Abstract
Bone marrow adipose tissue (BMAT) is different from fat found elsewhere in the body, and only recently have some of its functions been investigated. BMAT may regulate bone marrow stem cell niche and plays a role in energy storage and thermogenesis. BMAT may be involved also in obesity and osteoporosis onset. Given the paramount functions of BMAT, we decided to better clarify the human bone marrow adipogenesis by analyzing the role of the retinoblastoma gene family, which are key players in cell cycle regulation.
Our data provide evidence that the inactivation of RB1 or RB2/P130 in uncommitted bone marrow stromal cells (BMSC) facilitates the first steps of adipogenesis. In cultures with silenced RB1 or RB2/P130, we observed an increase of clones with adipogenic potential and a higher percentage of cells accumulating lipid droplets.
Nevertheless, the absence of RB1 or RB2/P130 impaired the terminal adipocyte differentiation and gave rise to dysregulated adipose cells, with alteration in lipid uptake and release. For the first time, we evidenced that RB2/P130 plays a role in bone marrow adipogenesis.
Our data suggest that while the inactivation of retinoblastoma proteins may delay the onset of last cell division and allow more BMSC to be committed to adipocyte, it did not allow a permanent cell cycle exit, which is a prerequisite for adipocyte terminal maturation.
Keywords: retinoblastoma gene family, marrow stromal stem cells, differentiation, adipocytes, bone marrow
Submitted: 10/30/2013; Revised: 11/15/2013; Accepted: 11/19/2013
Introduction
The physiology and role of adipose tissue is more complex than previously thought. Indeed, besides the white adipose tissue (WAT), which is the most abundant human fat, there are 2 other adipose tissue types that differ significantly from WAT. Brown adipose tissue (BAT)’s main role is the regulation of thermogenesis through burning of energy rather than its storage. Bone marrow adipose tissue (BMAT) is third fat depot that has similarities to both WAT and BAT. Fat occupies a significant portion of bone cavity; however, its role is largely unknown. The BMAT was traditionally thought to have no function and has been overlooked or ignored for long time.1,2
Recently, BMAT have become the focus of several investigations, since it has been acknowledged that cells in the bone marrow niche communicate with each other and are essential for the maturation of mesenchymal and hematopoietic stem cells. Evidence is mounting that BMAT is different from adipose tissue found elsewhere in the body, since some studies suggest that adipocytes in BMAT are responsive to signals that are different from those acting on WAT and BAT.
Some authors suggest that the adipocytes in bone marrow may assist local stem cells in some way, possibly by acting as placeholders until the stem cells differentiate into the cell type that is needed. Bone marrow fat may also play a role in energy storage and thermogenesis. Indeed, recent studies have shown that BMAT is a dynamic tissue whose mass increases markedly during starvation, a state of chronic negative energy balance. In other words, even as subcutaneous and visceral WAT is lost throughout the rest of the body, bone marrow adipose reserves expand.1,2 In addition, it has been proposed that impaired functions of BMAT may influence bone remodeling through the secretion of cytokines that target bone, the production of signaling molecules that affect sympathetic impulses to bone, and also through the paracrine influences on adjacent skeletal cells.3
Generation of fat in the marrow is presumed to be identical to adipogenesis in other tissues. Adipogenesis in mice and humans follows a defined pathway that begins with mesenchymal stem cells that are pluripotent and are a subset of bone marrow stromal cells. Mesenchymal stem cells differentiate into mesenchymal tissues, such as bone, cartilage, and fat cells, but also support hematopoiesis and contribute to homeostatic maintenance of many organs and tissues and, therefore, also have a significant therapeutic potential for tissue regeneration.4,5
The great majority of studies on adipogenesis have been performed on precursors of WAT adipocytes, which are committed progenitors that, upon specific cues, may terminally differentiate into mature adipocytes. In these experimental models, adipogenesis is described as a cascade of gene expression events initiated by the nuclear hormone transcription factor PPARγ in concert with CCAAT enhancer-binding proteins and sterol regulatory element-binding protein-1/adipocyte determination differentiation-dependent factor 1 (SREBP-1/ADD1) transcription factors. A few studies have described the complete adipogenesis process that from stem cells leads to mature adipocytes.
Even if some aspects of in vitro adipocyte differentiation are controversial, it is now clear that some cell cycle checkpoint proteins, such as the retinoblastoma gene family, influence adipogenesis.6,7 The classic role for the retinoblastoma family genes RB1, RB2/P130, and P107 is the regulation of the cell cycle G1/S transition through the negative modulation of the E2F family of transcription factors. In addition, this protein family plays an important role in regulating other cellular processes, such as terminal differentiation and senescence. Initial observations suggested that retinoblastoma family proteins showed overlapping functions, and that RB2/P130 and P107 had an ancillary role. However, several studies have evidenced functional differences among these genes.8,9 Recently, it has become clear that the role of RB1, RB2/P130, and P107 depends on several parameters, such as animal species under investigation, cell type, and cell status (stem cell, progenitor, differentiated cell).10-13
Pioneering studies showed that retinoblastoma proteins are differentially regulated during the adipogenic differentiation of pre-adipocyte cell lines.14,15 The sequestration of retinoblastoma family members by SV40 large T antigen inhibits adipocyte differentiation.16 In addition, RB1−/− fibroblasts are unable to differentiate without exogenous PPAR-gamma activators.17 In contrast to these studies, Auwerx’s group has recently suggested an inhibitory role of RB1 at particular stages of adipocyte differentiation, through the recruitment of the histone deacetylase HDAC3 to the nuclear receptor peroxisome proliferator-activated receptor gamma.18 Consistent with these findings, transgenic mice overexpressing Rb1 show a significant decrease in paraovarian fat pad mass.19
To reconcile these studies, it should be emphasized that retinoblastoma proteins could play a complex role in adipogenesis: they may be dispensable during the adipocyte cell commitment of multipotent stem cells, whereas they could further promote the differentiation process in committed cells. It is evident that the role of the retinoblastoma family in adipocyte differentiation should be investigated in more depth.
