Abstract
Kinesin-14 motor proteins play a variety of roles during metaphase and anaphase. However, it is not known whether members of this family of motors also participate in the dramatic changes in mitotic spindle organization during the transition from telophase to cytokinesis. We have identified the minus-end-directed motor, KIFC3, as an important contributor to central bridge morphology at this stage. KIFC3’s unique motor-dependent localization at the central bridge allows it to congress microtubules, promoting efficient progress through cytokinesis. Conversely, when KIFC3 function is perturbed, abscission is delayed, and the central bridge is both widened and extended. Examination of KIFC3 on growing microtubules in interphase indicates that it caps microtubules released from the centrosome, both in the region of the centrosome and in the cell periphery. In line with other kinesin-14 family members, KIFC3 may guide free microtubules to their destination at the bridge and/or may slide and crosslink central bridge microtubules in order to stage the cells for abscission.
Keywords: kinesin-14, KIFC3, cytokinesis, abscission, mitotic kinesin
Introduction
Motors of the kinesin-14 family of minus-end-directed microtubule motors have been implicated, together with plus-end kinesins, in the formation, maintenance and dynamic remodeling of the bipolar mitotic spindle during the course of mitosis. Their function can be attributed to the fact that in addition to the C-terminal motor domains, which bind microtubuless in an ATP-dependent manner, their N-terminal globular tail possesses a second, ATP-insensitive microtubule, binding site that allows kinesin-14s to crosslink and slide microtubules.1,2 Furthermore, all mitotic kinesin-14 motors characterized so far, including Drosophila melanogaster Ncd, Saccharomyces cerevisiae Kar3, Saccharomyces pombe Pkl1, Xenopus laevis XCTK2, and mammalian KIFC1/HSET possess active nuclear import signals that restrict their function to the stages of mitosis where the nuclear envelope is absent, i.e., from pro-metaphase to early telophase.3-7 To date, no minus-end-directed motors have been implicated in the dramatic remodeling of the central spindle that precedes abscission when the daughter nuclei have reformed and mitotic kinesin-14 proteins are once again sequestered from the cytoplasm.
Mammalian cells express 2 additional kinesin-14 proteins, KIFC2 and KIFC3, which unlike KIFC1/HSET, have cytoplasmic functions in interphase cells.8 Previous work on KIFC3 in particular has centered on its role in targeting of proteins to the apical surface in polarized epithelial cells and its localization at the ends of microtubules at the adherens junctions.9-11 We present here a novel localization for KIFC3 at the edges of the central spindle in cytokinesis and delineate its contributions to efficient progression through cell division and abscission. Our findings extend the involvement of kinesin-14 motors from the organization of the meta- and anaphase spindle to the congression of the midzone microtubules at the final stages of cytokinesis.
Results and Discussion
To test the hypothesis that KIFC3, similar to its closest homologs Drosophila Ncd and mammalian KIFC1/HSET, functions as a motor on the mitotic spindle, we performed a careful analysis of its localization during the cell cycle. Utilizing an antibody raised against the KIFC3 cargo-binding tail domain, we established that endogenous KIFC3 was restricted to the centrosome of HeLa cells in interphase, which is in agreement with the previously reported localization of N-terminally GFP-tagged KIFC3.11,12 To validate the specificity of our KIFC3 antibody (referred to as “Lab” antibody) we co-stained HeLa cells with KIFC3 and γ-tubulin antibodies in the presence of either GST, or the GST coupled KIFC3 peptide antigen against which the antibody was raised. The centrosomal