Abstract
Many proteins undergo a sharp decrease in chain dimensions during early stages of folding, prior to the rate-limiting step in folding. However, it remains unclear whether compact states are the result of specific folding events or a general hydrophobic collapse of the poly-peptide chain driven by the change in solvent conditions. To address this fundamental question, we extended the temporal resolution of NMR-detected H/D exchange labeling experiments into the microsecond regime by adopting a microfluidics approach. By observing the competition between H/D exchange and folding as a function of labeling pH, coupled with direct measurement of exchange rates in the unfolded state, we were able to monitor hydrogen-bond formation for over 50 individual backbone NH groups within the initial 140 microseconds of folding of horse cytochrome c. Clusters of solvent-shielded amide protons were observed in two α-helical segments in the C-terminal half of the protein while the N-terminal helix remained largely unstructured, suggesting that proximity in the primary structure is a major factor in promoting helix formation and association at early stages of folding while the entropically more costly long-range contacts between the N- and C-terminal helices are established only during later stages. Our findings clearly indicate that the initial chain condensation in cytochrome c is driven by specific interactions among a subset of α-helical segments rather than a general hydrophobic collapse.
Keywords: protein folding, kinetics, rapid mixing, microfluidics, NMR
INTRODUCTION
One of the most powerful and enduring paradigms in biology has been the notion that the amino acid sequence encodes the structure of a protein, which in turn gives rise to biological function. Indeed, most proteins have evolved the remarkable ability to fold into a specific and essentially unique three-dimensional structure, despite the countless alternative conformations accessible to a disordered chain. We have learned a great deal in recent years about the mechanisms of folding from experimental and computational studies of small globular proteins and isolated domains.1–5 This progress has been closely linked with technological advances, including the ability to monitor individual amide probes by NMR- or mass-detected hydrogen exchange labeling methods,6–14 improved techniques for rapid initiation of folding reactions, including turbulent and laminar mixing,15–21 laser-induced photolysis, temperature and pressure jump techniques,22–27 single-molecule spectroscopy,28 and mechanical unfolding.29,30 Our theoretical and conceptual understanding of protein folding has benefited greatly from more advanced computational models and their joint application with biophysical studies.31–35 However, a full understanding of sequence-encoded rules for folding remains elusive, as illustrated by the fact that ab initio methods still lag substantially behind knowledge-based computational methods for predicting 3D protein structure.36
A long-standing challenge in the protein folding field has been to determine whether the compact states populated during early stages of folding of many proteins are the result of specific folding or non-specific chain condensation events. Time-resolved spectroscopic and small-angle X-ray scattering (SAXS) studies on numerous proteins have shown that compact states, often containing significant levels of secondary structure, accumulate during the initial stages of folding, prior to the rate-limiting step in the formation of the native structure.2,3,37–40 It has been proposed that a general hydrophobic collapse of the polypeptide chain triggered by the rapid change from denaturing to aqueous solvent conditions gives rise to a dynamic ensemble of compact conformations that lack persistent long-range interactions.41–43 As in the case of a homopolymer in a poor solvent,44 such compact ensembles are formed in a continuous (multi-state) transition, lack specific secondary and tertiary structure, and can be viewed as the denatured state under non-denaturing solvent conditions. Arguing against this scenario are observations that significant free energy barriers separate early compact intermediates from the initial unfolded ensemble,45,46 indicating that they represent distinct thermodynamic states, as well as evidence for selective secondary structure formation and non-uniform chain collapse on the sub-millisecond time scale.37,38,40,47 However, a more definitive resolution of this controversy will require site-specific structural information on the ensemble of states formed during the initial collapse of a protein.
In this study we coupled NMR-detected H/D exchange with ultra-rapid mixing techniques to gain detailed structural insight into the ensemble of states populated within the initial 100 µs of folding of horse ferricytochrome c (cyt c). We have designed and built a highly efficient microfluidic mixer to extend the temporal resolution of these H/D exchange/folding competition experiments well into the microsecond time range. This advance makes it possible to directly observe the formation of individual hydrogen bonds on the submillisecond time scale. A detailed analysis of the pH-dependence of amide protection for cyt c at a folding time of 140 µs showed that amide protons in three α-helical segments in the C-terminal half of the protein (the 60s, 70s and C-terminal helices) were preferentially protected from solvent exchange within 140 µs of initiating the folding reaction. At the same time the N-terminal α-helix remained largely unprotected, indicating that sequence-local helix-helix contacts are formed preferentially during early stages of folding whereas long-range (N- to C-terminal) become important only during the later stages of folding (> 3 ms). Careful calibration of amide exchange rates from the initial, urea-unfolded, state, using amide protection measurements as a function of time (0.1 to 2.5 ms) made it possible to reliably measure even modest levels of solvent protection, and thus enabled accurate detection of individual hydrogen bonds in marginally stable early folding intermediates.
EXPERIMENTAL
Materials
Phenyl chloroacetate (PCA) was obtained from Apollo Scientific, Ltd., Denton, Manchester M34 3SG, UK (custom synthesis). Horse heart cytochrome c (>95% pure) used for fluorescence-detected kinetic measurements was from Sigma-Aldrich (St. Louis, MO). Ultra-pure urea was obtained from MP Biomedicals (Solon, OH). Other chemicals were reagent grade.
Protein Expression and Purification
The methods used for expression and purification of isotope-labeled horse cyt c were based on published protocols48 with some modifications to enhance yields. The H33N variant was chosen to minimize possible complications due to formation of a non-native His-heme ligand.49 Large quantities (>100 mg) of uniformly 15N-labeled and mg quantities of double-labeled (13C and 15N) protein were prepared by co-expressing the genes for the H33N mutant of cyt c and yeast heme lyase in the Rosetta 2 (DE3) host, a derivative of BL21 designed to enhance the expression of eukaryotic cDNA in E. coli. By supplementing the bacteria with the heme precursor 0.3mM δ-aminolevulinate we were able to further raise the overall yield of cyt c production to at least 20 mg of purified protein per liter of minimal medium.
Hydrogen-exchange Labeling
The protein was initially unfolded in D2O (pD 2.0, 3 M urea) and rapidly mixed with a 4-fold excess of H2O refolding buffer at alkaline pH. Aging times ranging from 90 µs to 2.1 ms were achieved by selecting various combinations of inlet and outlet ports on the microfluidic mixer chip (Fig. 1). The exchange reaction was then quenched by adding a 50% excess of quench buffer (0.2 M sodium acetate and 0.05 M sodium ascorbate, pH 4.8), lowering the pH to 5. These conditions result in dramatically slower H/D exchange while allowing folding to continue. Addition of ascorbate further stabilizes the protein by reducing the heme iron. The pH at each stage of the exchange experiment was determined by manual mixing. Solutions were immediately concentrated at 4°C using Millipore Ultra 10K concentrators and repeatedly washed with a D2O buffer, pD 5.3 (25 mM d4-acetic acid and 10–15 mM sodium ascorbate). The samples were concentrated to 400 µl and flash frozen for later NMR data collection.
