression rather than the underlying mechanism being a local effect. Finally, an alternative explanation based on population structure seems highly unlikely as we found no evidence for distinct genetic clusters within the RAD dataset (Fig. S8).
Although many reported HFCs yield highly significant P values, the proportion of variance explained tends to be small, typically of the order of 1–5% (34). This could be interpreted as meaning that heterozygosity only weakly affects fitness. Alternatively, heterozygosity could have a large effect on fitness, but because a small panel of microsatellites provides a poor estimate of f, these studies could have limited power to detect such effects. Theory predicts that, if inbreeding is the primary mechanism and thousands of markers could be deployed, the proportion of fitness explained by HFCs would rise considerably (15, 16, 18). This is exactly what we found in wild harbor seals, where the deviance in lungworm infection status explained by heterozygosity increased from around 10% based on 27 microsatellites to nearly 50% with 14,585 SNPs. This is not only qualitatively, but also quantitatively consistent with the theoretical predictions. Under the hypothesis that inbreeding is responsible for HFCs, when switching from microsatellites to RAD SNPs, the variance in infection status explained by heterozygosity (r2sMLH, infection) should increase in proportion to the predicted squared correlation coefficient between heterozygosity and f. Based on the g2 and variance in sMLH in microsatellites and SNPs estimated in young seals, the expected r2sMLH,f is 0.148 for microsatellites and 0.709 for SNPs. This expected 4.8-fold difference is in line with the observed 5-fold increase in deviance explained by the HFC. Thus, our results are consistent with inbreeding theory.
Our study clearly demonstrates that at least some HFCs may explain more variation in fitness than previously thought. However, we compared cases with controls, and it is possible that the former could be enriched for a small subset of unusually inbred individuals. Natal philopatry and breeding site fidelity can also be extremely strong in pinnipeds (35, 36) and might combine with polygyny to increase the risk of inbreeding. Moreover, historical changes in the structure of a population, including bottlenecks or population admixture, may also create variance in inbreeding in a broad sense (16). It therefore remains to be seen whether inbreeding depression could be responsible for HFCs more generally, as suggested by Szulkin et al. (16). This represents a fertile area for future research.
Finally, within a genome one expects that heterozygosities should be more correlated between closely linked loci than between unlinked ones (37, 38). Linkage is expected to result in stronger HFCs in some linkage groups than in others (i.e., local effects). However, in our study, models ignoring linkage strongly predict survival and parasite infection status in harbor seals, suggesting that local effects contribute very little to these HFCs, as predicted by theoretical arguments (16). The observed increase in variance explained when thousands of loci are deployed at least rules out the possibility that these relationships arise from local linkage disequilibria between microsatellites and one or a few phenotypically important loci in the absence of genome-wide inbreeding.
In conclusion, the fact that microsatellite heterozygosity is often poorly correlated with f in natural populations has been used as an argument for obtaining more and better pedigrees (39). However, we show that RAD sequencing is capable of generating enough SNP data to accurately estimate inbreeding, in this particular case even more accurately than from a pedigree of reasonable depth. Our approach is also powerful for studying heterozygosity because it can be applied to virtually any species without the need for prior genomic information.
Methods
Oldfield Mouse.
A population of oldfield mice was founded at Brookfield Zoo from 26 wild-caught individuals. These mice were paired to produce offspring with a range of inbreeding coefficients (0–0.453) over six generations of laboratory breeding and the resulting pedigree was recorded. Using 179 of these mice, Dasmahapatra et al. (20) reported a significantly negative correlation between pedigree-based inbreeding coefficient and heterozygosity estimated from 12 microsatellite loci. We selected 40 of these individuals at random for RAD sequencing, 36 of which yielded data of sufficient quality for analysis (details in SI Methods). The breeding study and the animal care protocols for the oldfield mice were approved by the Institutional Animal Care and Use Committee of the Chicago Zoological Society.
Harbor Seal.