KIFC3 staining seen in the presence of the GST protein alone was abolished by the competing GST-KIFC3 peptide (Fig. 1A). We observed similar centrosomal staining in MDCK cells, as well as in HeLa cells, when utilizing a commercial antibody (labeled as “Pierce” antibody; Fig. S1B). To determine whether this centrosomal localization requires the KIFC3 motor domain, we compared the localization of mCherry-tagged WT-KIFC3 with that of a recombinant protein lacking the motor domain (see Fig. 1B for domain structure). Motor-less KIFC3 was previously shown to interfere with KIFC3 function in a dominant-negative fashion9 and is therefore referred to here as mCherry-DN-KIFC3. Both recombinant proteins localized at centrosomes in the absence of polymerized microtubules (Fig. 1C). However, when an area around the centrosome was bleached in cells with a complete microtubule array, only WT-KIFC3 but not DN-KIFC3-fluorescence recovered on microtubules surrounding the centrosome (Fig. 1D). Taken together, this data indicated that KIFC3 localized to centrosomes in part by minus-end-directed microtubule transport, but in addition by binding to centrosomal proteins via its tail/neck domain(s). The nature of KIFC3’s obligate binding partner(s) at the centrosome is controversial at present: Welburn and Cheeseman reported that the centrosomal localization of KIFC3 is dependent on CEP170, a recently identified centrosomal protein localized to the mother centriole during interphase, while CEP170 localization was KIFC3-independent.13 However, Maliga et al. found the opposite, namely, a role for KIFC3 in the recruitment of CEP170 to the centrosome. KIFC3 might be part of several protein complexes at the centrosome, one containing CEP170.12
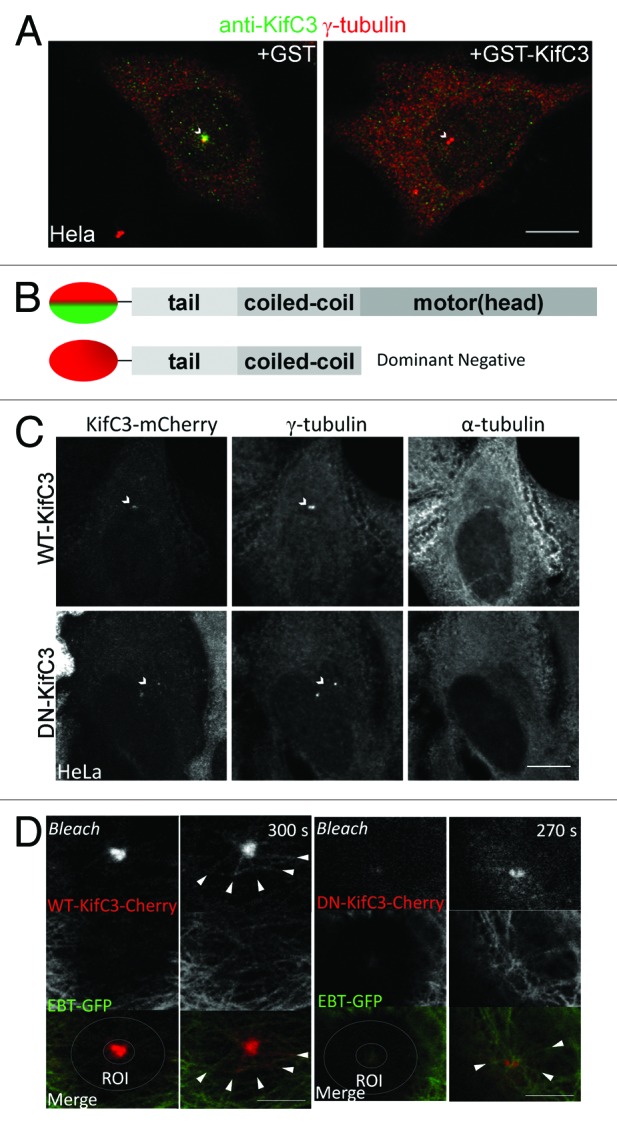
Figure 1. KIFC3 localizes to the centrosome in interphase. (A) HeLa cells were co-stained with anti-KIFC3 antibody (Lab antibody; green) and anti γ-tubulin (red). Primary antibody was incubated with either GST alone or GST-KIFC3 peptide antigen. Scale bar = 10 μm (B) Domain maps of WT-KIFC3 (top) and DN-KIFC3 with GFP or mCherry tag at the N terminus. (C) Microtubules were depolymerized in HeLa cells transfected with WT- or DN-KIFC3 mCherry (red) by incubation on ice and cells were stained with γ-tubulin (green) to visualize the centrosome and α-tubulin to confirm microtubule depolymerization (white). Scale bar = 10 μm (D) Photobleaching and recovery for MDCK cells expressing WT- or DN-KIFC3 (red) and EBT-GFP (green). Bleached area is indicated by a circle. Scale bar = 5 μm.
Throughout mitosis, KIFC3 was maintained at the spindle poles, but also displayed stage-specific localization changes. At metaphase, KIFC3 localized to regions of the spindle adjacent to the centrosomes (Fig. S1C and E). In contrast to KIFC1, KIFC3 did not flank the minus-ends of the nascent central spindle during anaphase (Fig. S1D and F), indicating that it did not mimic KIFC1’s activity in mediating congression of microtubules at the start of telophase.3 However, once cells were established in telophase, KIFC3 transited to the minus-ends of the microtubules of the central bridge. We established this localization for endogenous and GFP/mCherry-tagged forms of KIFC3 in both HeLa (Fig. 2A and C; Fig. S1G) and polarized MDCK cells (Fig. 2B and E). A localization at the flanking regions of the central bridge during telophase and cytokinesis has so far only been described for a limited number of other proteins including γ-tubulin and Asp, a microtubule-associated protein found in Drosophila.14-18 It was not observed for dynein, the other minus-end directed motor that we found at the central spindle at this late stage of mitosis (Fig. 2F). Unlike its centrosomal localization, accumulation of KIFC3 at the flanking regions of the bridge required the motor domain, as DN-KIFC3 did not localize to this region (Fig. 2D).