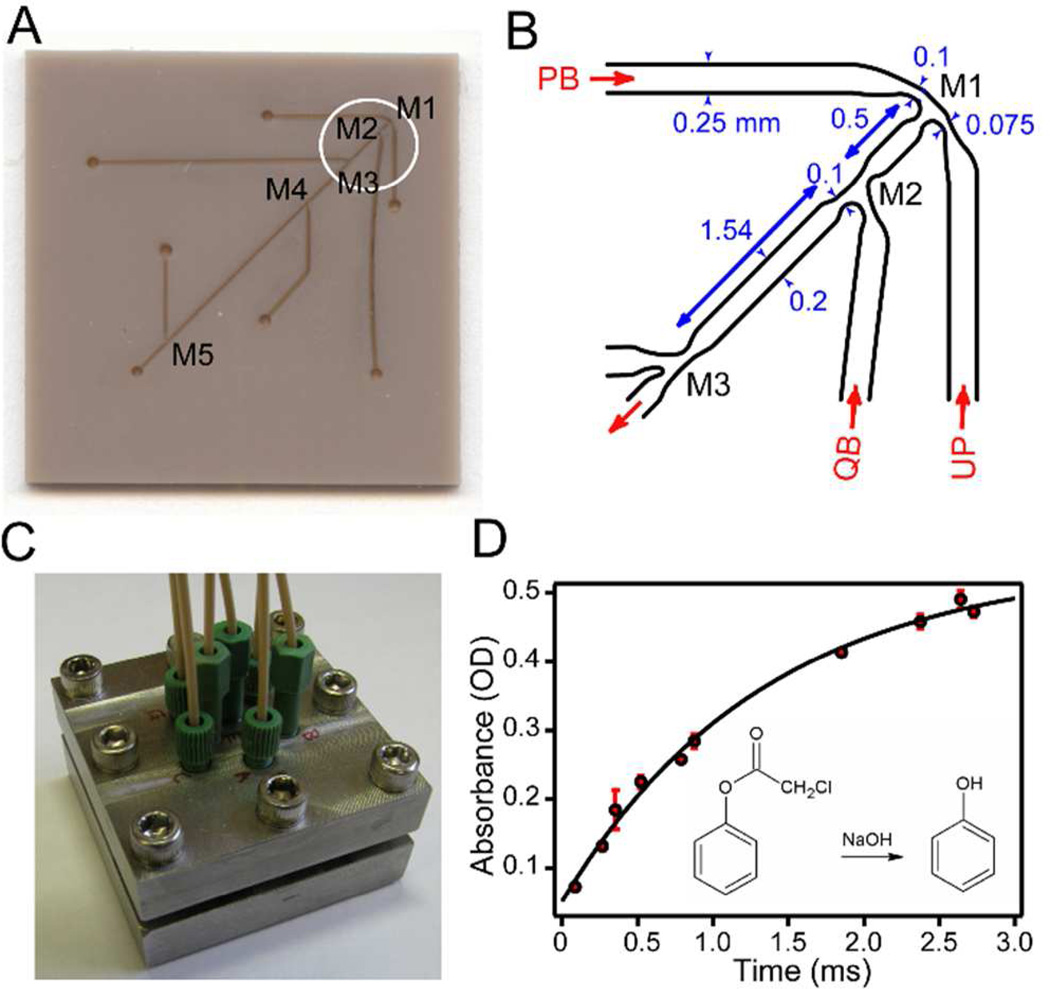
Figure 1.
Microfluidic mixing device. A. Photograph of PEEK chip (25×25 mm2) showing laser microfabricated surface features, including mixing regions M1 through M5 and access channels. B. Schematic of an expanded region (circled in A) covering mixing regions M1 through M3. PB: high-pH pulse buffer in H2O; UP: unfolded protein in D2O; QB: low-pH quench buffer in H2O. C. Photograph of the steel holder containing the multi-mixer chip illustrated in A. D. Calibration of the mixing device using alkaline hydrolysis of phenol chloroacetate (PCA; inset). The absorbance of the product (phenol) was measured at 270 nm vs. aging time.
NMR Measurements
NMR spectra were collected at 15 °C on a Bruker Avance II 600 MHz spectrometer equipped with a TCI CryoProbe. Published 1H assignments for WT horse cyt c were used as a reference for assigning the 1H–15N cross-peaks50 and confirmed by using standard heteronuclear 3D NMR methods.51 For each time delay or labeling pH proton occupancies were measured for ~50 individual NH groups by recording 2D NMR spectra, using the sensitive 1H-15N heteronuclear single-quantum correlation (HSQC) pulse sequence.51 HSQC experiments were run with 256 experiments in the 15N dimension consisting of 40 scans and 4096 data points in 1H dimension. Felix (Accelrys, San Diego, CA) was used for data processing and cross peak integration.
RESULTS
Design and Calibration of a Microfluidic Quenched-flow Mixing Device
H/D exchange labeling experiments coupled with NMR detection are a primary source of structural information on protein folding intermediates at the level of individual residues.6–8,10,11,52,53 Moreover, information on local structural stability of folding intermediates can be obtained by systematic variation of the labeling conditions.9,10,12,54,55 These experiments have for the most part relied on commercial quenched-flow equipment to carry out two or three sequential mixing steps with minimal delays of a few ms. One exception is a study by Uzawa et al., who used a microfluidic mixer with a minimal aging time of 0.4 ms for a detailed quenched-flow H-D exchange analysis of early folding steps in apomyoglobin.12 Previously, we have also been able to access sub-millisecond reaction times by using a quartz capillary mixer to generate a free-flowing jet of the protein/buffer mixture, which is injected into quench buffer after a variable delay.56 This simple device can reach aging times as short as 60 µs, but suffers from limited reproducibility, frequent mixer blockage and sample loss due to foaming. For this study, we developed a microfluidic mixing device that overcomes these limitations without sacrificing time resolution. Our design (Figure 1) consists of a series of 5 mixing chambers (M1-M5) connected through channels of different lengths (0.5 and 11 mm) to achieve aging times ranging from 90 µs to several ms (depending on flow rate). These features were etched onto the surface of a 25×25×1 mm3 polymer (PEEK) chip (Figure 1A), using laser micro-fabrication techniques (Potomac Photonics, Inc., Lanham, MD). Channel widths and depths range from ~0.1 mm in the mixing regions to 0.2 mm in the aging and access channels (Figure 1B). The mixer chip is covered with a smooth PEEK film and sandwiched between Teflon gaskets in a steel holder (Figure 1C). Trough-holes (0.63 mm diameter) at the end of each channel provide solution access via a tubing manifold. Solutions are delivered to the mixer via three Hamilton syringes driven by a servomotor actuator with a total flow-rate of 0.5 – 1 ml/sec. At 1 ml/s approximate flow velocities are 67 m/s at the exit of M1, 16 m/s in the aging channel and 100 m/s at the exit of the final mixer, ensuring highly turbulent flow conditions at every stage.
To determine the dead-time (minimal aging time) of the device we used alkaline hydrolysis of PCA as a test reaction.57 Rapid hydrolysis was achieved by mixing a PCA solution with 4-fold excess of 1 M sodium hydroxide. The hydrolysis reaction was subsequently quenched by adding 6-fold excess of hydrochloric acid (1 M, final pH 2.80), and the concentration of the reaction product, phenol, was measured by recording absorbance at 270 nm. By changing the combination of access channels for injecting the solutions, ten reaction times ranging from 90 µs to 2.1 ms were achieved (Figure 1D). The data fit well to a single-exponential function; extrapolation to y=0 corresponding to the absorbance prior to hydrolysis indicates that the minimal aging time achieved with the M1-M2 mixer combination corresponds to 90 ± 10 µs.