Moreover, data on the function of retinoblastoma family members in the control of adipogenesis have been biased by the fact that the majority of authors have focused their attention mainly on the RB1 protein. Data on the role of P107 and RB2/P130 in adipocyte differentiation are very scant, and few studies addressed the function of retinoblastoma proteins in bone marrow adipogenesis.
For these reasons, we analyzed the contribution of the retinoblastoma gene family in adipocyte differentiation of BMSC. An important aspect of our research also resides in the fact that we analyzed the complete adipocyte differentiation process, whereas most studies have used cell lines, such as the mouse cell lines 3T3-L1, 3T3-F442A, and C3H10T1/2, and human pre-adipocytes, which have a restricted potential to differentiate.7
Results
BMSC cultivation and gene silencing
Several surface markers (Stro-1, CD29, CD49a, CD73, CD90, CD105, CD166, CD44, CD146, and CD271) have been proposed to isolate mesenchymal stem cells from bone marrow.20 Among these, Stro-1 and CD146 have been largely used to identify and isolate clonogenic bone marrow stromal cell progenitors in adult human bone marrow.21,22
While positive selection to identify a progenitor population in human marrow can be used to select for cells capable of osteo/adipo/chondrocyte differentiation, each of the markers that have been used to identify marrow mesenchymal populations may be expressed on distinct subsets of marrow mesenchymal cells.20 For example, recently Battula and coworkers isolated a subset of BMSC that were CD56 positive and have higher clonogenic properties compared with the negative cell population. Nevertheless, CD146 expression was restricted to CD56(−) population.23
To avoid the selection of a specific subset of BMSC we used the conventional method of plastic adherence purification, which is the gold standard method for GMP isolation and large-scale expansion of bone marrow-derived mesenchymal stem cells.24 We cultivated BMSC with bFGF to stimulate cell proliferation, but we used low bFGF dosage to avoid the trigger of osteogenic program.25-27
After initial plating, BMSC cultures were expanded to 70–80% confluency. On these cells (P0), we performed silencing experiments (see flowchart of experimental plan in Fig. 1). We verified that, in our experimental conditions, BMSC cultures fulfilled the 3 proposed criteria to define BMSCs28 and were positive for CD14622 (Fig. S1).
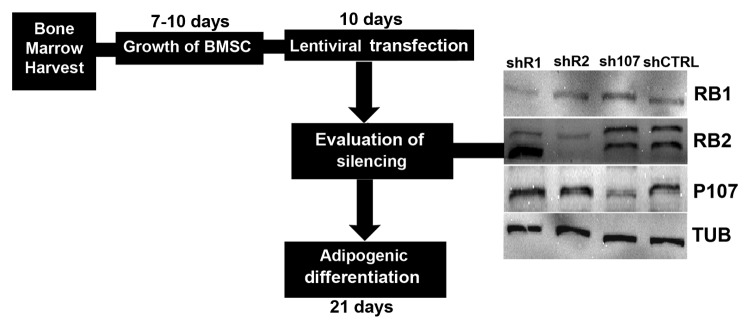
Figure 1. Experimental plan. Bone marrows were collected from healthy patients, and mononuclear cell fractions were used to have bone marrow stromal cultures containing MSC. Cultures were propagated for 7–10 d. Then cultures were transduced with lentivirures carrying shRNAs targeting Retinoblastoma gene family. Cultures were then incubated in adipogenic differentiation media for 21 d and the differentiation process was evaluated. In the picture is shown western blot analysis of BMSC that were tested for RB1, RB2/P130, and P107 knockdown after lentiviral transductions and puromycin selection. Cell expressing shRNAs against RB1, RB2/P130, and P107 were indicated as shR1, shR2, and sh107, respectively. Cells expressing scrambled shRNAs were indicated as shCTRL. The RB2 lane shows 2 bands, which correspond to hyperphosphorylated inactivated form (upper band) and active form (lower band). The protein levels were normalized with respect to α-tubulin, the loading control.
Human BMSC were tested for RB1, RB2/P130, and P107 knockdown after lentiviral transductions and puromycin selection. Cell expressing shRNAs against RB1, RB2/P130, P107 were indicated as shR1, shR2, and sh107, respectively. Cells expressing scrambled shRNA were indicated as shCTRL. The selected shRNAs were effective in silencing RB1, RB2/P130, and P130 (Fig. 1), as we already demonstrated (Alessio 2013).
Role of retinoblastoma family in adipocyte commitment
The role of retinoblastoma proteins has been evaluated on committed pre-adipocytes and not on multipotent precursors such as BMSC. To have a more complete insight on the role of these proteins in adipogenesis, we isolated, expanded, and characterized clones from control BMSC and from cultures with silenced retinoblastoma proteins to evaluate the osteo-chondro-adipogenic (OCA) potential.