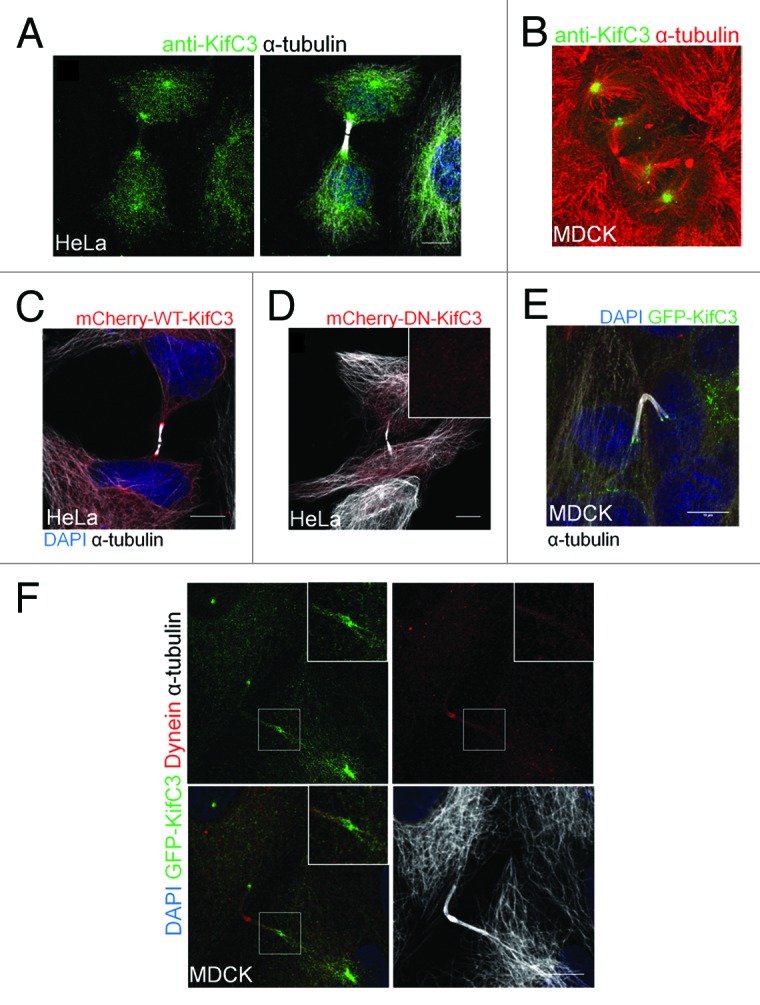
Figure 2. KIFC3 flanks the central spindle during cytokinesis. (A and B) Staining of endogenous KIFC3 (A) HeLa cells were stained with anti-KIFC3 (Lab; green), DAPI (blue), and α-tubulin (white). (B) MDCK cells were stained with anti-KIFC3 (green) and α-tubulin (red). (C–E) Only WT-KIFC3 flanks the central bridge. (C) HeLa cells were transfected with WT- (C) or DN-KifC3 mcherry (D) (red) and stained for DAPI (blue; only C) and α-tubulin (white). (E) MDCK cells stably expressing WT-KIFC3 GFP (green) were stained for DAPI (blue) and α-tubulin (white). (F) MDCK cells stably expressing KIFC3-GFP were stained with DAPI (blue), dynein (red), and α-tubulin (white). Region of interest is enlarged in box on upper left of each panel. (A–E) Maximum projections are shown, (F) single slice is shown. Scale bars = 10 μm
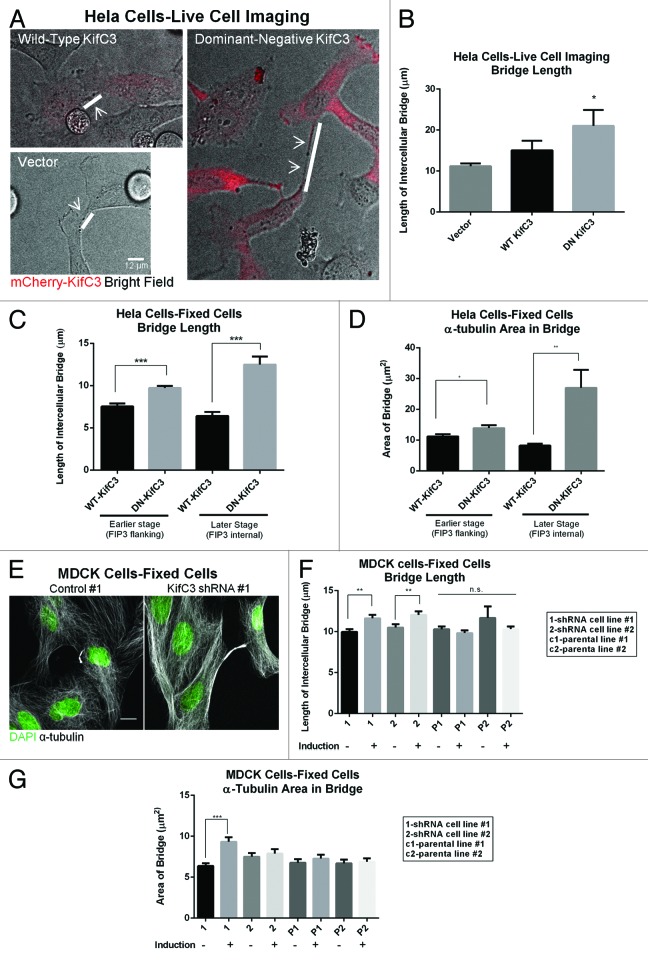
Detailed analysis of the effect of KIFC3 perturbation on mitosis was performed by both live cell imaging and analysis of fixed HeLa cells expressing either an empty vector, mCherry-tagged WT-, or DN-KIFC3. Overexpression or inhibition of KIFC3 with the dominant-negative construct did not interfere with the completion of mitosis, and we did not see an increase in aberrant spindle morphology or in multinucleated cells (multinucleation was 6% with vector control, 7% with WT-KIFC3, 6% with DN-KIFC3). However, HeLa cells expressing DN-KIFC3 displayed significantly longer intercellular bridges prior to abscission than was visible in the controls or cells expressing WT-KIFC3 at similar levels (Fig. 3A–C). We determined this by quantitating the length of the α-tubulin stained bridge in fixed cells (Fig. 3C). To ensure that we compared cells at similar stages of cytokinesis, we took advantage of a HeLa cell line stably expressing the endosomal marker FIP3-GFP. At earlier stages of telophase FIP3 flanks the central bridge, while at later stages it moves internal to the bridge as it delivers important factors for abscission.19 mCherry-KIFC3 and FIP3-GFP colocalized at the flanking regions of the central bridge (Fig. S2). At both stages, we observed an increase in central spindle length in cells expressing DN-KIFC3, which contrasts with the bridge shortening over time observed in cells expressing WT-KIFC3 (Fig. 3C). Similarly, we observed that in cells expressing WT-KIFC3 the spindle area decreased as cells progressed from the stage where FIP3 flanks the central bridge to the point where it is internal to the bridge (Fig. 3D), consistent with the previously reported narrowing of the central bridge over the course of telophase.20 However, in cells expressing DN-KIFC3 we observed an increased spindle area due to the presence of wider spindles in these cells (Fig. 3D). Live cell imaging by brightfield microscopy of cytokinesis progression until the completion of abscission confirmed the data obtained with the fixed cells (Fig. 3A and B). Figure 3B demonstrates that the bridge at its longest stage prior to abscission differed between the DN-KIFC3-expressing and control cells. Therefore, the increased bridge length in DN-KIFC3 cells did not merely reflect a kinetic block at a certain stage of abscission, but rather the generation of longer intercellular bridges upon perturbation of KIFC3.