NMR-detected Quenched-flow H/D Exchange
Amide protons are often found to become protected from H/D exchange within the dead-time of quenched-flow mixing (2–10 ms), indicating that hydrogen bonded structure can form within milliseconds of refolding.9–11,52,53 The development of a microfluidic quenched-flow mixing device with a dead-time of less than 100 µs (Figure 1) now makes it possible to directly observe the formation of individual hydrogen bonds on the sub-millisecond time scale. The protein was initially unfolded in D2O (p2H 2.0, 3 M urea) and rapidly mixed with an H2O refolding buffer at alkaline pH, which sets up a competition between refolding and hydrogen exchange.6,58 Aging times ranging from 90 µs to 2.1 ms were achieved by selecting various combinations of inlet and outlet ports on the microfluidic mixer chip (Figure 1). The exchange reaction was then quenched by lowering pH to 5, conditions that result in dramatically slower H/D exchange while allowing folding to continue. For each time delay, proton occupancies were measured for ~50 individual NH groups by recording 2D NMR spectra, using the sensitive 1H-15N heteronuclear single-quantum correlation (HSQC) pulse sequence.51 By comparing the pH and time-dependence of the proton occupancies with those measured under denaturing condition, we can thus characterize very short-lived folding intermediates in terms of the formation of individual hydrogen bonds.
HD Exchange/folding Competition vs. pH
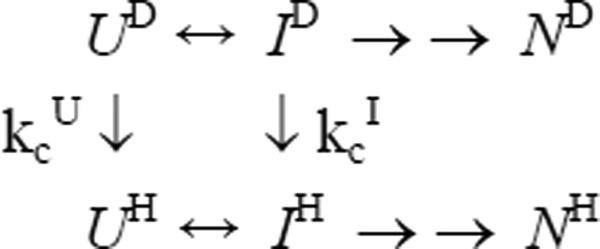
Continuous-flow fluorescence experiments revealed that a compact intermediate of cyt c, I, accumulates in an initial folding phase with a time constant of ~60 µs.45 To maximize accumulation of I, we allowed D to H exchange and structure formation to compete with each other for a period of 140 µs and measured the proton occupancy as a function of pH ranging from 7 (where no labeling occurs within the short competition period) to 12.5 (where even strongly protected NH groups are fully labeled). The use of such a short competition time makes it possible to quantify the protection factors in the early intermediate, even if amide protons are only marginally protected. Figure 2 shows amide protection data for a few representative residues spread throughout the sequence of cyt c (the complete set of data can be found in Supplemental Material, Figure S1). The contribution of an early intermediate to amide protection is described by Scheme 1: UH/D, IH/D and NH/D represent protonated/deuterated forms of the unfolded, intermediate and native states, and kcU and kcI are the exchange rates of a given NH group in the U and I states, respectively.9 If the I-state is well populated (kUI/kIU>>1) and direct exchange from the I-state is negligible compared to that from U-state (kcI << kcU), the proton occupancy, fH, as a function of labeling pH (Figure 2) or pulse time, tp, is described by Eq. 1:
kUIloc and kIUloc are local (residue-specific) folding/unfolding rate constants of the intermediate, kfglob and kuglob are global folding/unfolding rate constants due to alkaline denaturation (defined below), kc = kcU is the intrinsic exchange rate from the unfolded state (measured directly for the unfolded protein; see below) and tp is the duration of the exchange/folding competition period. The first term in the exponent of Eq. 1 accounts for both EX2 (exchange-limited) as well as EX1 (opening-limited) exchange from an early folding intermediate;59 the second term represents the global exchange of all amide protons due to alkaline unfolding of the protein, which sets in at pH values above 11.5.60 To determine these global rate constants, we carried out fluorescence-detected continuous-flow measurements of the kinetics of folding over a wide range of pH (Figure S2). The kinetics of folding over the whole pH range exhibits a major initial decay in Trp59 fluorescence with a time constant of ~30 µs, which has been attributed to Trp-heme energy transfer as the chain collapses into a compact early intermediate, I.45 This conclusion is supported by time-resolved SAXS measurements at folding times as short as 160 µs, which showed that formation of the I-state from the acid-unfolded state is accompanied by a 4 Å decrease in radius of gyration, corresponding to 40% of the overall change37. The initial rate of folding/collapse, kUI, remains essentially constant at a value of 33,000 ± 4,000 s−1 from pH 4.9 to 10.7 and increases to ~50,000 s−1 above pH 10.7 (Figure S2B). This provides clear evidence that the I-state accumulates even at very high pH. The decrease in the amplitude of the fast phase with increasing pH above 11 (Figure S2C) reflects the decreasing population of the I-state as it undergoes alkaline unfolding, a cooperative transition characterized by a pKa of 12.1 and a Hill coefficient of 2. With the well-founded assumption that the U to I transition is a two-state process,45,46 we can determine the pH-dependence of the unfolding rate constant, kIU (dashed line in Figure S2C), from the pH-independent folding rate, kUI = 50,000 s−1, and the amplitude of the fast phase (solid green line in Figure S2C), which represents the fractional population of the I-state.
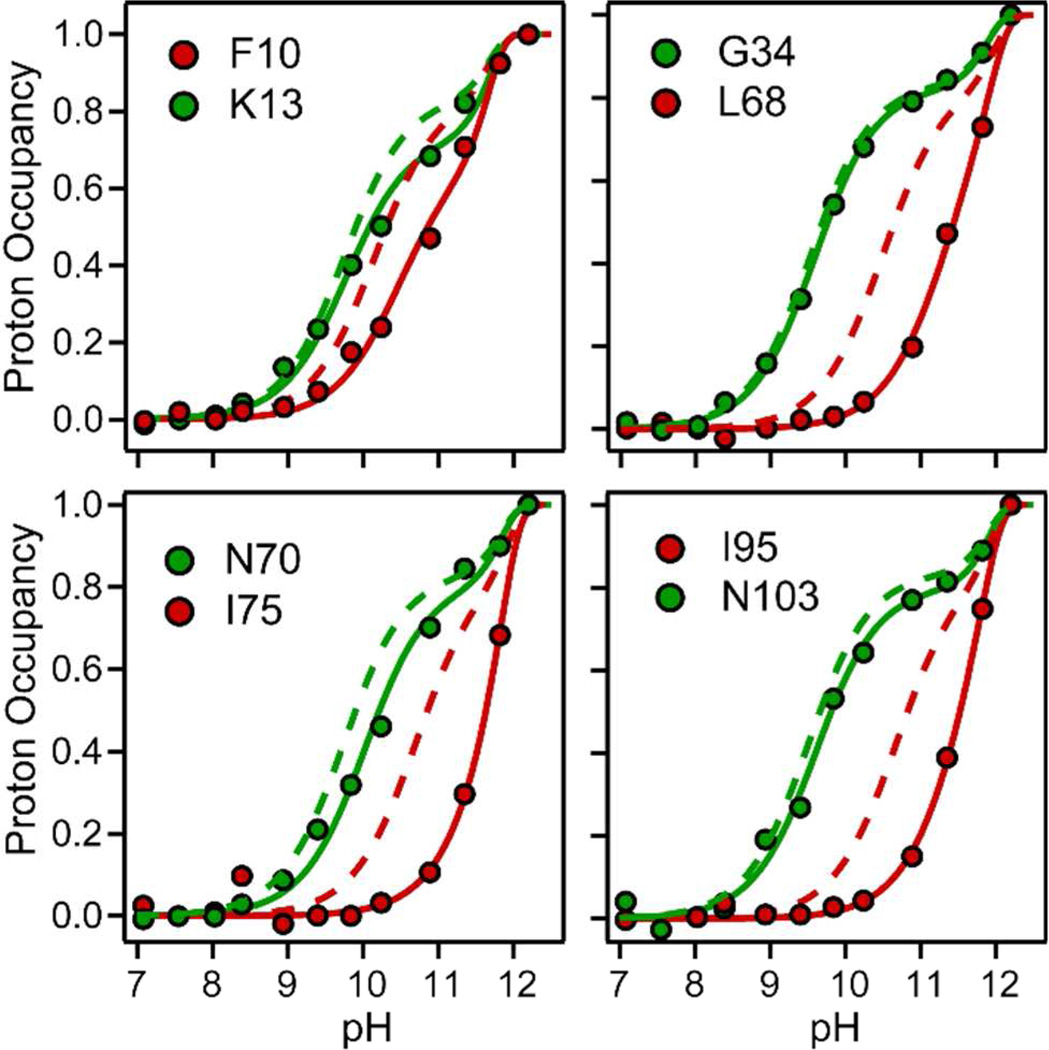
Figure 2.