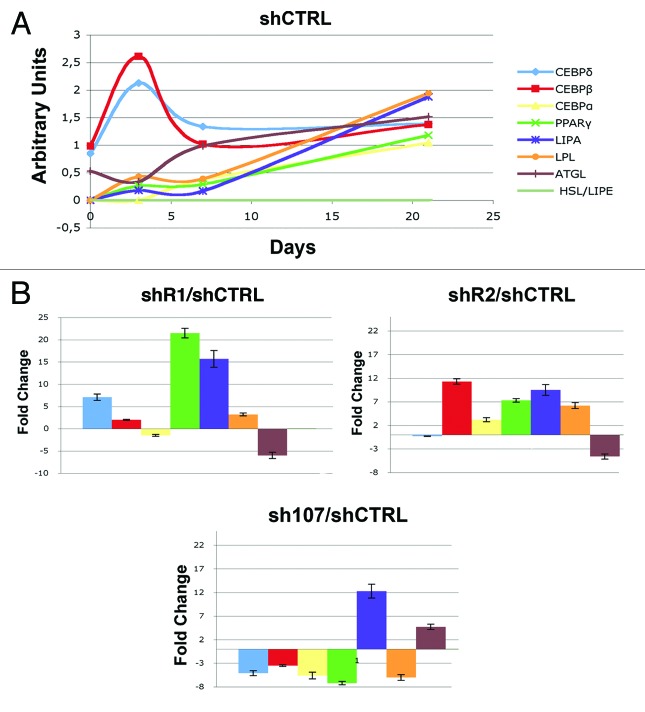
BMSC clones are heterogeneous and may have tri- bi- and uni-potential differentiation capability (Fig. S2).29 In our experimental condition, some clones were able to commit into the three lineages we analyzed, whereas other clones were bipotent (Osteo/Adipo; Osteo/Chondro; Adipo/Chondro) or unipotent. Cultures with silenced RB1 or RB2 gave 3 and 2.6 times more adipocyte-unipotent clones compared with control. This result suggested that silencing of RB1 or RB2 led to bias toward adipocyte commitment of BMSC.
Role of retinoblastoma family in adipocyte differentiation
Following silencing of retinoblastoma proteins, BMSC were stimulated for 3 wk in hMSC mesenchymal stem cell adipogenic differentiation medium (Lonza), and adipocyte formation was evaluated by Oil Red O staining. We observed a strong increase in the percentage of adipocytes in cells with silenced RB1 or RB2 (Fig. 2). We confirmed this result with Bodipy (Molecular Probes), which stains neutral lipids and is very specific for cellular lipid droplets (Fig. 2).
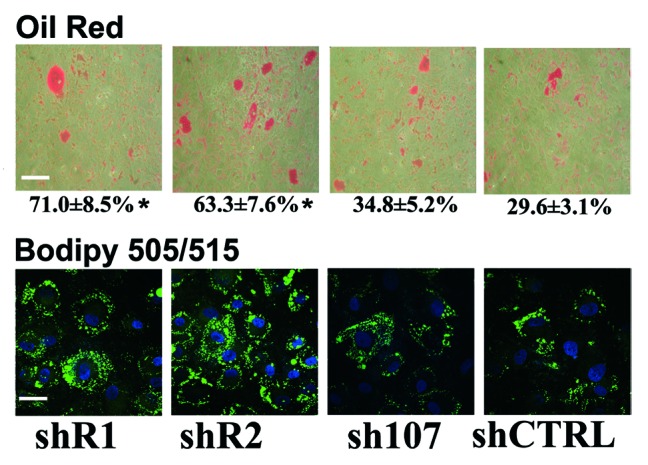
Figure 2. BMSC differentiation into adipose cells. The picture shows representative microscopic fields of BMSC induced to differentiate into adipocytes and stained with Oil Red O or with Bodipy 505/515. Scale bar: 80 µM The percentage of Oil Red O-positive cells is indicated. Data are expressed as mean values with standard deviations (*P < 0.05). shR1, shR2, shP107 are cells with silenced RB1, RB2/P130, and P107, respectively. Control cultures are indicated as shCTRL. Scale bar: 30 µM.
To gain further insight into adipogenesis of BMSC in the absence of retinoblastoma proteins, we determined the expression of genes involved in adipogenesis such as C/EBPß and C/EBPδ (early differentiation markers) and PPARγ, C/EBPα, LPL, and ATGL (late differentiation markers).7 To begin approaching such issues, we performed a molecular follow-up of BMSC adipogenesis by looking at the expression levels of adipocyte differentiation markers in basal conditions without the silencing of retinoblastoma proteins (Fig. 3). Expression of the early differentiation markers C/EBPß and C/EBPδ showed a typical bimodal expression profile, with a burst in expression during the first stage of differentiation and then a decline (Fig. 3). The other genes showed a progressive increase in their expression as the differentiation proceeded (Fig. 3). The temporal expression of these factors during adipocyte differentiation of BMSC is in agreement with the known cascade of molecular events that occur in adipogenesis, whereby early induction of C/EBPβ and C/EBPδ leads to induction of C/EBPα and then of other transcription factors, such as to several adipocyte promoters during differentiation, such as PPARγ.7 At 21 d post-induction of adipocyte differentiation, we detected a significant upregulation of almost all the markers in cells with silenced RB1 or RB2 compared with control (Fig. 3). On the opposite, in cells lacking P107, we observed a decrease of several differentiation markers (Fig. 3). These data are in agreement with the Oil Red O and Bodipy staining, suggesting that an absence of RB1 or RB2 may promote adipogenesis. Nevertheless, the sustained strong upregulation of early differentiation markers (C/EBPδ in shR1 and C/EBPβ in shR2 cells) in late phase of adipocyte maturation may either suggest that the differentiation process is dysregulated, or that this increased expression is to be ascribed to the greater percentage of adipocytes present in cultures with silenced RB1 or RB2. To distinguish between these 2 options we tried to roughly determine the mean expression level of C/EBPδ per cell in control and shR1 cultures by dividing RT-PCR expression values with the percentage of adipocytes in differentiated cultures as determined with Oil Red O. In cells lacking RB1, the mean cellular C/EBPδ was 2.6 times higher than in the control. We applied the same procedure for determining C/EBPβ in shR2 cells and the corresponding control. The mean expression level per cell was 5.6× greater in cells lacking RB2 compared with the control (statistical evaluation for these analyses are in Fig. S3). These data suggest that in absence of RB1 or RB2, the adipogenesis occurred in a dysregulated fashion.