Figure 3. Perturbation of KIFC3 increases intercellular bridge length and bridge area. (A) Single frames from live cell imaging of HeLa cells expressing WT- or DN-KIFC3 mCherry (red) or an empty vector. White bars and arrows highlight the bridge. Scale bar = 12 μm. (B) Quantitation of (A), n > 170 cells for each condition. (C and D) Quantitation of bridge length (C) and bridge area (D) from fixed HeLa-FIP3 GFP cells expressing WT or DN KIFC3, n > 30 cells for each condition. (E) MDCK cells uninduced (control) or induced to express an shRNA against KIFC3. (F and G) Quantitation of bridge length (F) and bridge area (G) of uninduced and induced cells expressing KIFC3 shRNA and parental controls, n > 60 cells for each condition from 2 independent experiments. Scale bar = 10 μm. Error bars in (B) are s.d., all other error bars are s.e.m. *P < 0.05, **P < 0.005, ***P < 0.0001.
To further test whether KIFC3 indeed has an essential role in the regulation of this final step of mitosis, we generated MDCK cell clones expressing 2 different shRNAs against KIFC3 under an inducible promoter. Both lines exhibited an inducible ~50% KIFC3 depletion that we determined by immunoblotting (Fig. S3) and qPCR (not shown). We examined the extended bridge phenotype in fixed cells by assessing α-tubulin staining at the bridge. Indeed, both cell lines showed a significant increase in central spindle length when KIFC3 was knocked down (Fig. 3E–G). The increase in bridge area was only statistically significant in one of the MDCK clones, indicating that the 2 phenotypes of increased bridge area and bridge length are separable, and the increase in area is not solely due to an increase in bridge length, and therefore reflective of increased width of microtubules due to splaying of bridge microtubules.
The extended bridge length correlated with an increase in the duration of mitosis. We imaged cells expressing an empty vector, WT-, or DN-KIFC3-mCherry by live cell imaging in brightfield microscopy, and counted the number of frames elapsed between when HeLa cells rounded up to enter metaphase and pinched the furrow indicating entry into telophase (M-T1), between entry into telophase and the start of late telophase indicated by cell spreading on the substratum (T1-T2) and between the start of cytokinesis and when the cells either broke their bridge or separated from their partner indicating abscission (T2-A) (Fig. 4A). The population of cells expressing DN-KIFC3 displayed an increased duration in time to abscission relative to cells expressing WT-KIFC3 or an empty vector. This abscission delay is also reflected as an increase in percentage of cells in late telophase in a population of cycling HeLa cells when DN-KIFC3 was expressed (Fig. 4B and C). This delay may be directly related to the increase in bridge length, as it was recently reported that in cytokinetic cells with longer bridges, the bridges were under more tension, and this increase in tension induced an abscission delay.21
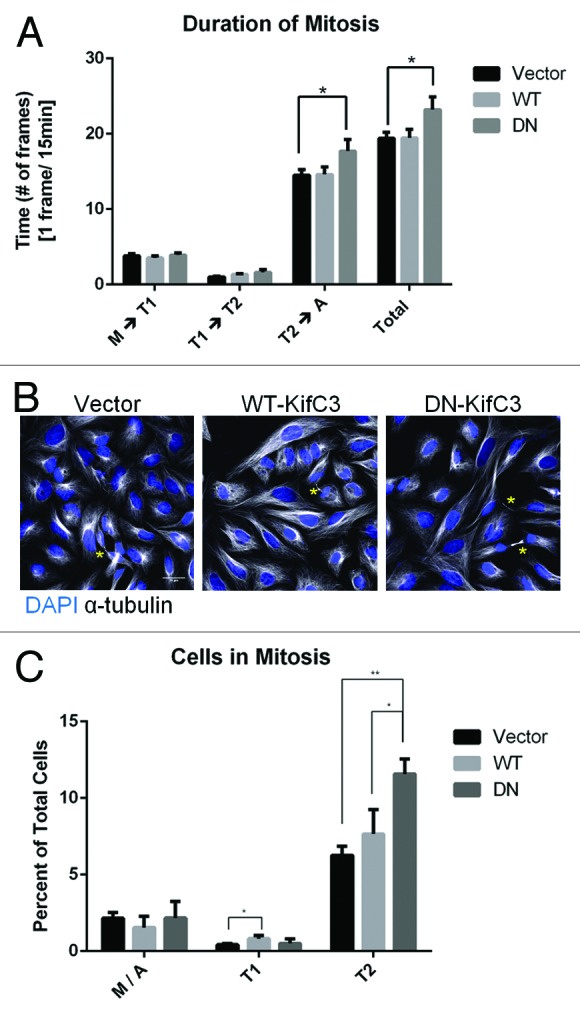
Figure 4. Increased bridge length is correlated with a delay in abscission. (A) HeLa cells expressing WT- or DN-KIFC3 or an empty vector were imaged live and the duration of each mitotic stage was quantified, see text for details. Error bars are s.e.m and n > 40 cells for each condition from two independent experiments. (B) HeLa cells expressing an empty vector or WT- or DN-KIFC3 were stained with DAPI (blue) and α-tubulin (white). Cells in telophase are marked with an asterisk. (C) Quantitation of (B). Error bars are s.d. and n > 1800 cells for each condition from 3 independent experiments. *P < 0.05, **P < 0.005. Scale bar = 30 μm.