pH-dependent competition between D-H exchange and folding of cyt c at constant time (140 µs, 22 °C). Normalized proton occupancies for representative residues (symbols, solid lines) are compare with the pH profiles predicted for the corresponding residues in the unfolded state (dashed lines). Solid lines represent fits of Eq. 1 to the data.
Scheme 1.
H/D exchange/folding competition
Intrinsic Exchange Rates and Time-dependent HD Exchange
Interpretation of these pH-dependent competition data requires accurate values for the rate of exchange of each amide proton in the unfolded state. In a previous study of the competition between HD-exchange and folding of cyt c at a longer competition time (2 ms), we compared the observed profiles of fH vs. pH with those under moderately denaturing conditions (in the presence of 2.5 M guanidine HCl) where any partially folded intermediates are destabilized.9 In order to avoid possible pitfalls due to residual structure formation at this relatively low denaturant concentration we chose here to directly measure the kinetics of HD exchange from the unfolded state, using our multi-mixer chip (Figure 1) to vary the exchange time (0.1 and 2.5 ms). In a first set of experiments we adopted the following time-dependent competition protocol first introduced by Roder and Wüthrich.6 Unfolded cyt c in D2O (pH 2, 3 M urea) was first mixed with a 4-fold excess of refolding buffer in H2O at pH 9.8 to set up a competition between refolding and D to H exchange. After a variable time period the exchange reaction was quenched by mixing in acetate buffer to lower pH to 5. HSQC spectra were recorded on the refolded protein samples to determine the relative degree of protonation (proton occupancy fH) for individual backbone amide groups. In a second set of experiments we directly measured the D to H exchange reaction in the unfolded state by using the same protocol, except that the acetate refolding buffer was replaced with a 6 M urea solution buffered at pH 9.8 (Figure S3). Figure 3 compares the results obtained under fully denaturing and folding conditions for a representative set of amide protons. In both cases fH increase exponentially with reaction time as D to H exchange proceeds. In 6 M urea (open symbols, dashed lines) fH approaches 1 at long times, indicating that exchange goes to completion. An exponential fit of the data directly yields exchange rates for individual residues in the urea-unfolded protein. By contrast, in the competition experiment at low urea concentration (filled symbols, solid lines), the proton occupancy at long reaction times levels off at a value well below 1 for most residues, indicating that the competing folding process (in this case, the initial U to I transition in Scheme 1) protects amide protons from exchange. If the I-state is well populated (kUI/kIU>>1) and a given NH group is protected in the intermediate (kcI << kcU), the time dependence of the proton occupancy, fH, can be approximated as follows:
| (Eq. 2) |
fH initially rises at a rate kcU+kf, where kf is an effective rate-constant for folding/protection of a given amide proton, and approaches at long times a steady-state level determined by the ratio of kf and kcU. Under conditions where exchange and folding rates are comparable, both kcU and kf can be determined independently by fitting Eq. 2 to time-resolved competition data.6
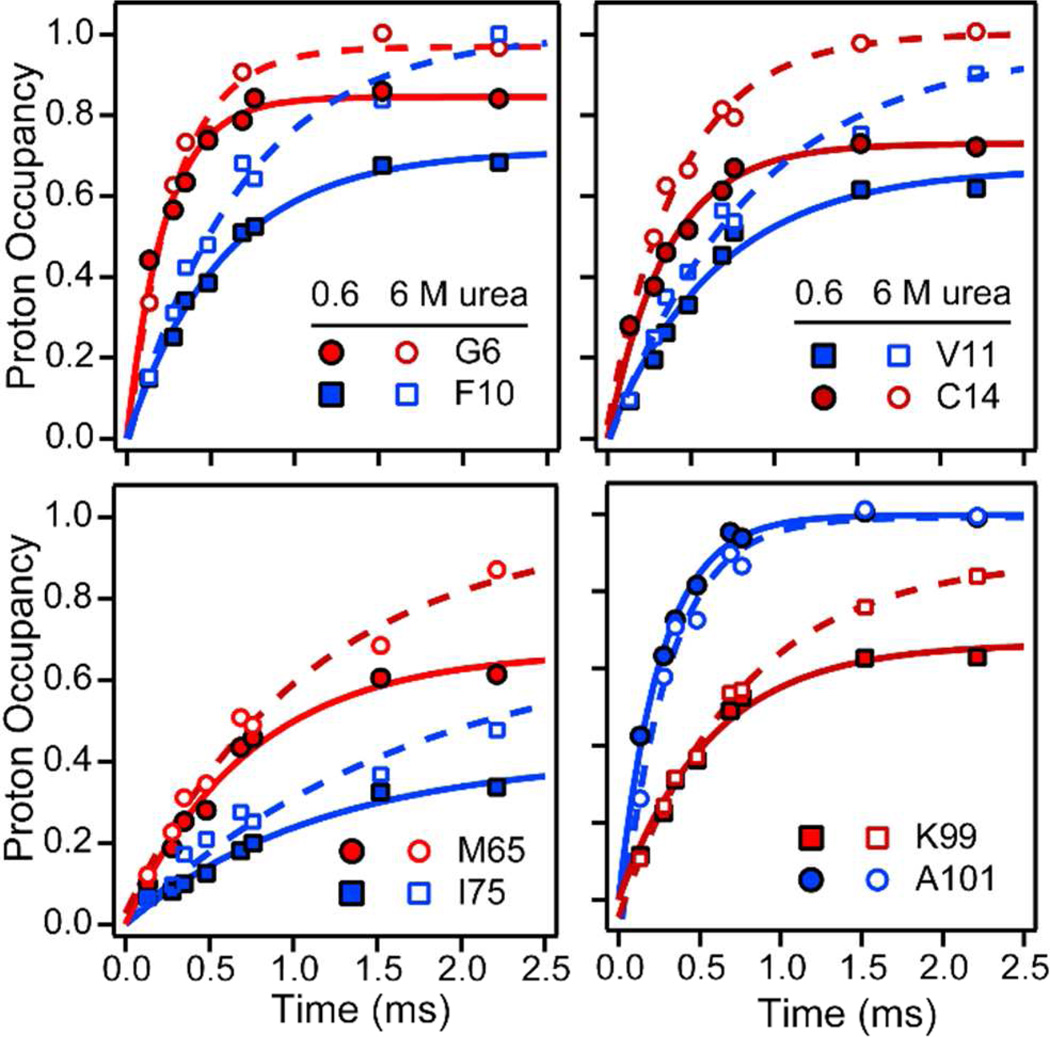
Figure 3.
Time-dependence of D-H exchange/folding competition at a urea concentration of 0.6 M (filled symbols) and intrinsic exchange kinetics in the urea-unfolded state (open symbols) for representative residues (pH 9.8, 22 °C). Solid lines represent fits of Eq. 2 to the competition data and dashed lines are single-exponential fits of the exchange data at 6 M urea.