Figure 3. RT-PCR expression analysis of early and late adipocyte differentiation markers. (A) The graph represents the expression follow-up of differentiation markers of BMSC induced to differentiate into adipocytes. mRNA levels were normalized with respect to GAPDH, which was chosen as an internal control. Data are expressed as arbitrary units. (B) In cells expressing either shR1, shR2, or sh107, the mRNA levels of genes under analysis were normalized with respect to GAPDH, chosen as internal control. The histogram shows the ratio of gene expression between treated and control cells (shCTRL). The mean expression values (± SD, n = 3; * P < 0.05) of each gene are presented. Data are expressed as arbitrary units.
The absence of P107, while it did not modify the percentage of adipocytes as detected by lipid droplet stains, reduced the expression of differentiation markers. This may suggest that differentiation process is stalled or delayed in shP107 cultures.
Alterations in adipogenesis may be associated with the critical role the retinoblastoma proteins play in controlling cell cycle. In fact, it is well established that pre-adipocytes must be in a growth-arrested post-confluent state to terminally differentiate.7,15 We analyzed cell cycle profiles of BMSC at the end of adipocyte differentiation in the absence of retinoblastoma proteins. At 21 d post-induction of adipocyte differentiation, we did not detect cells in S phase in control cultures, while in shR2 cultures, a significant percentage of cells were still cycling. Also, in cultures with silenced RB1 and P107, we detected some cells in S phase (Fig. 4).
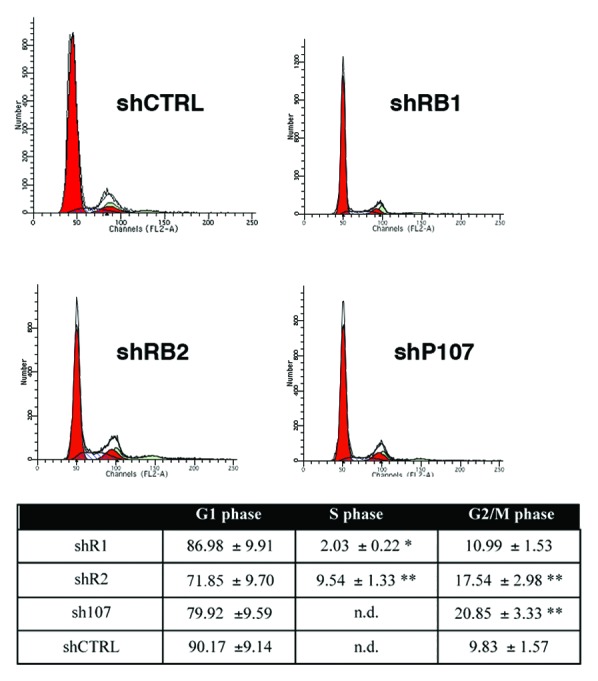
Figure 4. Flow cytometry analysis. The table shows the percentage of differentiated BMSC in different cell cycle phases. Mean expression values of flow cytometry analysis are reported for cells expressing either shR1, shR2, sh107, shCTRL (± SD, n = 3; * P < 0.05; ** P < 0.01). n.d. means not detected. A representative flow cytometry analysis is shown for each experimental condition.
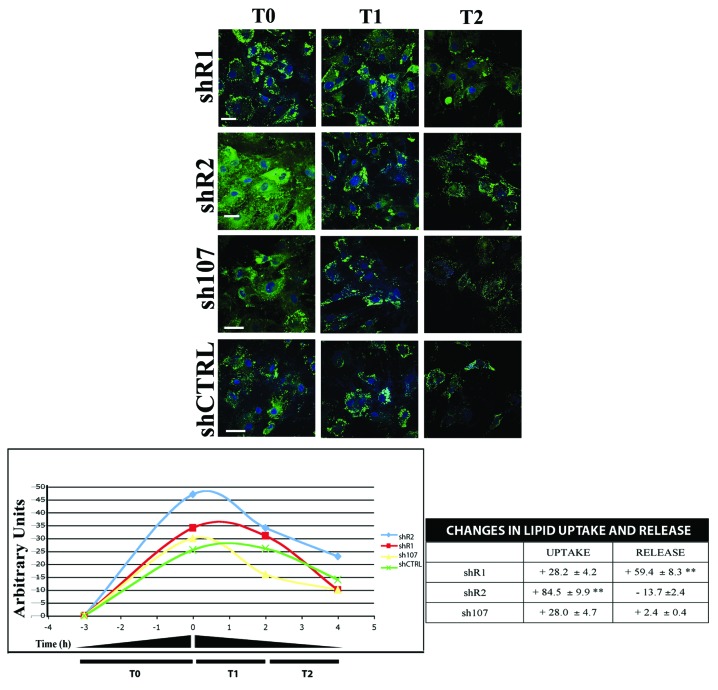
Adipose tissue stores excess energy intake in the form of triglycerides and mobilizes this storage in the form of free fatty acids (FFA) to meet the energy demands of the organism. Understanding the molecular mechanisms underlying the uptake of lipids and release of FFA from adipose tissue stores is key to the development of appropriate interventions for diabetes, obesity, and related disorders.30 We decided to evaluate whether the dysregulated adipogenesis in the absence of retinoblastoma proteins is coupled with an alteration in uptake and release of lipids. Adipocytes with silenced RB2 showed a higher lipid uptake compared with control cells (shCTRL), while adipocytes lacking RB1 showed a higher capacity in lipid release (Fig. 5). Cells lacking P107 did not show statistically significant changes in lipid uptake and release as compared with control cultures (Fig. 5).