Rijks et al. (21) reported a negative association between heterozygosity at 27 microsatellites and lungworm burden in dead harbor seals stranded on the Dutch Wadden Sea coast. However, this was only statistically significant for young seals, defined as being less than 1 y of age, which were more likely to carry lungworms than older seals (21). We therefore RAD sequenced all 43 of the young seals from this study, together with 37 older seals selected at random from the 161 available. The sampling locations of these individuals are shown in Fig. S9. RAD sequence data of adequate quality were obtained for 30 of the young seals, of which 23 were infected with lungworms, plus 30 older seals, none of which carried lungworm infections (SI Methods).
RAD Genotyping.
RAD libraries were constructed from whole genomic DNA using the protocol of Baird et al. (19) with minor modifications (details in SI Methods), and each library was 100-bp paired-end sequenced on an Illumina HiSeq2000 flow cell. Sequences have been deposited in the Short Read Archive (accession no. PRJEB5164). Separately for the mice and seals, Stacks (40) was used to quality filter reads, demultiplex samples, and assemble RAD stacks together with their associated paired-end contigs. After further stringent filtering steps, RAD contigs were used as a reference against which to map back the raw reads using BWA (41), followed by maximum-likelihood genotype calling using the GATK UnifiedGenotyper (42). Individual heterozygosity and pairwise relatedness were calculated using tag sequences containing only one biallelic SNP per tag. A detailed description of the bioinformatics pipeline used is provided in SI Methods.
Data Analyses.
For all analyses of the microsatellite and RAD data, individual heterozygosity was expressed as sMLH, which is defined as the total number of heterozygous loci in an individual divided by the sum of average observed heterozygosities in the population over the subset of loci successfully typed in the focal individual (10). Pairwise relatedness (RAD allele sharing) was calculated as the total number of identical alleles between individuals (zero, one, or two per tag) divided by twice the number of tags considered. The two-locus heterozygosity disequilibrium g2 was measured following David et al. (22) with the method of computation modified to analyze many thousands of loci in a reasonable computing time (details in SI Methods). Sensitivity of this estimate to the number of loci was explored by randomly selecting different-sized subsets of loci (between 50 and 15,000) and recalculating g2 100 times. Distributions of RAD sMLH under the hypothesis g2 = 0 were obtained by shuffling genotypes at each locus randomly across individuals 1,000 times; as the resulting distribution is very close to normal and the mean is one according to the definition of sMLH, we extracted the variance from these 1,000 simulations and used it to represent the expected distribution as a normal curve. The mean correlation in heterozygosity across loci based on 1,000 random samples was calculated using Rhh (43).
Statistical analyses were conducted using GLMs within R (44). A linear error structure was used for oldfield mouse body mass and a binomial error structure was used for harbor seal age (coded as 0 = young and 1 = old) and lungworm burden (coded as 0 = uninfected and 1 = infected). Where models included multiple predictor variables, standard deletion testing procedures using F tests were used to sequentially remove each term unless doing so significantly reduced the amount of deviance explained (deviance is analogous to sums of squares in standard regression analysis).
Supplementary Material
Acknowledgments
This research was supported by a Marie Curie FP7-Reintegration grant within the Seventh European Community Framework Programme (PCIG-GA-2011-303618), a Deutsche Forschungsgemeinschaft standard grant, and a Biotechnology and Biological Sciences Research Council grant (BB/G006903/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Short Read Archive database (accession no. PRJEB5164).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318945111/-/DCSupplemental.