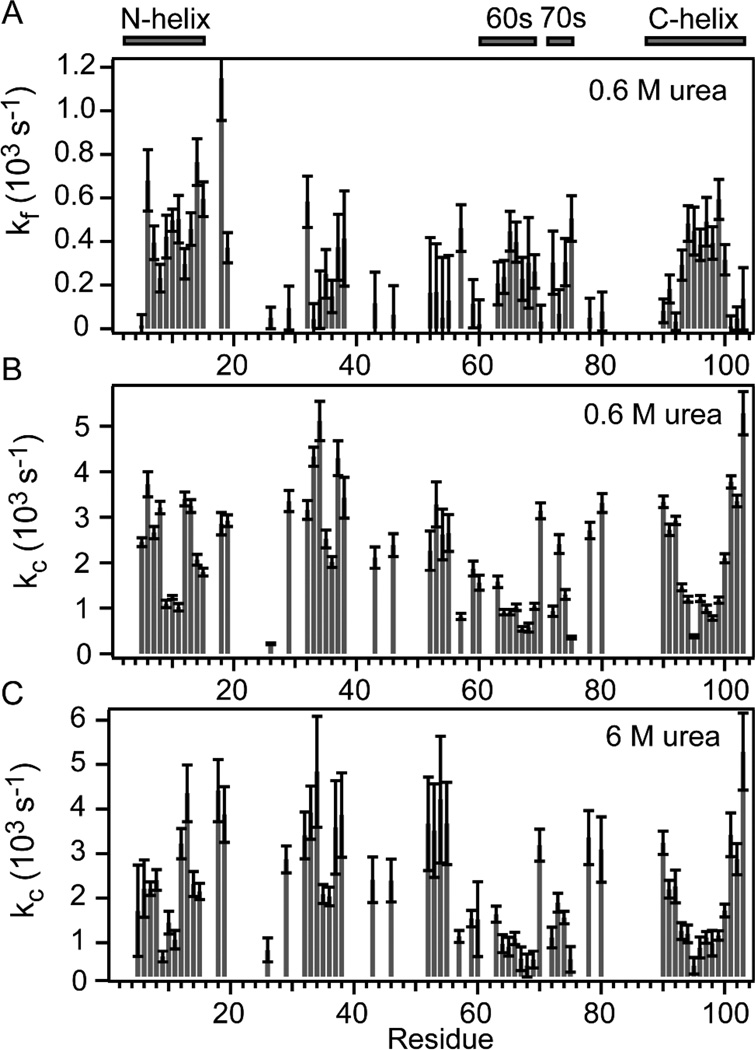
The time-dependent competition and direct exchange results are summarized in Figure 4. The effective rate of amide protection and exchange rates measured in the competition experiment at low urea concentration are shown in panels A and B, and the exchange rates for the unfolded protein in 6 M urea are shown in panel C (all at pH 9.8). Exchange rates in model peptides were shown to be largely independent of urea concentration.61 Indeed, the patterns of exchange rates vs. residue number we observed at low urea concentration (Figure 4B) and in 6 M urea (Figure 4C) are very similar. For the majority of residues the rates range from 1,000 to 5,000 s−1.
Figure 4.
Sequence dependence of time-dependent competition and exchange results. The bar graphs in panels A and B show effective protection rates (kf) and intrinsic exchange rates (kcU), respectively, at 0.6 M urea obtained from least-squares fitting of Eq. 2 to the competition data in Figure S3. Panel C shows direct exchange rates for unfolded cyt c at 6 M urea vs. residue number.
DISCUSSION
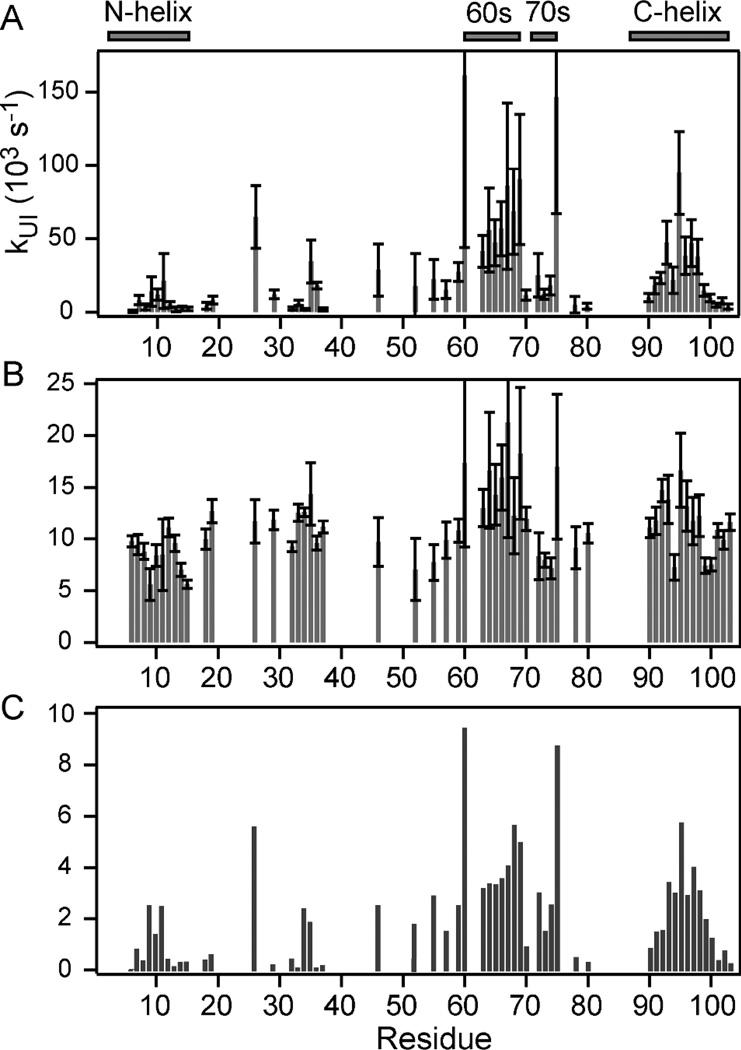
The exchange rates we measured for unfolded cyt c at both low and high urea concentrations (Figure 4) allow us to reliably calculate fH vs. pH profiles expected if no structure forms on the 100 µs time scale (dashed lines in Figure 2), which serve as a reference for interpretation of the pH-dependent folding/exchange competition data (solid lines). If a given amide group becomes protected from the solvent within the first 140 µs of refolding (generally due to formation of hydrogen-bonded structure62), we expect clear deviations in the fH vs. pH profile from the unfolded-state reference. Depending on the dominant exchange mechanism, the profile can either shift towards basic pH (EX2 mechanism) and/or level off at a lower plateau near pH 11, where exchange can approach an opening-limited (EX1) regime prior to the onset of global alkaline unfolding.10,60 For the quantitative analysis of the pH-dependent exchange/folding competition data (Figures 2 and S2), we used the kc values at low urea concentration (Figure 4B) as a constraint in fitting the data to Equation 1. With additional restraints to account for alkaline unfolding (see Figure S2), we were able to accurately describe the pH profiles (solid lines in Figures 2 and S2) with only two free fitting parameters, kUIloc and kIUloc (Figure 5). For different residues the initial folding rate, kUIloc, varies over two orders of magnitude with values ranging from ~1,500 s−1 to 150,000 s−1 (Figure 5A). In contrast, the rate constants in the unfolding direction, kIUloc, are much more uniform, ranging from ~5,000 to 20,000 s−1 over the whole cyt c sequence. The local folding rates span the overall rate of folding, kUI = 33,000 s−1 at pH 9.8, measured by continuous-flow fluorescence (Figure S2). Apparently, the effective folding/protection rate for weakly protected residues is much slower and that of well protected amides somewhat faster than the global rate of formation of the early collapsed intermediate. Figure 5C shows a bar graph of the local folding equilibrium constants, KUIloc = kUI-loc/kIUloc, which provide a measure of the local structural stability in the first compact state populated during folding of cyt c. While the uncertainty in the individual rate constants can be readily determined from the fitting errors, coupling between these parameters makes it difficult to determine errors for equilibrium constant. In the limiting cases of very weak or very strong protection the individual rates are poorly determined, but the equilibrium constant can still be measured reliably. Thus, for the structural interpretation of our results we will focus on local equilibrium constants (Figure 5C), as illustrated in Figure 6A.