Figure 5. Uptake and release of FFAs Differentiated BMSC were serum-starved for 16 h, then cells were treated with 10 µM BODIPY FL C16 for 3 h (T0 starting point). Subsequently, cultures were incubated for 2 and 4 h (T1 and T2) with complete medium (α-MEM containing 10%FBS and bFGF) in presence of 3 µg/mL of ritrodrine chlorohydrate. The picture shows representative fields of BMSC containing fluorescent palmitate (BODIPY FL C16) at different time points. The graph shows the uptake levels of palmitate in different experimental conditions. Data are expressed as arbitrary units. The table shows the percentage of released palmitate that was calculated as the ratio between the BODIPY level at 2 or 4 h and the starting point (T1/T0 or T2/T0). Scale bar: 30 µM.
Discussion
Adipogenesis in bone marrow is presumed to be identical to those in other tissues. However, the function of BMAT is uncertain, since little attention has been paid to fat in the marrow. For several years, it was believed that marrow adipocytes under most physiological conditions were metabolically inert. Bone marrow adipogenesis was considered a default mechanism whereby mesenchymal stem cells enter the fat lineage because of their inability to differentiate into muscle, bone, or chondrocytes. Now is evident that BMAT is different from adipose tissue found elsewhere in the body, since some studies suggest that adipocytes in BMAT are metabolically active and responsive to signals that are different from those acting on WAT and BAT. The function of BMAT is still not well understood, and also the molecular mechanisms regulating cell entry into the adipocytic lineage are poorly investigated. We decided to study the role of retinoblastoma gene family in BMAT adipogenesis, given the key role that cell cycle checkpoint proteins, such as the retinoblastoma proteins, have in adipocyte differentiation.2,7
Our interest arose from the observation that previous studies provided conflicting results. These discrepancies may derive from studies performed on different tissues sources (WAT, BAT, and BMAT), or on cells at different maturation stages (uncommitted vs. committed). While there is general agreement on the key role RB1 plays in adipogenesis, it is not clear whether this gene is fundamental for all the phases of adipocyte differentiation, since there are studies claiming that its role lies mainly in terminal differentiation.
The development of distinct cell types from multipotent precursors can be viewed as a 2-step process. In the first step, termed commitment or determination, the stem cell becomes limited to one particular lineage. In the second step, terminal differentiation, the cell develops along its determined lineage to become a functional specialized cell, such as an adipocyte.8,9,31 Studies on the role of RB1 in adipogenesis have been performed on mouse committed precursors (pre-adipocytes). There are no data on the role of RB1 on the entire adipocyte differentiation process starting from human stem cells (BMSC). This issue should be clearly addressed, since there are several data showing that molecular events of human BMSC adipogenesis did not completely overlap those of the mouse pre-adipocytes.6 Moreover, we clearly demonstrated that the role of retinoblastoma proteins depends on several parameters, such as animal species under investigation, cell type, and cell status (stem cell, progenitor, differentiated cell) (Alessio 2013). The lack of data on the possible role of the other 2 members of the retinoblastoma gene family (RB2/P130 and P107) in human BMSC adipogenesis is another caveat in the study of adipocyte commitment and differentiation.
In the current study we addressed all these issues, and we evidenced that lack of RB1 or RB2/P130 increased the percentage of BMSC committed toward adipocyte phenotype (Fig. 2). Nevertheless, the adipocytes appeared not to reach a terminal differentiation or, alternatively, showed dysregulated functions. Indeed, in adipocytes lacking RB1 or RB2, the mean cellular level of early differentiation markers (C/EBPδ and C/EBPβ, respectively) persisted at a high level (Fig. 3). The uptake and release of lipids is a key function of adipose cells. Adipocytes lacking RB2 showed a higher propensity in lipid uptake compared with control cells, while adipocytes lacking RB1 showed a higher capacity in lipid release (Fig. 5).
Overall, these results suggest that RB1 and RB2/P130 are not required for all steps of the adipocyte differentiation. Commitment of BMSC to adipocyte lineage is facilitated by a lack of RB1 and RB2. This may be due to the role these genes play in controlling the cell cycle. Indeed, in recent years, great efforts have been dedicated to addressing the contribution of cell cycle exit to the acquisition of cell identity. The differentiation of multipotent progenitor cells into mature differentiated cells occurs over a protracted period and appears to be accompanied by a progressive restriction in the range of fates available to individual cells. The fate of certain classes of cells appears to be restricted several divisions before a definitive cell cycle exit; however, for other cell types, this restriction occurs much closer to the last mitosis, and, for yet others, only after they have reached the postmitotic stage.8,9,31 In this setting, a lack of RB1 or RB2 may delay the onset of last cell division and allow more BMSC to be committed to adipocyte. On the other hand, a lack of RB1 or RB2 does not permit a careful control of cell cycle and/or a definitive exit from cell cycle as our flow cytometry result evidenced (Fig. 4). This event, associated with previous research showing that RB1 promotes differentiation of pre-adipocyte to mature cells by interacting with adipogenesis-related transcription factors, indicates that RB1 and RB2 are indispensable in the second step of adipocyte differentiation (from committed precursors to mature cells).