References
- 1.Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- 2.Jimenez JA, Hughes KA, Alaks G, Graham L, Lacy RC. An experimental study of inbreeding depression in a natural habitat. Science. 1994;266:271–273. doi: 10.1126/science.7939661. [DOI] [PubMed] [Google Scholar]
- 3.Meagher S, Penn DJ, Potts WK. Male-male competition magnifies inbreeding depression in wild house mice. Proc Natl Acad Sci USA. 2000;97:3324–3329. doi: 10.1073/pnas.060284797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller LF. Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia) Evolution. 1998;52:240–250. doi: 10.1111/j.1558-5646.1998.tb05157.x. [DOI] [PubMed] [Google Scholar]
- 5.Walling CA, et al. Inbreeding depression in red deer calves. BMC Evol Biol. 2013;11:318. doi: 10.1186/1471-2148-11-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansson B, Westerberg L. Heterozygosity-fitness correlations within inbreeding classes: Local or genome-wide effects? Conserv Genet. 2007;9(1):73–83. [Google Scholar]
- 7.David P. Heterozygosity-fitness correlations: New perspectives on old problems. Heredity. 1998;80:531–537. doi: 10.1046/j.1365-2540.1998.00393.x. [DOI] [PubMed] [Google Scholar]
- 8.Hansson B, Westerberg L. On the correlation between heterozygosity and fitness in natural populations. Mol Ecol. 2002;11:2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. [DOI] [PubMed] [Google Scholar]
- 9.Coltman DW, Bowen WD, Wright JM. Birth weight and neonatal survival of harbour seal pups are positively correlated with genetic variation measured by microsatellites. Proc R Soc Lond B Biol Sci. 1998;265:803–809. doi: 10.1098/rspb.1998.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coltman DW, Pilkington JG, Smith JA, Pemberton JM. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- 11.Slate J, Kruuk LEB, Marshall TC, Pemberton JM, Clutton-Brock TH. Inbreeding depression influences lifetime breeding success in a wild population of red deer (Cervus elaphus) Proc R Soc Lond B Biol Sci. 2000;267:1657–1662. doi: 10.1098/rspb.2000.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seddon N, Amos W, Mulder RA, Tobias JA. Male heterozygosity predicts territory size, song structure and reproductive success in a cooperatively breeding bird. Proc R Soc Lond B Biol Sci. 2004;271:1823–1829. doi: 10.1098/rspb.2004.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall RC, Buchanan KL, Catchpole CK. Sexual selection and individual genetic diversity in a songbird. Proc R Soc Lond B Biol Sci. 2003;270:248–250. doi: 10.1098/rsbl.2003.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman JI, Forcada J, Trathan PN, Amos W. Female fur seals show active choice for males that are heterozygous and unrelated. Nature. 2007;445:912–914. doi: 10.1038/nature05558. [DOI] [PubMed] [Google Scholar]
- 15.Balloux F, Amos W, Coulson T. Does heterozygosity estimate inbreeding in real populations? Mol Ecol. 2004;13:3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- 16.Szulkin M, Bierne N, David P. Heterozygosity-fitness correlations: A time for reappraisal. Evolution. 2010;64:1202–1217. doi: 10.1111/j.1558-5646.2010.00966.x. [DOI] [PubMed] [Google Scholar]
- 17.Charlesworth D. Balancing selection and its effects on sequences in nearby gene regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman JI, Forcada J, Amos W. Exploring the mechanisms underlying a heterozygosity-fitnesss correlation for canine size in the Antarctic fur seal Arctocephalus gazella. J Hered. 2010;101:539–552. doi: 10.1093/jhered/esq046. [DOI] [PubMed] [Google Scholar]
- 19.Baird NA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dasmahapatra KK, Lacy RC, Amos W. Estimating levels of inbreeding using AFLP markers. Heredity. 2007;100:286–295. doi: 10.1038/sj.hdy.6801075. [DOI] [PubMed] [Google Scholar]
- 21.Rijks JM, Hoffman JI, Kuiken T, Osterhaus ADME, Amos W. Heterozygosity and lungworm burden in harbour seals (Phoca vitulina) Heredity. 2008;100:587–593. doi: 10.1038/hdy.2008.18. [DOI] [PubMed] [Google Scholar]