Kinesin-14 motor dimers can bind microtubules directly via both their motor and tail domains, and, by virtue of their microtubule-crosslinking ability, they can organize parallel microtubule arrays in vitro.1 In vivo evidence, however, suggested that kinesin-14 motors preferentially associate via microtubule-binding proteins with either the microtubule plus- or minus-ends rather than with the microtubule lattice. Thus, Ncd has been shown to track in an EB1-dependent manner on microtubule plus-ends.4 Kar3 also displayed a preference for microtubule ends and depolymerized microtubule plus-ends.22 Pkl1 and HSET, by contrast, appeared to bind via microtubule-nucleating factors (γ-tubulin and augmin) that were associated with microtubule minus-ends.3,6
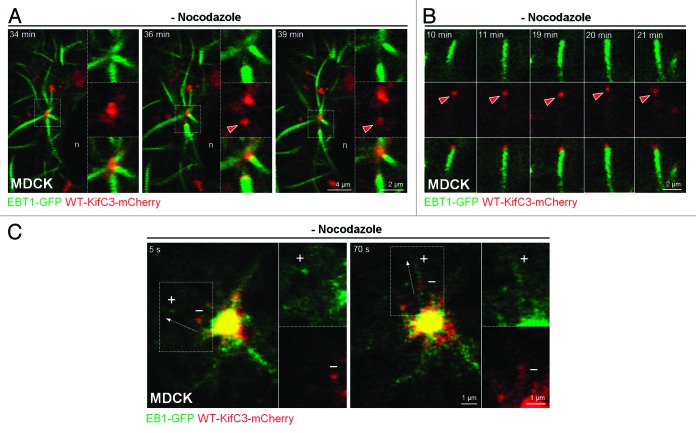
To determine whether KIFC3 likewise tracked with moving microtubule ends, we examined mCherry-KIFC3 localization relative to growing microtubules in MDCK cells during the recovery from microtubule depolymerization by nocodazole. In these cells, microtubules are nucleated at the centrosome but released and captured elsewhere to give rise to the characteristic non-centrosomal microtubule organization of polarized epithelial cells.23 We observed by live cell imaging that KIFC3 capped the minus-ends of microtubules released from the centrosome (Fig. 5A; Video S1) and was also visualized on the ends of short microtubules at the periphery (Fig. 5B; Video S2). Co-expression of KIFC3 with the plus-end marker EB1 indicated that the 2 proteins did not co-localize as microtubules were released from the centrosome, supporting the notion that KIFC3 is at the minus-ends of released microtubules (Fig. 5C; Video S3). Since KIFC3 is reportedly associated with proteins of the epithelial adherens junctions, where non-centrosomal microtubules are localized, the KIFC3 cap might guide centrosomal nucleated and released microtubules to their final position at the lateral domain.11 Analagous to the interphase situation, KIFC3 might also cap the minus-ends of microtubules at the central bridge to guide them to their appropriate destination in the central bridge. Interestingly, S. pombe kinesin-14 Pkl1 promoted severing of γ-tubulin-anchored microtubules from the centrosome, which led the authors to suggest that microtubules released from the spindle poles contributed to central spindle organization.6
Figure 5. KIFC3 localizes to the minus-end of centrosomal-derived microtubules. (A and B) Time-lapse sequences that show growing microtubules in MDCK cells expressing WT-KIFC3-mCherry (red) and EBT1-GFP (green) after nocodazole washout. Red arrowheads indicate progressive movement of KIFC3-capped microtubules away from the centrosome (A) or in the cell periphery (B). n, nuclei. (C) Growing microtubules in MDCK cells expressing WT-KIFC3-mCherry (red) and microtubule plus-end protein EB1-GFP (green) after nocodazole washout. Note the segregation between KIFC3 and EB1 vesicular structures. “+” and “−” refer to the relative location of the presumed plus and minus-ends of a microtubule.