Our hypothesis is in agreement with the findings of Lee’s research team, who demonstrated that in vivo RB1 loss favors adipogenesis over osteogenesis to the extent it can reduce the levels of calcified bone and greatly increase the level of brown fat.32
The novelty of our research resides on the fact that for the first time we demonstrated that RB2/P130 and not only RB1 play a role in adipogenesis. Moreover, our studies were performed on human cells and not in a mouse model, which could also explain why we did not evidence any switch toward brown adipogenesis in the absence of RB1 (data not shown) as some other groups reported.14
In conclusion, our research demonstrated that RB1 and RB2/P130 play a key role in the correct lineage commitment of BMSC and that their functions are important for a proper adipogenesis.
Materials and Methods
BMSC cultures
Bone marrow was obtained from healthy donors after informed consent. We separated cells on 1.073 g/mL Ficoll density gradient (GE Healthcare) to minimize loss of BMSC during separation steps.33 The mononuclear cell fraction was collected and washed in PBS. We seeded 1–2.5 × 105 cells/cm2 in α-MEM containing 10% lot selected FBS and 2 ng/ml bFGF. We performed FBS selection according to Kuznetsov.34 After 72 h, non-adherent cells were discarded, and adherent cells were further cultivated to confluency (Passage 0–P0). Cells were then incubated for 7–10 d to reach confluence and were further propagated for our experimental plain. All experiments were performed P3/P4 cultures. All cell culture reagents were obtained from Euroclone Life Sciences and Hyclone unless otherwise stated.
Silencing
shRNAs targeting the human RB1, RB2/P130, and P107 mRNA, as well as scrambled control shRNAs, were designed following the procedure described by the RNAi consortium of Broad Institute. We used the following pLKO.1 vectors to express the shRNAs, available at the RNAi consortium: clone TRCN0000010418 to silence RB1 (NCBI Reference Sequence: NM_000321.2); clone TRCN0000039923 to silence RB2/P130 (NCBI Reference Sequence: NM_005611.3); clone TRCN0000040022 to silence P107 (NCBI Reference Sequence: NM_183404.1).
To generate knockdown cells, lentiviral particles were produced as described (http://www.broadinstitute.org/genome_bio/trc/publicProtocols.html).10
Cell cycle analysis
For each assay, cells were collected and re-suspended in a hypotonic buffer containing propidium iodide. Samples were acquired on a FACSCalibur flow cytometer using the Cell Quest software. They were then analyzed with a standard procedure using Cell Quest and ModFitLT software.
Clonal committment
We prepared BMSC clones from P0 cultures. Clones were obtained by limiting dilution: briefly, 10 cells were plated in each well of a 96-multiwell plate, and 4–5 plates were prepared for each experimental condition. From the initial seeding, clones were cultured in standard medium and were examined daily for the appearance of colonies. Wells containing more than one colony were not considered. All the clones that reached confluence were detached with 0.05% trypsin-0.01% EDTA (Sigma) and each of them plated in 4 wells (replicas) of a 24-multiwell plate. Out of the 4 replicas of each clone, 3 were stimulated for committment experiments when cells reached confluence, and 1 was cultivated undifferentiated (Fig. S2).
Adipogenic commitment and differentiation
BMSC were incubated for 3 wk in hMSC mesenchymal stem cell adipogenic differentiation medium (Lonza). Lipid droplets were revealed by staining with Oil red O or with BODIPY 505/515 (D-3921) (Invitrogen), which stains neutral lipids and is very specific for cellular lipid droplets.
Uptake and release of free fatty acid (FFA)
To evaluate the FFA uptake, differentiated BMSC were serum starved for 16 h, then cells were incubated in RPMI with 10 µM of the green fluorescent fatty acid, BODIPY FL C16 (Invitrogen), for 3 h (T0 starting point). Subsequently, cells were washed 3 times with PBS and incubated for different time periods with complete medium (α-MEM containing 10% FBS and bFGF) in the presence or absence of 3 µg/mL of ritrodrine chlorohydrate (adrenoceptor agonist).
Osteogenic commitment
BMSC were incubated for 2 wk stimulated in hMSC mesenchymal stem cell osteogenic differentiation medium (Lonza). Staining with Alizarin red revealed a calcium deposit.
Chondrogenic commitment
BMSC were incubated for 2 wk stimulated in hMSC mesenchymal stem cell chondrogenic differentiation medium (Lonza). Differentiatied chondrocytes were revealed by staining with Fast Green and Safranin O.
RNA extraction and RT-PCR
Total RNA was extracted from cell cultures using TRI REAGENT (Molecular Research Center Inc) according to the manufacturer’s protocol. The mRNA levels of the genes analyzed were measured by RT-PCR amplification, as previously reported.35
Sequences for mRNAs from the nucleotide data bank (National Center for Biotechnology Information) were used to design primer pairs for RT-PCR reactions (Primer Express, Applied Biosystems) (Table 1). Appropriate regions of GAPDH cDNA were used as controls. PCR cycles were adjusted to have linear amplification for all the targets. Each RT-PCR reaction was repeated at least 3 times. A semi-quantitative analysis of mRNA levels was performed by the “GEL DOC UV SYSTEM” (Biorad Company). Primer sequences were designed with Primer express software.