- 22.David P, Pujol B, Viard F, Castella V, Goudet J. Reliable selfing rate estimates from imperfect population genetic data. Mol Ecol. 2007;16:2474–2487. doi: 10.1111/j.1365-294X.2007.03330.x. [DOI] [PubMed] [Google Scholar]
- 23.Miller JM, et al. Estimating genome-wide heterozygosity: Effects of demographic history and marker type. Heredity. 2014;112:240–247. doi: 10.1038/hdy.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeWoody YD, DeWoody JA. On the estimation of genome-wide heterozygosity using molecular markers. J Hered. 2005;96(2):85–88. doi: 10.1093/jhered/esi017. [DOI] [PubMed] [Google Scholar]
- 25.Santure AW, et al. On the use of large marker panels to estimate inbreeding and relatedness: Empirical and simulation studies of a pedigreed zebra finch population typed at 771 SNPs. Mol Ecol. 2010;19:1439–1451. doi: 10.1111/j.1365-294X.2010.04554.x. [DOI] [PubMed] [Google Scholar]
- 26.Forstmeier W, Schielzeth H, Mueller JC, Ellegren H, Kempenaers B. Heterozygosity-fitness correlations in zebra finches: Microsatellite markers can be better than their reputation. Mol Ecol. 2012;21:3237–3249. doi: 10.1111/j.1365-294X.2012.05593.x. [DOI] [PubMed] [Google Scholar]
- 27.Lachance L, Tishkoff SA. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. Bioassays. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2013;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynn WJ. 2000. Social organization and burrow-site selection of the Alabama beach mouse (Peromyscus polionotus ammobates). MSc thesis (Auburn University, Auburn, AL)
- 30.Curtis C, Stewart BS, Karl SA. Pleistocene population expansions of Antarctic seals. Mol Ecol. 2009;18:2112–2121. doi: 10.1111/j.1365-294X.2009.04166.x. [DOI] [PubMed] [Google Scholar]
- 31.Matthee CA, Fourie F, Oosthuizen WH, Meyer MA, Tolley KA. Mitochondrial DNA sequence data of the Cape fur seal (Arctocephalus pusillus pusillus) suggest that population numbers maybe affected by climatic shifts. Mar Biol. 2006;148:899–905. [Google Scholar]
- 32.Kappe AL, Bijlsma R, Osterhaus ADME, VanDelden W, VandeZande L. Structure and amount of genetic variation at minisatellite loci within the subspecies complex of Phoca vitulina (the harbour seal) Heredity. 1997;78:457–463. doi: 10.1038/hdy.1997.73. [DOI] [PubMed] [Google Scholar]
- 33.Slate J, Pemberton JM. Comparing molecular measures for detecting inbreeding depression. J Evol Biol. 2002;15:20–31. [Google Scholar]
- 34.Chapman JR, Nakagawa S, Coltman DW, Slate J, Sheldon BC. A quantitative review of heterozygosity-fitness correlations in animal populations. Mol Ecol. 2009;18:2746–2765. doi: 10.1111/j.1365-294X.2009.04247.x. [DOI] [PubMed] [Google Scholar]
- 35.Hoffman JI, Forcada J. Extreme natal philopatry in female Antarctic fur seals (Arctocephalus gazella) Mamm Biol. 2012;77(1):71–73. [Google Scholar]
- 36.Hoffman JI, Trathan PN, Amos W. Genetic tracking reveals extreme site fidelity in territorial male Antarctic fur seals Arctocephalus gazella. Mol Ecol. 2006;15:3841–3847. doi: 10.1111/j.1365-294X.2006.03053.x. [DOI] [PubMed] [Google Scholar]
- 37.Leutenegger AL, et al. Estimation of the inbreeding coefficient through use of genomic data. Am J Hum Genet. 2003;73:516–523. doi: 10.1086/378207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill WG, Weir BS. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet Res. 2011;93(1):47–64. doi: 10.1017/S0016672310000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pemberton J. Measuring inbreeding depression in the wild: The old ways are the best. Trends Ecol Evol. 2004;19:613–615. doi: 10.1016/j.tree.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3. 2011;1(3):171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alho J, Valimaki K, Merila J. Rhh: An R extension for estimating multilocus heterozygosity and heterozygosity-heterozygosity correlation. Mol Ecol Res. 2010;10:720–722. doi: 10.1111/j.1755-0998.2010.02830.x. [DOI] [PubMed] [Google Scholar]
- 44.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.