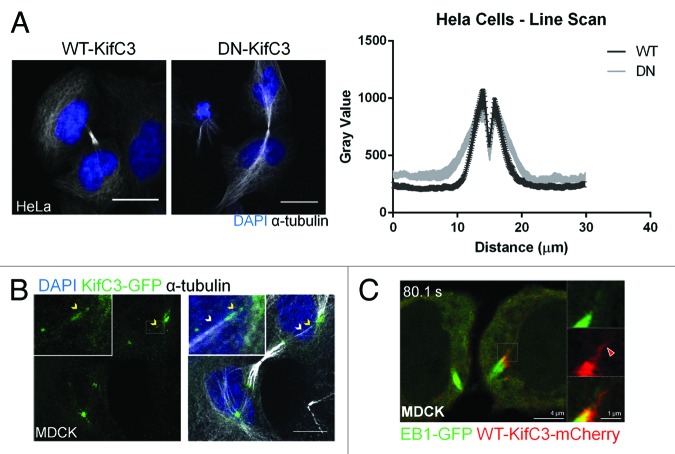
We found that the longer and wider central bridges in DN-KIFC3 cells contained more microtubules that extended beyond the border of the central bridge than those in WT-KIFC3-expressing cells (Fig. 6A). We were unable to track their origins in most cases, but we detected a number of microtubules anchored at the centrosome and bundled with the central bridge (Fig. 6B) that contained KIFC3-positive punctae (Fig. 6B, yellow arrowheads). Employing live cell imaging, we also observed mcherryKIFC3 punctae extending from the bridge-flanking region outwards (Fig. 6C; Video S4). Together, these observations suggest that KIFC3 straddles centrosomal and bridge microtubules during cytokinesis. Although the minus-ends of microtubules at the central bridge are often portrayed as unanchored to any structures in the daughter cell cytoplasm, we propose that some bridge microtubules originate from the 2 daughter centrosomes and might serve as templates for the nucleation of the bridge microtubules in telophase akin to the situation in anaphase. At that stage, kinetochore microtubules, which originate from the spindle poles, serve as templates for the assembly of the central spindle.24 Although DN-KIFC3 did not dramatically alter the microtubule-regrowth pattern in interphase MDCK cells (data not shown), it is conceivable that KIFC3 promotes microtubule release from centrosomes in cytokinesis, which would reduce the number of such nucleation templates and, hence, promote the thinning of the mitotic spindle in preparation for abscission. Support for such a model comes from the effects resulting from depleting the microtubule-severing protein spastin, which, similar to KIFC3-inhibition, caused an extended spindle phenotype, accompanied by a delay in abscission without the accumulation of multinucleated cells.25 According to one model for spastin function during bridge remodeling prior to abscission, spastin is required for microtubule release from the centrosome at this step, since spastin-depleted cells in this study also featured prominent centrosome-anchoring of central bridge microtubules.26 As we propose for the centrosomal population, KIFC3 flanking the bridge could provide a signal to release and disassemble microtubules from the central bridge at the onset of cytokinesis. Alternatively to regulating microtubule minus-end capping and release from nucleating/anchoring complexes, KIFC3 could remove, bundle, or re-position bridge microtubules from or in the midbody region and support microtubule destabilization via microtubule-sliding mechanisms as established as a mode of action for kinesin-14 motors in vitro.1,2,20,26 While future work is needed to elucidate the mechanisms by which KIFC3 contributes to microtubule organization in preparation for abscission, our study has established KIFC3 as a novel mitotic motor that takes the baton from KIFC1 at the spindle after cells progress from anaphase to telophase/ cytokinesis. Our findings furthermore contribute to the emerging notion that kinesin-14 motors function as microtubule-tip-binding proteins.
Figure 6. KIFC3 promotes incorporation of centrosomal microtubules into the central bridge. (A) HeLa cells expressing WT- or DN-KIFC3 were stained with DAPI (blue) and α-tubulin (white). Sum projections are shown. Scale bar = 20 µm. For quantitation a line was drawn, which was centered on the midbody and extended on both sides of the central bridge and into the cytoplasm. The intensity of α-tubulin staining across the line was measured in ImageJ. Average value and s.d. is shown for each point, n > 30 cells per condition. (B) MDCK cells in telophase stably expressing KIFC3-GFP (green) and cytokeratin-19 (not shown) were stained with DAPI (blue) and α-tubulin (white). White arrowhead indicates linking microtubule and yellow arrowhead points to KIFC3 punctae on that microtubule. Region of interest is enlarged in box on upper left of each panel. Maximum projections are shown. Scale bars = 10 μm. (C) Detail of KIFC3 structures that extend outward of the central bridge of MDCK cells expressing WT-KIFC3-mCherry and EB1-GFP during cytokinesis. Red arrowhead points to KIFC3 punctae outside of the central bridge.
Materials and Methods
Cell lines
HeLa and MDCK cells were grown in DMEM with additional 10% FBS, 10 mM HEPES, 2 mM glutamine, and non-essential amino acids (Corning). HeLa-FIP3 GFP cells were generated by Rytis Prekeris’ group (University of Colorado, Aurora) and were grown as above with 5% FBS. EB1-MDCK cell line was a kind gift of Ching-Hwa Sung (Cornell University, Medical College). MDCK tet-off cell lines were a gift from Keith Mostov (University of California, San Francisco), and MDCK tet-on cell lines were a gift from Thomas Weimbs (University of California, Santa Barbara). The canine KIFC3-targeting sequences for shRNA construction were: #1: 5′-CCAATGCTGT GACCTTTGAT T-3′ and #2: 5′-CCCTCACCAA TGACTACAAT T-3′ and were cloned into the pSUPERIOR vector (Oligoengine) and stable MDCK-tet-off lines selected for inducible expression of shRNA#1 and MDCK-tet-on lines for shRNA#2 expression. Cells were induced by either addition (MDCK-tet-on) or withdrawal (MDCK-tet-off) of 200 ng/ ml doxycycline for 2 d, and replated in induction media and grown for another day prior to fixation or live imaging. HeLa cells were transfected with Fugene 6 (Roche) or X-tremeGENE HP DNA Transfection Reagent (Roche http://www.roche-applied-science.com/webapp/wcs/stores/servlet/ProductDisplay?catalogId=10001&partNumber=3.5.3.18.1.11#tab-0) according to the manufacturer’s protocol. MDCK cells were transfected with Amaxa™ nucleofection. To enrich for mitotic cells, cells were synchronized with 2 mM thymidine for 16–20 h and fixed 12 h post-washout. Microtubules were depolymerized on ice for 30 min (HeLa) or with 33 µM nocodazole for 1 h at 37 °C (MDCK).