Figure 5.
Summary of pH-dependent exchange/folding competition data for the compact intermediate, I, of cyt c populated at a folding time of 140 µs. Panels A and B show bar graphs of the local rate constants for formation (kUIloc) and unfolding (kIUloc) of the I-state vs. residue number obtained by fitting Eq. 1 to the data in Figure S2. The corresponding equilibrium constants, KUIloc = kUIloc/kIUloc, are plotted in panel C.
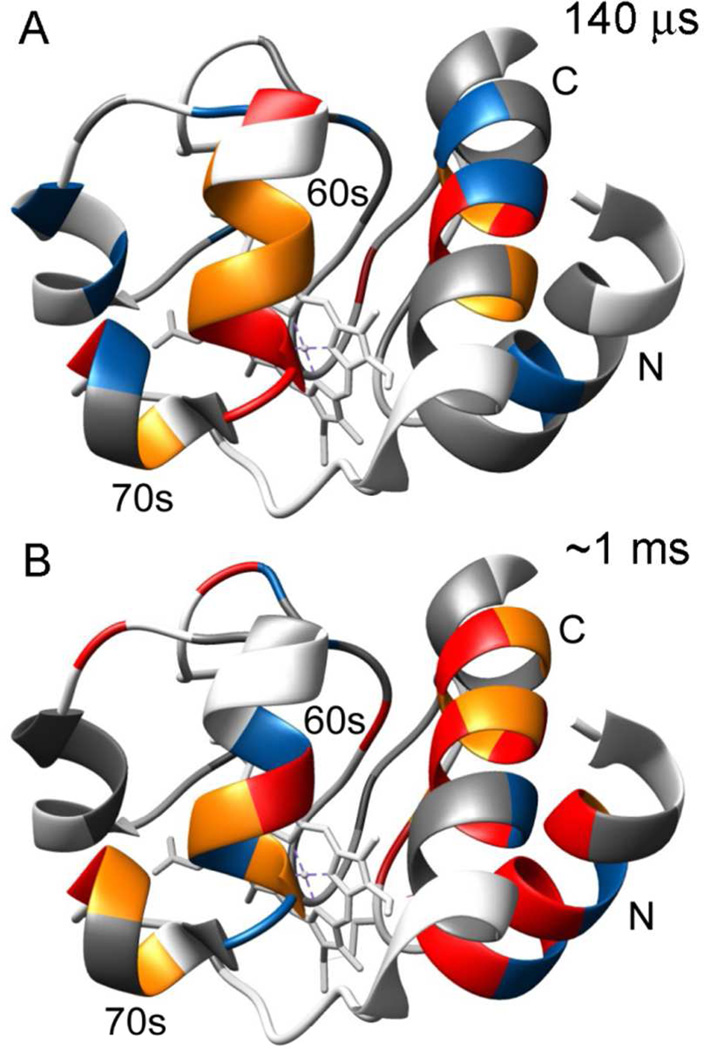
Figure 6.
Ribbon diagrams of the X-ray structure of native horse cytochrome c (pdb 1hrc) color-coded to show amide protection patterns at folding times of 140 µs (A) and ~1 ms (B). In both panels, residues that are protected in the native state, but unprotected in the I-state (KUIloc < 2; kfloc <200) are shown in gray. The pH-dependent competition results (Figure 5C) are summarized in panel A, where residues with low (2 ≤ KUIloc < 3), intermediate (3 ≤ KUIloc < 4) and high (KUIloc ≥ 4) are colored blue, orange and red, respectively. The time-dependent competition data (Figure 4A) are summarized in panel B, where residues with low (200 ≤ kfloc < 300), intermediate (300 ≤ kfloc < 400) and high (kfloc ≥ 400) are colored blue, orange and red, respectively.
The majority of residues in the N-terminal half of cyt c show little or no protection in the early folding intermediate (KUIloc ranging from less than 1 to about 2.5). The only exception is His 26 with KUIloc ≅ 6, which can be attributed to a tendency of this His side chain, the only potential non-native heme ligand in this H33N variant of the cyt c, to interact with the heme iron in the denatured state.49 The average KUIloc for the N-helix (residues 2–14) is less than 1, indicating that this segment is still largely disordered. Significantly stronger protection indicative of more persistent hydrogen bonded structure is observed for most residues of the 60s helix (residues 60–69), Ile 75 at the end of the short 70s helix (residues 71–75) and the central residues of the C-helix (residues 87–104). The C-terminal region shows a striking bell-shaped pattern with little or no protection at both ends and higher local stability constants in the middle (KUIloc ≥ 2.9 for residues 93–98, a segment dominated by hydrophobic side chains). The presence of a contiguous stretch of well-protected residues in both the 60s and 90’s clearly shows that within 140 µs of refolding both segments form α-helical secondary structure similar to that found in the native structure (Figure 6A). There are strong indications that the helices are not formed in isolation, but stabilize each other by mutual long-range interactions: (i) The local folding equilibrium constants for the core regions of the 60s and C-terminal helices are similar (KUIloc ~4 to 6), which supports the presence of mutual interactions stabilizing both helices. (ii) Time-resolved SAXS measurements at tf = 160 µs indicate that the I-state is a non-uniformly collapsed state containing a compact (helical) core surrounded by extended segments.37 (iii) Although the stability of the 60s and C-terminal helices is modest at this early stage of folding, it is substantially higher than that typically observed for helical model peptides. In particular, based on the CD spectrum of a synthetic peptide corresponding to the C-terminal helix of cyt c (residues 87–104), we previously concluded that the average helix content at 10 °C, pH 7, was approximately 15%,63 which corresponds to an average folding equilibrium constant of 0.2. In contrast, the average of the KUIloc values in Figure 5C over the C-terminal helix is 2.3 with a maximum value of 5.7 for the central Ile95. This increase in overall and local stability of the helix can be readily explained by helix-helix interactions with the 60s helix involving primarily the hydrophobic core residues, similar to those present in the native cyt c structure. It is difficult to compare our amide protection data with time-resolved CD measurements because of differences in dead time and/or sample conditions (denaturant concentration, pH, oxidation state, etc.). Continuous-flow CD measurements with a dead time of 400 µs showed evidence for a burst-phase intermediate with a moderate amount of helical secondary structure.64 CD measurements of folding of reduced cyt c triggered by photolysis of a CO ligand or photoreduction can reach the 100 ns time range, but require highly denaturing conditions that disfavor formation of compact in-termediates.39
The hydrogen exchange rates we measured here for the unfolded states at low and high urea concentrations (Figure 4) do not correlate well with the intrinsic exchange rates predicted on the basis of model-peptide data.65 Whereas the rates in short model peptides are deter-mined by the local amino acid sequence, our results exhibit patterns related to the secondary structure of the folded protein. The predominantly hydrophobic residues found at the center of all major α-helices (labeled in Figure 4) exhibit slower exchange rates than the residues at the helix termini and in non-helical regions of the protein. These patterns can be readily explained if the unfolded state of cyt c contains marginally stable (nascent) helical structure in segments coinciding with the major α-helices of the native structure. The dynamic nature of these helices is most apparent for the C-terminal helix (residues 90–104), which shows relatively low rates (400 – 1,200 s−1) for the hydrophobic residues in the center (res. 94–99) and up to an order of magnitude faster rates towards either end of the helix indicative of a nascent helical core with fraying ends. A more detailed discussion of denatured-state exchange rates and comparison with peptide-based intrinsic rates will be presented elsewhere.