Table 1. Primers for RT-PCR gene expression analysis.
| Gene | Primer | Sequence | Annealing | Amplicon | ID |
|---|---|---|---|---|---|
| T (°C) | Length (bp) | ||||
| RB1 | 2443 | 5′-TTGCAGTATG CTTCCACCAG G-3′ | 59 | 107 | 5925 |
| 2549 | 5′-ATGTTCCCTC CAGGAATCCG T-3′ | ||||
| RB2 | 3958 | 5′-TGACAAAGGA GAGCACACCC A-3′ | 60 | 134 | 5934 |
| 4091 | 5′-TGCACACCCG AACAATCAAG-3′ | ||||
| P107 | 2949 | 5′-TGATGGATGC TCCACCACTC T-3′ | 59 | 125 | 5933 |
| 3073 | 5′-TGGTGTAAGG CCTGACCCAT T-3′ | ||||
| PPAR-γ | 844 | 5′-TCGACCACGT CAATCCAGAG T-3′ | 59 | 102 | 5468 |
| 945 | 5′-TCGCCTTTGC TTTGGTCAG-3′ | ||||
| ATGL | 618 | 5′-ATCCAGGCCA ATGTCTGCA-3′ | 62 | 103 | 57104 |
| 720 | 5′-GGTTGTCTGA AATGCCACCA T-3′ | ||||
| HSL | 556 | 5′-CGCACCACAG CAATCAACCT TA-3′ | 60 | 118 | 3991 |
| 673 | 5′-TCTCAATGCT GGCTCCTGTT G-3′ | ||||
| LIPF | 968 | 5′-ATGATCAGTC CCAACCTCCC T-3′ | 59 | 116 | 8513 |
| 1083 | 5′-TGGAAGCAAA AAGGCCAACA-3′ | ||||
| LIPJ | 1062 | 5′-CACAACCCAG CAGGAACATC T-3′ | 55 | 108 | 142910 |
| 1237 | 5′-TTCCAAATTG CAGTTGCCAC-3′ | ||||
| LIPK | 304 | 5′-TTGGCTTTCC CTCTGGCAGA-3′ | 58 | 104 | 340654 |
| 407 | 5′-TCCGGTGATT TCGGTGACA-3′ | ||||
| LIPM | 443 | 5′-TGTGGTGTTA CTGCAGCATG G-3′ | 60 | 109 | 643414 |
| 551 | 5′-CCACACGTCA AAACCAGCAT C-3′ | ||||
| LIPN | 541 | 5′-GTAGCCTTTT CCACCATGCC T-3′ | 57 | 104 | 643418 |
| 644 | 5′-AACCTGGTAA AAATGCCCGT G-3′ | ||||
| LPL | 413 | 5′-ATGGCTGGAC GGTAACAGGA A-3′ | 59 | 107 | 4023 |
| 519 | 5′-TGACAGCCAG CTCAGCACAA T-3′ | ||||
| GAPDH | 406 | 5′-TTCACCACCA TGGAGAAGGG T-3′ | 61 | 107 | 2597 |
| 512 | 5′-TGGTTCACAC CCATGACGAA C-3′ |
For each gene the table shows the sequences of primers, annealing temperature, amplicon length and NCBI gene identification number (ID) (http://www.ncbi.nlm.nih.gov/gene/).
Western blotting
Cells were lysed in a buffer containing 0.1% Triton X-100 for 30 min at 4 °C. Lysates were centrifuged for 10 min at 10 000 g at 4 °C. After centrifugation, 10–40 μg of each sample was loaded, resolved by electrophoresis on a polyacrylamide gel, and electroblotted onto a nitrocellulose membrane. Primary antibodies were used according to the manufacturer’s instructions.
Immunoreactive signals were detected with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz) and reacted with ECL plus reagent (GE Healthcare).
Statistical analysis
Statistical analysis was performed with GraphPad Prism-version 5.01 statistical software package (GraphPad Software Inc). Statistical significance was evaluated using ANOVA analysis followed by Student t and Bonferroni tests.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was partially funded by the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement n° 323267 to UG, by Progetto PON - “Ricerca e Competitività 2007-2013” - PON01_00802 dal titolo: “Sviluppo di molecole capaci di modulare vie metaboliche intracellulari redox-sensibili per la prevenzione e la cura di patologie infettive, tumorali, neurodegenerative e loro delivery mediante piattaforme nano tecnologiche” to GP, by Human Health Foundation, Italy (www.hhfonlus.com) and Sbarro Health Research Organization and Commonwealth of Pennsylvania , USA (www.shro.org).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27275
References
- 1.Devlin MJ. Why does starvation make bones fat? Am J Hum Biol. 2011;23:577–85. doi: 10.1002/ajhb.21202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen CJ, Ackert-Bicknell C, Rodriguez JP, Pino AM. Marrow fat and the bone microenvironment: developmental, functional, and pathological implications. Crit Rev Eukaryot Gene Expr. 2009;19:109–24. doi: 10.1615/CritRevEukarGeneExpr.v19.i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai M, de Paula FJ, Rosen CJ. New insights into osteoporosis: the bone-fat connection. J Intern Med. 2012;272:317–29. doi: 10.1111/j.1365-2796.2012.02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyer Nardi N, da Silva Meirelles L. Mesenchymal stem cells: isolation, in vitro expansion and characterization. Handb Exp Pharmacol 2006:249-82. [PubMed] [Google Scholar]
- 5.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 6.Qian SW, Li X, Zhang YY, Huang HY, Liu Y, Sun X, Tang QQ. Characterization of adipocyte differentiation from human mesenchymal stem cells in bone marrow. BMC Dev Biol. 2010;10:47. doi: 10.1186/1471-213X-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–96. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 8.Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–6. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- 9.Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22:5208–19. doi: 10.1038/sj.onc.1206558. [DOI] [PubMed] [Google Scholar]
- 10.Alessio N, Bohn W, Rauchberger V, Rizzolio F, Cipollaro M, Rosemann M, Irmler M, Beckers J, Giordano A, Galderisi U. Silencing of RB1 but not of RB2/P130 induces cellular senescence and impairs the differentiation potential of human mesenchymal stem cells. Cell Mol Life Sci. 2013;70:1637–51. doi: 10.1007/s00018-012-1224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera RE, Sah VP, Williams BO, Mäkelä TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–7. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003;23:1044–53. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–94. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen JB, te Riele H, Kristiansen K. Novel function of the retinoblastoma protein in fat: regulation of white versus brown adipocyte differentiation. Cell Cycle. 2004;3:774–8. doi: 10.4161/cc.3.6.908. [DOI] [PubMed] [Google Scholar]