Recombinant cDNAs
Mouse KifC3 cDNA was provided by Larry Goldstein (UC California, San Diego).27 cDNAs of full-length (amino acids 1–709) or the motorless form (amino acids 1–400) were N-terminally tagged with EGFP or mCherry. Microtubule polymerization was monitored with EMTB, the microtubule-binding domain of Ensconsin coupled to GFP, a kind gift of J. Chloe Bulinski (Columbia University).28
Antibodies, immunofluorescence, and live cell imaging
Cells were fixed with methanol at −20 °C, and blocked with 1% BSA/5% FBS in PBS. KIFC3 rabbit polyclonal antibodies were raised against a GST-fusion protein encompassing amino acids 92 to 125 of mouse KifC3 and affinity purified. Other antibodies used were KIFC3 (Pierce PA5-30074 http://www.pierce-antibodies.com/KIFC3-antibody-Polyclonal-PA530074.html), Dynein IC (Santa Cruz sc-9115), γ-tubulin (Sigma-T6557 http://www.sigmaaldrich.com/catalog/product/sigma/t6557?lang=en®ion=US), and α-tubulin (YL1/2-Abcam ab6160 http://www.abcam.com/tubulin-antibody-yl12-loading-control-ab6160.html). For the GST-blocking experiment in Figure 1A coverslips were incubated for 1 h with anti-KifC3 and 1 µg/µl GST or GST-KifC3 peptide. Secondary antibodies used were anti-mouse and anti-rabbit coupled to Alexa Fluor -488, or -568 and anti-rat antibodies coupled to Alexa-Flour -647 or -649 (Jackson). Samples were post-fixed in 2% PFA in PBS and mounted with mowiol or glycerol/2.5% DABCO. Confocal microscopy was performed on a TCS SP5; Leica with either an HCX Plan Apo CS 63.0 × 1.40 NA oil (Figs. 1, 2, 3E, 5, and 6) or 40.0 × 1.25 NA oil objective (Figs. 3A and 4B). For live cell imaging cells were grown on MatTek dishes with additional 15 mM HEPES added to the growth media. Cells were placed on a motorized stage heated to 37 °C in a CO2-enriched atmosphere. In Figure 3A, time-lapse images were taken every 15 min for 12 h, with an open pinhole, in stacks of 3 axial scans per frame. For FRAP experiments (Fig. 1D) a ROI around the centrosome containing EBT1-GFP and KIFC3-mCherry was photo-bleached using 488 and 594 laser lines at full power.
Western blot
Lysates were made in SDS-sample buffer (150 mM Tris pH 8.8, 10% glycerol, 2% SDS) with 100 mM DTT added and resolved on a 9% SDS-PAGE gel and transferred to nitrocellulose. Blots were incubated with anti-KIFC3 (Pierce PA5–30074 http://www.pierce-antibodies.com/KIFC3-antibody-Polyclonal-PA530074.html) and anti-actin (AbCam ab8227 http://www.abcam.com/beta-actin-antibody-ab8227.html) and secondary antibodies (DyLight 680-anti-rabbit and anti-mouse http://www.pierce-antibodies.com/targets/t/DyLightFluorSecondaryAntibodies.cfm) and the signal visualized in a laser scanner (FLA-9000 [Fujifilm]).
Statistical analysis
Quantitation and processing of images was performed with ImageJ or LAS AF software (Leica). Bridge length in Figure 3B was measured by tracing the brightfield image of the bridge between the daughter cells using the scale bar tool in LAS AF software (Leica). For Figure 3C and F image stacks were converted to sum projections and the length of the bright α-tubulin staining was traced with a segmented line and measured in ImageJ. For Figure 3D and G, image stacks were converted to sum projections, and bright α-tubulin staining was thresholded; the thresholded area was selected with the wand function in ImageJ and the area was measured. For Figure 6A, image stacks were converted to sum projections. A line was drawn, which was centered on the midbody and extended on both sides of the central bridge and into the cytoplasm. The intensity of α-tubulin staining across the line was measured using the plot profile function in ImageJ. Unpaired t tests were performed in GraphPad Prism 6.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs Lawrence Goldstein and Chloe Bulinski for providing cDNAs and we are grateful to Dr Ching-Hwa Sung for making her EB1-GFP-MDCK cell available to us and to Dr Thomas Weimbs for the MDCK TET-ON cells. This work was supported by NIH/NIDDK R01KD064842-07A1 and NIH/NCI 1R01CA1607901-01 to AM and by funds from the Sue Golding Graduate Program at the Albert-Einstein College of Medicine to JN.