The effective rates of amide protection (Figure 4A) obtained by fitting the time-dependent competition data (Figure 3, Figure S3) to a simple competition model (Eq. 2) provides qualitative information on structure formation for individual amide groups on the sub-millisecond time scale. Comparison with the degree of protection at a constant folding time, tf = 140 µs, observed in the pH-dependent competition experiments (Figure 5) shows both similarities and differences. This is illustrated by Figure 6, where the KUIloc values at tf =140 µs are mapped onto the native structure of cyt c (A) and compared with the NH protection patterns observed at longer times (B), based on the time-dependent competition data. The protection rates for predominantly hydrophobic core residues in the C-terminal helix (residues 93–100) range between 400 and 600 s−1, about an order of magnitude higher than those at the helix termini (residues 90–93 and 101–103). This pattern closely resembles the sequence variation of kUIloc and KUIloc in Figure 5. On the other hand, the N-terminal region (res. 6–19) shows higher than average protection rates in the time-dependent competition experiments (Figure 4A), whereas this segment exhibits lower local folding rates and equilibrium constants compared to other helices in the pH-dependent competition data (Figure 5). This striking difference is most likely due to the fact that the effective protection rates extracted from the time-dependent competition data at constant pH (Figure 3) are dominated by longer folding times (approximately 0.5 to 2.2 ms) while the pH-dependent competition experiment detects structure formation within the first 140 µs of refolding. We therefore conclude that the N-terminal segment is still poorly structured at 140 µs and becomes progressively stabilized at folding times in the 1 ms range (Figure 6). This conclusion is consistent with our earlier finding that the N-terminal helix is as well protected as the 60s and C-terminal helices at a folding time of 2 ms; the average protection factor (equivalent to KUIloc) for the N-, 60s and C-helices was 3.0, 3.5 and 3.9, respectively.9 Although the range of protection factors is similar in both studies, the patterns observed here at a 15-fold shorter folding time are qualitatively different with substantially lower levels of protection for the N-helix (<KUIloc> = 0.97) compared to the 60s and C-helices (<KUI-loc> = 4.7 and 2.3, respectively). The preferential contact formation between the 60s and C-terminal helices during the initial collapse phase can be attributed to their relatively short separation along the sequence (17 residues), which makes the entropic penalty for loop closure much smaller than that of the N- and C-terminal helices (82 residues).
Formally, we cannot rule out the possibility that hydro-phobic interactions or contacts with the covalently bound heme group can form rapidly in the absence of detectable protection of backbone amides. Such effects may be responsible for residual structure in denatured proteins involving local clusters of hydrophobic side chains66 and may explain the observation of rapid changes in the emission spectrum of Trp59 that precede the main collapse phase of cyt c.20 However, the global collapse of the protein is likely to be accompanied by formation of hydrogen-bonded secondary structure.
Our site-resolved amide protection data now allow us to revisit the question whether specific folding or nonspecific chain condensation events dominate early stages of folding. If the compact state of cyt c populated on the 100 µs time scale were the result of a non-specific hydro-phobic collapse,44 we would expect little or no variation in amide protection along the amino acid sequence. In particular, the degree of protection should be comparable for different hydrophobic residues. In contrast, we found that the local equilibrium constants for formation of the compact intermediate vary by up to two orders of magnitude (Figure 5C). Some of the hydrophobic residues are among the least protected with KUIloc<<1 (e.g., A15, L32, F36, M80, A102) while others are among the most strongly protected residues with KUIloc>5 (e.g., L68, I75, I95). These observations provide strong evidence that the compaction of the cyt c chain within the initial 140 µs of folding is driven by relatively specific structural events in preferred regions of the protein, including some, but not all of the helical segments. While this is the first study of sub-millisecond folding with single-residue resolution, our findings are consistent with previous evidence that the initial phase during cyt c folding is a barrier-limited (two-state) event rather than a continuous collapse process.45,46
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH Grant GM056250 and NSF grant MCB0744607 to HR, and NIH Grant CA06927 and an Appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. HF was supported by an NIH Postdoctoral Training Grant.
ABBREVIATIONS
- SAXS
small-angle X-ray scattering
- cyt c
cytochrome c
- HSQC
heteronuclear single-quantum correlation
- PEEK
polyether ether ketone
- PCA
phenyl chloroacetate
Footnotes
SUPPORTING INFORMATION
Supplementary figures. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Kubelka J, Hofrichter J, Eaton WA. Curr. Opin. Struct. Biol. 2004;14:76. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Roder H, Maki K, Cheng H. Chem Rev. 2006;106:1836. doi: 10.1021/cr040430y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilsel O, Matthews CR. Curr. Opin. Struct. Biol. 2006;16:86. doi: 10.1016/j.sbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Bowman GR, Voelz VA, Pande VS. Curr. Opin. Struct. Biol. 2011;21:4. doi: 10.1016/j.sbi.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosnick TR, Barrick D. Curr. Opin. Struct. Biol. 2011;21:12. doi: 10.1016/j.sbi.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roder H, Wüthrich K. Proteins: Struct., Funct., Genet. 1986;1:34. doi: 10.1002/prot.340010107. [DOI] [PubMed] [Google Scholar]
- 7.Roder H, Elöve GA, Englander SW. Nature. 1988;335:700. doi: 10.1038/335700a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Udgaonkar JB, Baldwin RL. Nature. 1988;335:694. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- 9.Sauder JM, Roder H. Folding & Design. 1998;3:293. doi: 10.1016/S1359-0278(98)00040-6. [DOI] [PubMed] [Google Scholar]