- 15.Richon VM, Lyle RE, McGehee RE., Jr. Regulation and expression of retinoblastoma proteins p107 and p130 during 3T3-L1 adipocyte differentiation. J Biol Chem. 1997;272:10117–24. doi: 10.1074/jbc.272.15.10117. [DOI] [PubMed] [Google Scholar]
- 16.Higgins C, Chatterjee S, Cherington V. The block of adipocyte differentiation by a C-terminally truncated, but not by full-length, simian virus 40 large tumor antigen is dependent on an intact retinoblastoma susceptibility protein family binding domain. J Virol. 1996;70:745–52. doi: 10.1128/jvi.70.2.745-752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 18.Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell. 2002;3:903–10. doi: 10.1016/S1534-5807(02)00360-X. [DOI] [PubMed] [Google Scholar]
- 19.Nikitin AYu, Shan B, Flesken-Nikitin A, Chang KH, Lee WH. The retinoblastoma gene regulates somatic growth during mouse development. Cancer Res. 2001;61:3110–8. [PubMed] [Google Scholar]
- 20.Mödder UI, Roforth MM, Nicks KM, Peterson JM, McCready LK, Monroe DG, Khosla S. Characterization of mesenchymal progenitor cells isolated from human bone marrow by negative selection. Bone. 2012;50:804–10. doi: 10.1016/j.bone.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gronthos S, Graves SE, Ohta S, Simmons PJ. The STRO-1+ fraction of adult human bone marrow contains the osteogenic precursors. Blood. 1994;84:4164–73. [PubMed] [Google Scholar]
- 22.Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG, Riminucci M, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–36. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Battula VL, Treml S, Bareiss PM, Gieseke F, Roelofs H, de Zwart P, Müller I, Schewe B, Skutella T, Fibbe WE, et al. Isolation of functionally distinct mesenchymal stem cell subsets using antibodies against CD56, CD271, and mesenchymal stem cell antigen-1. Haematologica. 2009;94:173–84. doi: 10.3324/haematol.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fekete N, Rojewski MT, Fürst D, Kreja L, Ignatius A, Dausend J, Schrezenmeier H. GMP-compliant isolation and large-scale expansion of bone marrow-derived MSC. PLoS One. 2012;7:e43255. doi: 10.1371/journal.pone.0043255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn HJ, Lee WJ, Kwack K, Kwon YD. FGF2 stimulates the proliferation of human mesenchymal stem cells through the transient activation of JNK signaling. FEBS Lett. 2009;583:2922–6. doi: 10.1016/j.febslet.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 26.Kizhner T, Ben-David D, Rom E, Yayon A, Livne E. Effects of FGF2 and FGF9 on osteogenic differentiation of bone marrow-derived progenitors. In Vitro Cell Dev Biol Anim. 2011;47:294–301. doi: 10.1007/s11626-011-9390-y. [DOI] [PubMed] [Google Scholar]
- 27.Lai WT, Krishnappa V, Phinney DG. Fibroblast growth factor 2 (Fgf2) inhibits differentiation of mesenchymal stem cells by inducing Twist2 and Spry4, blocking extracellular regulated kinase activation, and altering Fgf receptor expression levels. Stem Cells. 2011;29:1102–11. doi: 10.1002/stem.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 29.Muraglia A, Cancedda R, Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113:1161–6. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 30.Bjorndal B, Burri L, Staalesen V, Skorve J, Berge RK. Different adipose depots: their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. J Obes. 2011;2011:490650. doi: 10.1155/2011/490650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison SJ, Shah NM, Anderson DJ. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–98. doi: 10.1016/S0092-8674(00)81867-X. [DOI] [PubMed] [Google Scholar]
- 32.Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–4. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grisendi G, Annerén C, Cafarelli L, Sternieri R, Veronesi E, Cervo GL, Luminari S, Maur M, Frassoldati A, Palazzi G, et al. GMP-manufactured density gradient media for optimized mesenchymal stromal/stem cell isolation and expansion. Cytotherapy. 2010;12:466–77. doi: 10.3109/14653241003649510. [DOI] [PubMed] [Google Scholar]
- 34.Kuznetsov SA, Mankani MH, Bianco P, Robey PG. Enumeration of the colony-forming units-fibroblast from mouse and human bone marrow in normal and pathological conditions. Stem Cell Res. 2009;2:83–94. doi: 10.1016/j.scr.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galderisi U, Di Bernardo G, Cipollaro M, Jori FP, Piegari E, Cascino A, Peluso G, Melone MA. Induction of apoptosis and differentiation in neuroblastoma and astrocytoma cells by the overexpression of Bin1, a novel Myc interacting protein. J Cell Biochem. 1999;74:313–22. doi: 10.1002/(SICI)1097-4644(19990901)74:3<313::AID-JCB1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.