Glossary
Abbreviations:
- ATP
adensoine triphosphate
- DAPI
4',6-diamidino-2-phenylindole
- DN
dominant negative
- GFP
green fluorescent protein
- GST
glutathione S-transferase, MDCK, Madine Darby canine kidney
- s.d.
standard deviation
- s.e.m
standard error of the mean
- qPCR
quantitative polymerase chain reaction
- WT
wild-type
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27266
References
- 1.Braun M, Drummond DR, Cross RA, McAinsh AD. The kinesin-14 Klp2 organizes microtubules into parallel bundles by an ATP-dependent sorting mechanism. Nat Cell Biol. 2009;11:724–30. doi: 10.1038/ncb1878. [DOI] [PubMed] [Google Scholar]
- 2.Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol. 2009;11:717–23. doi: 10.1038/ncb1877. [DOI] [PubMed] [Google Scholar]
- 3.Cai S, Weaver LN, Ems-McClung SC, Walczak CE. Proper organization of microtubule minus-ends is needed for midzone stability and cytokinesis. Curr Biol. 2010;20:880–5. doi: 10.1016/j.cub.2010.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goshima G, Nédélec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus-end-directed motor proteins. J Cell Biol. 2005;171:229–40. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner MK, Haase J, Mythreye K, Molk JN, Anderson M, Joglekar AP, O’Toole ET, Winey M, Salmon ED, Odde DJ, et al. The microtubule-based motor Kar3 and plus-end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol. 2008;180:91–100. doi: 10.1083/jcb.200710164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olmsted ZT, Riehlman TD, Branca CN, Colliver AG, Cruz LO, Paluh JL. Kinesin-14 Pkl1 targets γ-tubulin for release from the γ-tubulin ring complex (γ-TuRC) . Cell Cycle. 2013;12:842–8. doi: 10.4161/cc.23822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walczak CE, Verma S, Mitchison TJ. XCTK2: a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J Cell Biol. 1997;136:859–70. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bananis E, Nath S, Gordon K, Satir P, Stockert RJ, Murray JW, Wolkoff AW. Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for Dynein and Kinesin. Mol Biol Cell. 2004;15:3688–97. doi: 10.1091/mbc.E04-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noda Y, Okada Y, Saito N, Setou M, Xu Y, Zhang Z, Hirokawa N. KIFC3, a microtubule minus-end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes. J Cell Biol. 2001;155:77–88. doi: 10.1083/jcb.200108042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Takeda S, Nakata T, Noda Y, Tanaka Y, Hirokawa N. Role of KIFC3 motor protein in Golgi positioning and integration. J Cell Biol. 2002;158:293–303. doi: 10.1083/jcb.200202058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng W, Mushika Y, Ichii T, Takeichi M. Anchorage of microtubule minus-ends to adherens junctions regulates epithelial cell-cell contacts. Cell. 2008;135:948–59. doi: 10.1016/j.cell.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 12.Maliga Z, Junqueira M, Toyoda Y, Ettinger A, Mora-Bermúdez F, Klemm RW, Vasilj A, Guhr E, Ibarlucea-Benitez I, Poser I, et al. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat Cell Biol. 2013;15:325–34. doi: 10.1038/ncb2689. [DOI] [PubMed] [Google Scholar]
- 13.Welburn JPI, Cheeseman IM. The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol Cell. 2012;23:4786–95. doi: 10.1091/mbc.E12-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julian M, Tollon Y, Lajoie-Mazenc I, Moisand A, Mazarguil H, Puget A, Wright M. gamma-Tubulin participates in the formation of the midbody during cytokinesis in mammalian cells. J Cell Sci. 1993;105:145–56. doi: 10.1242/jcs.105.1.145. [DOI] [PubMed] [Google Scholar]
- 15.Shu HB, Li Z, Palacios MJ, Li Q, Joshi HC. A transient association of gamma-tubulin at the midbody is required for the completion of cytokinesis during the mammalian cell division. J Cell Sci. 1995;108:2955–62. doi: 10.1242/jcs.108.9.2955. [DOI] [PubMed] [Google Scholar]
- 16.Riparbelli MG, Callaini G, Glover DM, Avides MdoC. A requirement for the Abnormal Spindle protein to organise microtubules of the central spindle for cytokinesis in Drosophila. J Cell Sci. 2002;115:913–22. doi: 10.1242/jcs.115.5.913. [DOI] [PubMed] [Google Scholar]
- 17.Wakefield JG, Bonaccorsi S, Gatti M. The drosophila protein asp is involved in microtubule organization during spindle formation and cytokinesis. J Cell Biol. 2001;153:637–48. doi: 10.1083/jcb.153.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 19.Schiel JA, Simon GC, Zaharris C, Weisz J, Castle D, Wu CC, Prekeris R. FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat Cell Biol. 2012;14:1068–78. doi: 10.1038/ncb2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elad N, Abramovitch S, Sabanay H, Medalia O. Microtubule organization in the final stages of cytokinesis as revealed by cryo-electron tomography. J Cell Sci. 2011;124:207–15. doi: 10.1242/jcs.073486. [DOI] [PubMed] [Google Scholar]
- 21.Lafaurie-Janvore J, Maiuri P, Wang I, Pinot M, Manneville J-B, Betz T, Balland M, Piel M. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science. 2013;339:1625–9. doi: 10.1126/science.1233866. [DOI] [PubMed] [Google Scholar]
- 22.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus-ends. Curr Biol. 2005;15:1420–7. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–63. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 24.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiel JA, Park K, Morphew MK, Reid E, Hoenger A, Prekeris R. Endocytic membrane fusion and buckling-induced microtubule severing mediate cell abscission. J Cell Sci. 2011;124:1411–24. doi: 10.1242/jcs.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Xia Ch, Roberts EA, Bush K, Nigam SK, Goldstein LS. Molecular cloning and functional analysis of mouse C-terminal kinesin motor KifC3. Mol Cell Biol. 2001;21:765–70. doi: 10.1128/MCB.21.3.765-770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulinski JC, Gruber D, Faire K, Prasad P, Chang W. GFP chimeras of E-MAP-115 (ensconsin) domains mimic behavior of the endogenous protein in vitro and in vivo. Cell Struct Funct. 1999;24:313–20. doi: 10.1247/csf.24.313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.