- 10.Krishna MM, Lin Y, Mayne L, Englander SW. J. Mol. Biol. 2003;334:501. doi: 10.1016/j.jmb.2003.09.070. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura C, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4765. doi: 10.1073/pnas.0409538102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzawa T, Nishimura C, Akiyama S, Ishimori K, Takahashi S, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13859. doi: 10.1073/pnas.0804033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konermann L, Pan J, Liu YH. Chem. Soc. Rev. 2011;40:1224. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 14.Hu W, Walters BT, Kan ZY, Mayne L, Rosen LE, Marqusee S, Englander SW. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7684. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan C-K, Hu Y, Takahashi S, Rousseau DL, Eaton WA, Hofrichter J. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1779. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi S, Yeh S-R, Das TK, Chan C-K, Gottfried DS, Rousseau DL. Nat. Struct. Biol. 1997;4:44. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 17.Shastry MCR, Luck SD, Roder H. Biophys J. 1998;74:2714. doi: 10.1016/S0006-3495(98)77977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollack L, Tate MW, Darnton NC, Knight JB, Gruner SM, Eaton WA, Austin RH. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10115. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilsel O, Kayatekin C, Wallace LA, Matthews CR. Rev. Sci. Instr. 2005;76:014302. [Google Scholar]
- 20.Lapidus LJ, Yao S, McGarrity KS, Hertzog DE, Tubman E, Bakajin O. Biophys J. 2007;93:218. doi: 10.1529/biophysj.106.103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapidus LJ. Curr. Opin. Struct. Biol. 2013;23:30. doi: 10.1016/j.sbi.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Jones CM, Henry ER, Hu Y, Chan C-K, Luck SD, Bhuyan A, Roder H, Hofrichter J, Eaton WA. Proc. Natl. Acad. Sci. U. S. A. 1993;90:11860. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruebele M, Sabelko J, Ballew R, Ervin J. Acc. Chem. Res. 1998;31:699. [Google Scholar]
- 24.Callender RH, Dyer RB, Gilmanshin R, Woodruff WH. Annu. Rev. Phys. Chem. 1998;49:173. doi: 10.1146/annurev.physchem.49.1.173. [DOI] [PubMed] [Google Scholar]
- 25.Wang T, Lau WL, DeGrado WF, Gai F. Biophys J. 2005;89:4180. doi: 10.1529/biophysj.105.068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumont C, Emilsson T, Gruebele M. Nat Methods. 2009;6:515. doi: 10.1038/nmeth.1336. [DOI] [PubMed] [Google Scholar]
- 27.Serrano AL, Waegele MM, Gai F. Protein Sci. 2012;21:157. doi: 10.1002/pro.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuler B, Hofmann H. Curr. Opin. Struct. Biol. 2013;23:36. doi: 10.1016/j.sbi.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 29.Cecconi C, Shank EA, Bustamante C, Marqusee S. Science. 2005;309:2057. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 30.Forman JR, Clarke J. Curr. Opin. Struct. Biol. 2007;17:58. doi: 10.1016/j.sbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Daggett V, Fersht AR. Trends Biochem. Sci. 2003;28:18. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 32.Onuchic JN, Wolynes PG. Curr. Opin. Struct. Biol. 2004;14:70. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Voelz VA, Singh VR, Wedemeyer WJ, Lapidus LJ, Pande VS. J. Am. Chem. Soc. 2010;132:4702. doi: 10.1021/ja908369h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piana S, Lindorff-Larsen K, Shaw DE. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17845. doi: 10.1073/pnas.1201811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prigozhin MB, Gruebele M. Phys Chem Chem Phys. 2013;15:3372. doi: 10.1039/c3cp43992e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Runthala A. J. Biomol. Struct. Dyn. 2012;30:607. doi: 10.1080/07391102.2012.687526. [DOI] [PubMed] [Google Scholar]
- 37.Akiyama S, Takahashi S, Kimura T, Ishimori K, Morishima I, Nishikawa Y, Fujisawa T. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1329. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uzawa T, Akiyama S, Kimura T, Takahashi S, Ishimori K, Morishima I, Fujisawa T. Proc. Natl. Acad. Sci. USA. 2004;101:1171. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldbeck RA, Chen E, Kliger DS. Int J Mol Sci. 2009;10:1476. doi: 10.3390/ijms10041476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arai M, Iwakura M, Matthews CR, Bilsel O. J. Mol. Biol. 2011;410:329. doi: 10.1016/j.jmb.2011.04.057. [DOI] [PubMed] [Google Scholar]
- 41.Agashe VR, Shastry MCR, Udgaonkar JB. Nature. 1995;377:754. doi: 10.1038/377754a0. [DOI] [PubMed] [Google Scholar]
- 42.Sosnick TR, Mayne L, Englander SW. Proteins: Struct., Funct., Genet. 1996;24:413. doi: 10.1002/(SICI)1097-0134(199604)24:4<413::AID-PROT1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Krantz BA, Mayne L, Rumbley J, Englander SW, Sosnick TR. J. Mol. Biol. 2002;324:359. doi: 10.1016/s0022-2836(02)01029-x. [DOI] [PubMed] [Google Scholar]
- 44.Ziv G, Thirumalai D, Haran G. Physical Chemistry Chemical Physics. 2008;11:83. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shastry MCR, Roder H. Nat. Struct. Biol. 1998;5:385. doi: 10.1038/nsb0598-385. [DOI] [PubMed] [Google Scholar]
- 46.Hagen SJ, Eaton WA. J. Mol. Biol. 2000;297:781. doi: 10.1006/jmbi.2000.3508. [DOI] [PubMed] [Google Scholar]
- 47.Mizukami T, Xu M, Cheng H, Roder H, Maki K. Protein Sci. 2013;22:1336. doi: 10.1002/pro.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latypov RF, Cheng H, Roder NA, Zhang J, Roder H. J. Mol. Biol. 2006;357:1009. doi: 10.1016/j.jmb.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 49.Colón W, Wakem LP, Sherman F, Roder H. Biochemistry. 1997;36:12535. doi: 10.1021/bi971697c. [DOI] [PubMed] [Google Scholar]
- 50.Liu W, Rumbley J, Englander SW, Wand AJ. Protein Sci. 2003;12:2104. doi: 10.1110/ps.03211303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR spectroscopy. San Diego: Academic Press; 1996. [Google Scholar]
- 52.Jennings PA, Wright PE. Science. 1993;262:892. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 53.Walkenhorst WF, Edwards JA, Markley JL, Roder H. Protein Sci. 2002;11:82. doi: 10.1110/ps.28202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elöve GA, Roder H. ACS Symp. Ser. 1991;470:50. [Google Scholar]
- 55.Gladwin ST, Evans PA. Folding & Design. 1996;1:407. doi: 10.1016/S1359-0278(96)00057-0. [DOI] [PubMed] [Google Scholar]
- 56.Roder H, Maki K, Cheng H, Shastry MC. Methods. 2004;34:15. doi: 10.1016/j.ymeth.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 57.Bökenkamp D, Desai A, Yang X, Tai Y-C, Marziuff EM, Mayo SL. Anal. Chem. 1998;70:232. [Google Scholar]
- 58.Schmid FX, Baldwin RL. J. Mol. Biol. 1979;135:199. doi: 10.1016/0022-2836(79)90347-4. [DOI] [PubMed] [Google Scholar]
- 59.Hvidt A, Nielsen SO. Advances in Protein Science. 1966;21:287. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 60.Roder H, Wagner G, Wüthrich K. Biochemistry. 1985;24:7396. doi: 10.1021/bi00346a055. [DOI] [PubMed] [Google Scholar]
- 61.Loftus D, Gbenle GO, Kim PS, Baldwin RL. Biochemistry. 1986;25:1428. doi: 10.1021/bi00354a036. [DOI] [PubMed] [Google Scholar]
- 62.Milne JS, Mayne L, Roder H, Wand AJ, Englander SW. Protein Science. 1998;7:739. doi: 10.1002/pro.5560070323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L, Laub PB, Elöve GA, Carey J, Roder H. Biochemistry. 1993;32:10271. doi: 10.1021/bi00089a050. [DOI] [PubMed] [Google Scholar]
- 64.Akiyama S, Takahashi S, Ishimori K, Morishima I. Nat Struct Biol. 2000;7:514. doi: 10.1038/75932. [DOI] [PubMed] [Google Scholar]
- 65.Bai Y, Milne JS, Englander SW. Proteins: Struct., Funct., Genet. 1993;17:75. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Science. 2002;295:1719. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.